Abstract
The genotype of nodule occupants in four annual clovers – balansa (Trifolium michelianum), Persian (Trifolium resupinatum), gland (Trifolium glanduliferum) and arrowleaf (Trifolium vesiculosum) – was investigated. The clovers were inoculated with the ALOSCA® group C granule preparation of strain WSM1325. A total of 224 strains were recovered from root nodules with between 55 and 58 strains for each clover species. Genotyping showed that no strains had fingerprints identical to strain WSM1325. The nodule occupants were diverse with 26, 35, 31 and 32 genotypes identified on arrowleaf, balansa, gland and Persian clovers, respectively. Arrowleaf clover had some specificity for genotype A with 43% of nodules occupied by this strain. The most dominant strain for the other three clovers ranged between 13%–18% occupancy. This work demonstrates a high diversity of naturalized rhizobia strains in New Zealand soils that had the ability to nodulate these top flowering annual clover species.
Introduction
Legumes, such as clover, are important components of New Zealand dryland pastures (Moot Citation2012). In symbiosis with rhizobia they fix nitrogen and provide forage with high nutritive quality. Annual clovers are widely used in dryland regions due to their faster growth in cool weather than perennial clovers (18%–25% crude protein and 60%–80% dry matter digestibility) (Allinson et al. Citation1985). Annual clovers differ from each other in aspects such as their growth habit, persistence, response to temperature and day length, and grazing tolerance. In this study, four top flowering annual clover species (balansa, Persian, arrowleaf and gland) were used.
In New Zealand, as in many parts of the world, pasture legumes are normally inoculated at sowing with commercial strains of rhizobia. Rhizobia inoculants are usually commercialized as either powder (peat moss), liquid or granular products (Stephens & Rask Citation2000). There has been increasing interest in the development of granular inoculants (Fouilleux et al. Citation1996) as they can reportedly maintain high rhizobial populations over time, retain water holding capacity, are chemically and physically uniform, have low toxicity and high environmental safety (Stephens & Rask Citation2000). Granular products, manufactured in Australia and other parts of the world, are typically produced under non-sterile conditions and contain a substantial number of other bacteria and fungi (Denton et al. Citation2009). In contrast, peat slurry inoculants are partially sterilized by gamma irradiation and rarely contain high concentrations of contaminants (Hartley et al. Citation2005). While peat slurry inoculation is considered the current benchmark for inoculation in Australia, granular inoculants potentially offer a range of advantages in the delivery of inoculants. The physical separation of granular inoculants from the seed means that they can be delivered without seed-applied chemicals that may be toxic to rhizobia (Stephens & Rask Citation2000). The granular formulations eliminate the task of applying inoculants to the seed coat close to the time of sowing, as is required with the standard peat slurry method (Denton et al. Citation2009).
Recent research reports a wide range of titres of naturalized rhizobia in standard New Zealand soils with current recommendations that inoculation is unnecessary for red and white clovers (Lowther & Kerr Citation2011). Commercial strains are usually added as they have been selected for their nitrogen-fixing efficacy, persistence and competitive nodulation ability. Despite this, there are few reported experiments in New Zealand on the nodule occupancy by commercial rhizobia strains deployed in soils with resident populations of rhizobia and none for annual clovers. As many of the commercial inoculants are derived from Australia and based on single strains of rhizobia, there is no guarantee that the inoculants are competitive in New Zealand soil environments or symbiotically effective for different clover species (Brockwell & Bottomley Citation1995).
The ALOSCA® Technologies Group C Dry granule contains a single Rhizobium leguminosarum bv. trifolii strain (WSM1325) (C Poole, ALOSCA Technologies, pers. comm.). The aim of this research was: (1) to determine whether inoculation with the ALOSCA® preparation of strain WSM1325 resulted in a high number of nodules containing that strain; and (2) to determine whether there was a difference in occupancy by WSM1325 between the four species of annual clover.
Materials and methods
Inoculant
The ALOSCA® group C dry granule inoculant was purchased in 2009, stored at 2.5 °C and used in February 2010. The ALOSCA® product contained R. leguminosarum bv. trifolii strain WSM1325. This strain was isolated from an annual clover growing near Livadi beach on the Greek Cyclades island of Serifos in 1993 (Howieson et al. Citation2005). It was demonstrated as an effective microsymbiont under competitive conditions for nodulation (Yates et al. Citation2008).
Clover species
Balansa clover (Trifolium michelianum Savi) is a native of the Mediterranean region and is a self-regenerating annual legume that may complement white clover (Trifolium repens L.) and subterranean clover (Trifolium subterraneum L.) as mixed pasture in New Zealand dryland regions (Craig & Ballard Citation2000; Ballard et al. Citation2002). Gland clover (Trifolium glanduliferum Boiss) is also native to the Mediterranean region (Gillet et al. Citation2001) and can be used to provide high quality fodder for livestock and is adapted to a wide range of soil types (Dear et al. Citation2001). Arrowleaf clover (Trifolium vesiculosum Savi) is a self-regenerating winter annual native to the Mediterranean region, Balkan peninsula, Crimea and Caucasus. In Australia, at latitudes similar to New Zealand, arrowleaf clover has been shown to persist in areas with low annual rainfall (Evans Citation2006; Evans & Mills Citation2008). Persian clover (Trifolium resupinatum L.), a native to central and southern Europe and southwest Asia, is adapted to most soil types, especially clay and alkaline (Zhang et al. Citation2004). Presently there is no work published on the compatibility of these clover species in New Zealand with commercial rhizobial inoculant.
Field experiment
A total of 12 plots of 2.1×10.0 m were planted with the four clover species, namely, balansa (T. michelianum), Persian (T. resupinatum), gland (T. glanduliferum), arrowleaf (T. vesiculosum) in a split-plot design experiment in Iversen field 9 at Lincoln University, New Zealand. Each clover species was replicated three times and contained plants at a density of 40 plants/m2 for arrowleaf clover, 47 plants/m2 for balansa clover, 63 plants/m2 for gland clover and 22 plants/m2 for Persian clover and were sown on 1 February 2010. At planting, seeds were sown with ALOSCA® group C granule inoculant according to the manufacturer's instructions.
Recovery of rhizobia from the commercial ALOSCA™ preparation
Method 1—Dilution plating: To extract the commercial inoculant strain from the ALOSCA® product, 10 g of ALOSCA® type C granules were added to each of two 90 mL bottles of sterile water and mixed for 10 min on a wrist action shaker (Griffin, Britain). The resultant suspensions were serially diluted five times (10−2 to 10−6) in water and 100 µL of each solution spread onto duplicate yeast mannitol agar (YMA; Yates Citation2008). Plates were incubated at 20 °C for 48 h after which colony numbers were counted.
Method 2—Recovery from nodules: Seeds of white clover ‘Grasslands Tribute’ were sterilized by immersion in 95% ethanol for 10 s, immersion in 1% sodium hypochlorite for 2 min, followed by three washes in sterile water and then plated onto water agar (15% w/v) to germinate. Plates were incubated at room temperature for up to 7 d for germination. Seedlings were transferred aseptically into sterile vermiculite contained in 150 mm high polyethylene terepthalate containers (diameter 8 cm) and wetted with 20 mL of nutrient media (Tan et al. Citation2012). Each of the three replicate plants was inoculated by the addition of 1 mL of an ALOSCA® granule slurry (10% w/v) in sterile water or 1 mL sterile water. The containers were sealed with cling film, placed in a controlled environment cabinet (Adaptis, Conviron) and exposed to a 16 h photoperiod (400 µmol photons m−2 s−1) at a constant 22 °C. After 6 weeks' growth, nodules were recovered from each of the inoculated plants and the colonizing bacteria recovered into culture.
Recovery of rhizobia from field grown plants
In August 2010, at least 10 plants (from the middle of the plot) were taken from each of the three replicate plots for each clover species. Plant roots were washed thoroughly to remove soil. Twenty nodules were collected from the top 50 mm of the tap root (average of two nodules per plant) from each species of clover from each of the three replicate plots. Each nodule was recovered by cutting the root at a distance of approximately 5 mm on either side of the nodule. The nodule and remaining root fragment were immersed intact for 5–10 s in 95% ethanol, sterilized in 5% bleach for 2 min and then rinsed four times in sterile water for 20 s each time. The nodules were then detached from the root fragments, crushed in a sterile Petri dish and a loop of the bacterial contents spread onto a YMA plate. The YMA plates were incubated at 20 °C for 48 h. Single colonies were subcultured at least once to ensure purity.
Isolation of DNA from rhizobia cultures recovered from the inoculant and nodules
A single loopful of each bacterial colony was used to inoculate 1.2 mL of yeast mannitol broth (YMB; Yates Citation2008) in a sterile 2 mL tube. This liquid suspension was incubated at 28 °C for 24–48 h at 220 rpm (LABNET 211 DS, Labnet International, US). DNA was extracted from the rhizobial suspension using the PUREGENE® (Gentra Systems, US) DNA extraction kit, according to the manufacturer's instructions.
PCR amplification of rhizobial DNA using ERIC primers
DNA fingerprinting of the recovered bacterial strains was done by polymerase chain reaction (PCR) with primers ERIC 1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC 2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) (Versalovic et al. Citation1991). Each PCR contained 1×buffer (FastStart, Roche, US), 200 µM of each dNTP, 50 µM of each primer, 50 ng of genomic DNA and 1 U of FastStart Taq polymerase (Roche, US) in a total volume of 25 µL. The thermal cycle parameters were 94 °C for 3 min followed by 35 cycles of 94 °C for 1 min, 52 °C for 1 min, 72 °C for 1 min with a final extension of 7 min at 72 °C. The resultant PCR products (7 µL) were separated by electrophoresis on a 1% agarose gel at 10 V/cm for 50 min in 1×TAE buffer (40 mM Tris acetate, 2 mM Na2EDTA, pH 8.5). The gels were stained by immersion for 15 min in 0.5 µg/mL ethidium bromide and visualized under ultraviolet light. After electrophoresis, strains with identical banding patterns, based on the molecular weight of the ERIC-PCR products, were identified and the frequency information was used to construct histograms.
PCR amplification of the 16S ribosomal subunit for DNA sequencing
For the strains recovered from the ALOSCA® granule and the main genetic groups observed on the ERIC-PCR fingerprints, amplification of the 16S ribosomal subunit was done using primers F27 (5′-AGRGTTTGATCMTGGCTCAG-3′) and R1494 (5′-GGTTACCTTGTTACGACTT-3′). The thermal cycle parameters, agarose gel electrophoresis and gel staining processes were as described for ERIC-PCR. The 16S ribosomal subunit PCR products were sequenced in one direction using primer R1494 at the Lincoln University DNA Sequencing Facility.
Results
Recovery of bacterial isolates from the ALOSCA® inoculant
Method 1—Serial dilution: An average (from four plates) of 13.5 bacterial colonies grew on the 10−4 serial dilution plate. Thus, the ALOSCA® granule contained approximately 1.4×105 CFU per gram of culturable bacteria. All colonies (n=17), regardless of appearance, from one 10−4 plate were isolated into pure culture and their 16S rRNA gene sequenced. No rhizobia were recovered. Comparison of the 600–800 bp sequence with those present on GenBank (http://www.ncbi.nlm.nih.gov/genbank/) indicated that most of the recovered bacteria were Pseudomonas spp. with Athrobacter sp. and Bacillus sp. also present.
Method 2—Recovery from nodules: All of the plants inoculated with a slurry of the ALOSCA® granules were nodulated and none of the control plants. This indicated that the inoculant did contain R. leguminosarum. Six bacterial strains were recovered from the inoculated plants (two per plant). All six strains had identical fingerprints generated by ERIC-PCR. One of these six strains was confirmed as R. leguminosarum by DNA sequencing. This strain was used as the positive control for ERIC-PCR.
Recovery of rhizobia from clover nodules
The major morphological characteristics of rhizobia are that they form round, flat to conical colonies. They grow rapidly on YMA and have colour and texture that ranges from white, opaque, milky to watery translucent. Based on these morphological characteristics, the majority of nodule inhabitants had characteristics consistent with rhizobia and were usually not co-purified with bacteria of other morphologies. The 16S rRNA region of the main genotypes from arrowleaf (genotypes A–C), balansa (A–D), gland (A–D) and Persian (A–C) were sequenced. Comparison of the 600–800 bp DNA sequence with those present on the GenBank database (http://www.ncbi.nlm.nih.gov/) showed that all strains were R. leguminosarum, except balansa genotype D, which was a Paenibacillus sp. Out of 60 nodules for each clover species, 55 strains characteristic of rhizobia were recovered for each of Persian and gland clover, 56 for balansa clover and 58 for arrowleaf clover.
DNA fingerprinting of isolates recovered from clover nodules
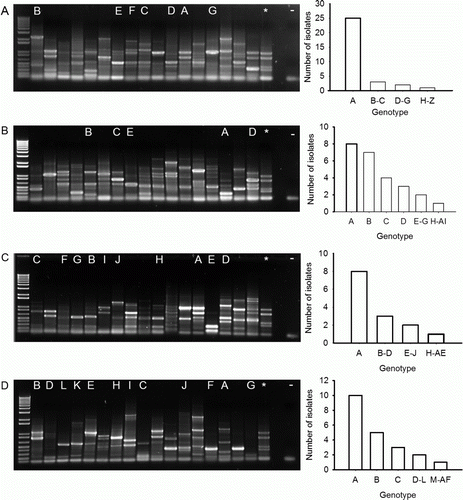
A total of 224 strains from the four clover species were subjected to DNA fingerprinting using ERIC-PCR. Of the 58 rhizobium strains recovered for arrowleaf clover, 26 unique banding patterns were observed (A). The most common banding pattern was present in 43% (n=25) of the recovered strains with 33% (n =19), 7% (n =4) and 3% (n=2) of the strains having banding patterns present as either single, duplicate or triplicates, respectively. Out of the 56 rhizobium strains recovered for balansa clover, 35 unique banding patterns were observed (B). The two most common banding patterns were present in 14 and 13% (n=8 and 7, respectively) of the recovered strains, 50% (n=28) and 5% (n=3) of the strains had banding patterns that were observed as single or duplicates, respectively. Out of the 55 rhizobium strains recovered for gland clover, 34 unique banding patterns were observed (C). The most common banding pattern was present in 15% (n=8) of the recovered strains whereas 40% (n=22), 15% (n=8), and 10% (n=3) of the strains had banding patterns that were observed as single, duplicate or triplicates, respectively. Of the 55 strains characteristic of rhizobia that were recovered from Persian clover, 32 different banding patterns were observed (D). The most common banding patterns were present in 18% (n=10) and 9% (n=5) of the recovered strains, 36% (n=20), 16% (n=9) and 3% (n=1) of the strains had banding patterns that were observed as single, duplicate or triplicates, respectively.
Figure 1. Agarose gel (1%) of up to 17 unique fingerprints generated by ERIC-PCR of DNA extracted from bacteria recovered from the nodules of four clover species. A, Arrowleaf; B, balansa; C, gland; D, Persian. The positive control (strain WSM1325; *), the negative control (-) and the code for each of the most common fingerprints is designated at the top of the gel. On the right hand side is a histogram showing the frequency of each fingerprint. The 1 kb plus DNA ladder (Invitrogen, Australia) is in the first lane of each gel.
Discussion
This research demonstrated that R. leguminosarum bv. trifolii from the ALOSCA® group C granule preparation of strain WSM1325 was not present in nodules recovered from the annual clover species balansa, Persian and gland clover in a field experiment at Lincoln University. In contrast, the dominant genotype (43%) in arrowleaf clover was a strain of R. leguminosarum that had similarity to strain WSM1325. This lack of nodule occupancy may be explained by either: (1) the inability of the commercial strain to compete effectively with naturalized strains present at the site; (2) the use of a granule inoculant that contained low numbers of effective bacteria; or (3) host specificity.
Nodules recovered from the four field grown annual clovers yielded 224 bacterial strains that were morphologically characteristic of rhizobia. DNA fingerprinting of these strains demonstrated a high genotypic diversity with between 26 and 35 different strains present within 55–58 nodules recovered from each of the four clover species. This is the first New Zealand study to demonstrate a large number of genotypes present in a single field using ERIC-PCR, however, similar results have been shown for surveys of naturalized rhizobia in Australian pastures (Ballard et al. Citation2002; Denton et al. Citation2002; Simon & Salava Citation2006). This work reinforces other work (using chromosomal and plasmid restriction fragment length polymorphism) done at the University of Otago, New Zealand, (Elliot Citation1997; Shah Citation2003) that demonstrated genotypic variation in rhizobia in New Zealand pastures. In previous surveys, a high titre of naturalized strains has been demonstrated (Greenwood Citation1965; Hale Citation1980; Rhys & Bonish Citation1981; Gaur & Lowther Citation1982) and this field site is typical of New Zealand lowland soils having a history of inoculation with commercially available strains of rhizobia.
The commercial strain was recovered by baiting the rhizobia with clover plants grown in sterile environments. Comparison of ERIC-PCR fingerprints demonstrated that none of the recovered nodules contained an identical genotype to the inoculant strain WSM1325. This was surprising as the nodules were harvested from the base of the taproot as this was the most likely site for recovery of the inoculant strain. It is possible that strain WSM1325 was unable to compete with the diverse population of site-adapted strains present at the field site. However, previous work has demonstrated that this strain is highly competitive when introduced to new sites and compatible with clover hosts derived from the Mediterranean (Yates Citation2008). Thus, it seems more likely that the lack of nodule occupancy was due either to low numbers of the commercial strains present in the granules or specificity for particular clover species.
Of the four annual clovers, arrowleaf showed the greatest specificity for a single strain with one rhizobial genotype occupying almost half of the nodules. The same genotype was also the most frequent nodule occupant in gland clover where it occupied 15% of the nodules. The genotype of this particular strain was similar to that of WSM1325, but lacked two low molecular weight bands observed in the commercial strain. It is likely that these strains were not WSM1325. However, it is possible that these small differences are the result of recombination between WSM1325 and naturalized strains during the 6 months between inoculation and harvest. There is little information available on the genetic stability of commercial strains once field deployed. If these strains are derived from the inoculant WSM1325, it indicates that the strain has much higher occupancy of arrowleaf clover nodules than the other three clover species tested here. This suggests that there is some host specificity towards arrowleaf clover shown by strain WSM1325. In contrast, the dominant genotypes in balansa and Persian clover were naturalized strains and occupied 14%–18% of nodules. Similar results were demonstrated by Denton et al. (Citation2002) who showed that dominant field strains could occupy up to 19% of clover nodules in competition with commercial inoculants in an alkaline Australian pasture soil. They hypothesized that the naturalized strains were competitive because they were more adapted to the edaphic conditions at the field site. It is unknown whether the dominant naturalized strains at the Canterbury site could fix N2 at levels similar to strain WSM1325. Research has shown that the symbiotic effectiveness of naturalized strains varies widely, between 2 and >100% of that demonstrated by highly effective commercial inoculants (Gaur & Lowther Citation1980, Citation1982; Rhys & Bonish Citation1981; Svenning et al. Citation2001).
Dilution plating of the ALOSCA® group C granule suggested that low numbers of bacteria were present in the granule, as rhizobia were not recovered by this method. In contrast, other species of bacteria, such as Pseudomonas, Arthrobacter and Bacillus were recovered. Granular products produced in Australia and other parts of the world are manufactured in non-sterile conditions, leading to the presence of numerous bacterial and fungal contaminants (Hartley et al. Citation2005; Denton et al. Citation2009). A field evaluation of granule preparations in Australia demonstrated that nodulation by rhizobial strains delivered in clay granules differed and was dependent on the competitive ability and titre of the naturalized population present in the field (Denton et al. Citation2009). The inability to recover R. leguminosarum bv. trifolii strain WSM1325 directly from the granular inoculants in this batch suggests that the strain was present at a concentration <105 colonies g−1 which is 100×lower than the generally accepted level of approximately 107 colonies g−1 (Denton et al. Citation2009). Further testing of a range of batches, with greater replication is recommended to determine whether this result is representative of commercial preparations of the ALOSCA® Technologies Group C Dry granule.
In conclusion, this research has shown that naturalized strains are present as diverse populations in a Canterbury soil and that they compete well for occupancy of new annual clover species. The tested batch of ALOSCA® product had low bacterial counts of which rhizobia were a minor component overall. Given the low levels of rhizobia in the product, it is likely that the WSM1325 present was unable to establish nodulation of the clovers against a high background of naturalized soil rhizobia. The similarity of ERIC-PCR fingerprints of the dominant genotype recovered from arrowleaf clover may indicate it is a recombinant of strain WSM1325. From a practical perspective, the resident rhizobial population has successfully colonized each of these annual clover species. The relative N2 fixation efficacy of these strains remains to be demonstrated. The high diversity of strains competitive for nodulation of these clover species suggests that selection of an effective naturalized strain may be possible.
Acknowledgments
The authors thank Hollena Nori for access to her field experiment. This work was funded by the Lincoln University Research Fund (LURF).
References
- Allinson DW , Speer GS , Taylor RW , Guillard K 1985 . Nutritional characteristics of kura clover (Trifolium ambiguum Bieb.) compared with other forage legumes . Journal of Agricultural Science 104 : 227 – 229 . doi: 10.1017/S0021859600043161
- Ballard RA , Craig AD , Charman N 2002 . Nodulation and growth of pasture legumes with naturalised soil rhizobia. 2. Balsana clover (Trifolium michelianum Savi) . Australian Journal of Experimental Agriculture 42 : 939 – 944 . doi: 10.1071/EA01092
- Brockwell J , Bottomley PJ 1995 . Recent advances in inoculant technology and prospects for the future . Soil Biology & Biochemistry 27 : 683 – 697 . doi: 10.1016/0038-0717(95)98649-9
- Craig AD , Ballard RA 2000 . Balansa clover (Trifolium michelianum)—a forage legume for temperate pastures . Cahiers Options Méditerranéennes 45 : 177 – 180 .
- Dear BS , Sandral GA , Nutt B , Wilson B , Rodham C , Taylor J 2001 . Gland clover—a new insect resistant legume . Proceedings of the 16th Annual Grasslands Society Conference , Gundagai New South Wales Australia . . Pp. 63 – 64 .
- Denton MD , Coventry DR , Murphy PJ , Howieson JG , Bellotti WD 2002 . Competition between inoculants and naturalised Rhizobium leguminosarum bv. trifolii for nodulation of annual clovers in alkaline soils . Australian Journal of Agricultural Research 53 : 1019 – 1026 . doi: 10.1071/AR01138
- Denton MD , Pearce DJ , Ballard RA , Hannah MC , Mutch LA , Norng S et al. 2009 . A multi-site field evaluation of granular inoculants for legume nodulation . Soil Biology & Biochemistry 41 : 2508 – 2516 . doi: 10.1016/j.soilbio.2009.09.009
- Elliot RM 1997 . Interaction of Caucasian clover rhizobia with white clover rhizobia . PhD thesis . University of Otago Dunedin
- Evans P 2006 . Arrowleaf clover. Agriculture notes . State of Victoria, Department of Primary Industries . 2 p. http://www.dpi.vic.gov.au/dpi/nreninf.nsf/v/2C74CA45607D05A2CA2574160076270F/$file/Arrowleaf%20Clover.pdf (accessed 10 September 2010).
- Evans PM , Mills A 2008 . Arrowleaf clover: potential for dryland farming systems in New Zealand . Proceedings of the New Zealand Grassland Association 70 : 239 – 244 .
- Fouilleux G , Revellin C , Hartmann A , Catroux G 1996 . Increase of Bradyrhizobium japonicum numbers in soils and enhanced nodulation of soybean (Glycine max (L) Merr.) using granular inoculants amended with nutrients . FEMS Microbial Ecology 20 : 173 – 183 . doi: 10.1111/j.1574-6941.1996.tb00316.x
- Gaur YD , Lowther WL 1980 . Distribution, symbiotic effectiveness, and fluorescent antibody reaction of naturalised populations of Rhizobium trifolii in Otago soils . New Zealand Journal of Agricultural Research 23 : 529 – 532 . doi: 10.1080/00288233.1980.10417878
- Gaur YD , Lowther WL 1982 . Competition and persistence of strains of Rhizobium trifolii in relation to inoculation level and lime pelleting on white clover sown into cultivated soil . New Zealand Journal of Agricultural Research 25 : 277 – 280 . doi: 10.1080/00288233.1982.10420925
- Gillet JM , Taylor NL , Collins M 2001 . The world of clovers . Ames , , US , Iowa State University Press . 457 p.
- Greenwood RM 1965 . Populations of rhizobia in New Zealand soils . Proceedings of the New Zealand Grassland Association 26 : 95 – 101 .
- Hale CN 1980 . Competition between strains of Rhizobium trifolii in the establishment of white clover . Proceedings of the New Zealand Grassland Association 41 : 138 – 145 .
- Hartley EJ , Gemmell LG , Slattery JF , Howieson JG , Herridge DF 2005 . Age of peat-based lupin and chickpea inoculants in relation to quality and efficacy . Australian Journal of Experimental Agriculture 45 : 183 – 188 . doi: 10.1071/EA03158
- Howieson JG , Yates RJ , O'Hara GW , Ryder M , Real D 2005 . The interactions of Rhizobium leguminosarum biovar trifolii in nodulation of annual and perennial Trifolium spp. from diverse centres of origin . Australian Journal of Experimental Agriculture 45 : 199 – 207 . doi: 10.1071/EA03167
- Lowther WL , Kerr GA 2011 . White clover seed inoculation and coating in New Zealand . Proceedings of the New Zealand Grassland Association 73 : 93 – 101 .
- Moot DJ 2012 . An overview of dryland legume research in New Zealand . Crop & Pasture Science 63 : 726 – 733 . doi: 10.1071/CP12103
- Rhys GJ , Bonish PM 1981 . Effectiveness of Rhizobium trifolii populations associated with Trifolium species in Taranaki . New Zealand Journal of Experimental Agriculture 9 : 327 – 335 .
- Shah S 2003 . Genetic analysis of rhizobial diversity in the symbiosis between Rhizobium leguminosarum biovar trifolii with white clover (Trifolium repens) . MSc thesis . University of Otago Dunedin
- Simon T , Salava J 2006 . New Rhizobium leguminosarum bv. trifolii isolates: Evaluation of competitiveness for clover nodule occupancy . Plant, Soil & Environment 52 : 441 – 448 .
- Stephens JHG , Rask HM 2000 . Inoculant production and formulation . Field Crops Research 65 : 249 – 258 . doi: 10.1016/S0378-4290(99)00090-8
- Svenning MM , Gudmundsson J , Fagerli I-L , Leinonen P 2001 . Competition for nodule occupancy between introduced strains of Rhizobium leguminosarum biovar trifolii and its influence on plant production . Annals of Botany 88 : 781 – 787 . doi: 10.1006/anbo.2001.1484
- Tan HW , Weir BS , Carter N , Heenan PB , Ridgway HJ , James EK et al. 2012 . Rhizobia with 16S rRNA and nifH similar to Mesorhizobium huakuii but novel recA, glnII, nodA and nodC are symbionts of New Zealand Carmichaelinae . PLOS ONE 7 : e47677 . doi: 10.1371/journal.pone.0047677
- Versalovic J , Koeuth T , Lupski JR 1991 . Distribution of repetitive DNA sequences in eubacteria and applications to fingerprinting of bacterial genomes . Nucleic Acids Research 19 : 6823 – 6831 . doi: 10.1093/nar/19.24.6823
- Yates R 2008 . Symbiotic interactions of geographically diverse annual and perennial Trifolium spp. with Rhizobium leguminosarum bv. trifolii . PhD thesis . Murdoch University Australia . 202 p.
- Yates RJ , Howieson JG , Reeve WG , Brau L , Speijers J , Nandasena K et al. 2008 . Host-strain mediated selection for an effective nitrogen-fixing symbiosis between Trifolium spp. and Rhizobium leguminosarum biovar trifolii . Soil Biology & Biochemistry 40 : 822 – 833 . doi: 10.1016/j.soilbio.2007.11.001
- Zhang X , Evans PM , Riffken PA 2004 . Performance of annual pasture legumes in cropping rotations in the cool temperate zone of south-eastern Australia . Australian Journal of Experimental Agriculture 44 : 863 – 871 .