Abstract
Fifty-five accessions from the National New Zealand Flax Collection were subjected to genotyping using eight nuclear and two plastid simple sequence repeats (SSRs) and compared with a selection of predominantly wild Phormium plants. Several groups of plants with different names but identical SSR genotypes were found and these largely correspond with groups previously identified by analysis of their morphology. Maori cultivars are genetically diverse and include plants with interspecific (Phormium tenax × Phormium cookianum) hybrid origin. Genotyping supports historical and archaeological evidence that Phormium was taken to the subantarctic islands from southern New Zealand by sealers, and probably from the Chatham Islands by Maori and Moriori. As well as such tests of relatedness among Phormium selections, our markers provide opportunities to study the process of speciation, evolution and interspecific hybridization in Phormium.
Introduction
The named varieties of flax are very numerous, but they are not generally and accurately distinguished even amongst the Natives themselves; and there is no doubt that the same variety has often a different name in different districts… There has been no opportunity yet of satisfactorily fixing the varieties; and it will be only when plants from all parts of the island have been collected at one place that they can be compared, and an accurate list compiled.
Report of the Flax Commissioners Citation1870. AJHR G-no4 p. 68
Phormium tenax and Phormium cookianum are herbaceous monocotyledons, known by the Maori names harakeke and wharariki, respectively, and collectively as New Zealand flax. Phormium tenax is native to New Zealand, Norfolk and Chatham Islands, and P. cookianum is an endemic New Zealand species. There is considerable variation in form within and between the species, often related to habitat features such as climate and soil conditions (Harris et al. Citation2005a; Wehi & Clarkson Citation2007), and both species hybridize (Allan & Zotov Citation1937). Primarily bird-pollinated (Craig & Stewart Citation1988), Phormium is capable of self-fertilization but in natural populations is almost totally outcrossing (Jesson et al. Citation2006; Houliston et al. Citation2008). All reported chromosome counts for Phormium are 2n=32 (Moore & Edgar Citation1970; Dawson Citation2000; de Lange & Murray Citation2002) and both species are presumed diploid. A comprehensive review of the biosystematics, chemistry, phenology, ecology, and cultural and economic uses of P. tenax has recently been completed by Wehi & Clarkson (Citation2007).
Prior to European arrival, Phormium was an essential economic plant for Maori, who used the leaves and extracted fibre to make containers, clothing, cordage, nets and mats. Plants with particular qualities (such as robustness, ease of fibre extraction, strength and length of fibre) were selected and cultivated by vegetative reproduction. The Europeans traded fibre mainly for use as cordage, developed mechanical processing techniques and in the 20th century used some traditional cultivars to breed plants for strong fibre and disease resistance. The Phormium fibre industry proved uneconomic by the 1970s, with competition from other natural fibres and synthetics. There is, however, a resurgence of interest in potential new commercial applications, and a vibrant community of weavers involved in traditional crafts and contemporary art, who all want information on and access to the best quality plants.
Distinguishing morphologically similar cultivars and determining their place of natural origin is a problematic issue for the proper curation of the National New Zealand Flax Collection (the Collection), held at Landcare Research, Lincoln. The Collection comprises named weaving cultivars of Phormium (primarily those collected since the 1950s by Rene Orchiston of Gisborne) and other accessions of historical and cultural interest such as selections used for commercial milling. Of particular focus in this article are Phormium collections from the New Zealand subantarctic islands where the species was introduced for its economic uses.
Names of cultivars or classes of Phormium were recorded in the 1800s by flax industry proponents (Flax Commissioners Citation1870; Hector Citation1872) and ethnographers (e.g. Best Citation1899, Citation1942). Many of the cultivars listed in these early accounts are not described in detail, making it mostly unfeasible to reliably match the name with extant cultivars, even though many of the names are still in use. BD Cross undertook a comprehensive and detailed study of variation in Phormium in 1910–11 (Cross Citation1912) and, although her classification system is still worthy of imitating in describing varieties of Phormium, it is not possible to correlate with confidence her descriptions of named selections with accessions in the Collection (with the exception of the variegated cultivar, Parekoritawa).
Information on where Orchiston collected the 50 cultivars in her collection is mostly limited to region (Scheele Citation2005). Because desirable Phormium selections were widely moved about by Maori, we cannot be sure of the original provenance of the plants. Some cultivars, such as Kohunga and Tapamangu, are well known by traditional weavers and reasonably distinctive in form and leaf properties. Other named cultivars, however, are less widely known and not easily distinguished; some share similar morphological and weaving characteristics and appear to be of the same genetic stock, albeit having different names.
Molecular genetic techniques have the potential to illuminate the relationships among accessions within the Collection, and to link those selections with wild populations and geographic areas. In earlier work, McBreen et al. (Citation2003) investigated a range of techniques and used sequenced characterized amplified region (SCAR) data to separate Orchiston and subantarctic accessions into two distinct clusters, although relationships within the groupings remained largely unresolved. More recent genotyping analyses of Phormium have used isozymes (Jesson et al. Citation2006) or AFLP DNA fingerprinting data (Smissen & Heenan Citation2007; Smissen et al. Citation2008). Levels of polymorphism in isozyme studies are generally low and they require fresh or frozen tissue, making them difficult to implement for clone or cultivar identification. AFLPs are highly informative in Phormium, and have been used for systematic analyses of wild populations, addressing issues such as hybridization and species circumscription. However, the generation of AFLP profiles is a complex technical procedure, reproducible only under tightly controlled conditions and requiring access to living tissue. Some subjectivity is also often involved in scoring AFLP profiles if conducted interactively, or, if scoring is automated, apparent polymorphisms may be arbitrarily introduced or excluded from a dataset as a result of technical ambiguity (Meudt & Clarke Citation2007). Also, the information content of AFLP profiles is limited by their dominant inheritance.
Therefore, we sought to characterize simple sequence repeat (SSR) loci from Phormium. Typically SSR markers provide a high level of reproducibility robust to changes in laboratory conditions and DNA quality, are inherited in co-dominant fashion and often show very high levels of polymorphism, making them ideal for discriminating between genetic individuals or identifying closely related individuals (e.g. paternity testing; Oliveira et al. Citation2006). Our aim was to develop a sufficient number of SSR markers to identify genetic groupings within the flax collection, and to test the genetic identity among duplicate cultivars within the collection and eventually plants in other collections.
We also assayed variation at two variable chloroplast homopolymer A/T repeat loci in the trnL intron and trnL–trnF intergenic spacer (see Smissen et al. Citation2008).
Methods
We used a plant collected from Norfolk Island (Landcare Garden accession 262/94A) as the source of DNA for SSR isolation. A genomic DNA library enriched for SSRs was constructed according to Glenn & Schable (Citation2005). DNA was extracted from freshly harvested vegetative shoot tissue (rito) using the CTAB extraction method (Doyle & Dickson Citation1987) as modified by Smissen & Heenan (Citation2007). Approximately 100 ng of genomic DNA was digested for 3 h with 10 units of RsaI (Fermentas #ER1121) and 10 units of PdmI (Fermentas #ER1531) restriction enzymes in 1× T4 ligase buffer in the presence of 400 cohesive end ligation units of T4 DNA ligase (Fermentas #EL0021) and 70 pmol of SuperSNX linker. Restriction/ligation products were amplified by PCR using the superSNX primer and then the PCR products were hybridized to biotinylated oligonucleotide probe mix 2 supplied by Savannah River Ecology Lab. Streptavidin-coated magnetic beads were then used to capture probes and hybridized DNA. Washed and eluted enriched SSR DNA was then amplified by a second round of PCR, cloned into an Invitrogen TOPO 4.0 plasmid, and transformed into Invitrogen TOP10 chemically competent cells, according to the manufacturer's instructions. Plasmids were sequenced using M13f and M13r primers. Primers for SSR candidate loci were designed without reference to a computer program and screened for interpretable banding patterns against a panel of 12 Phormium samples using silver-stained 6% polyacrylamide sequencing gels.
Primer pairs producing interpretable banding patterns in the 12-sample screen were fluorescently labelled with 6-FAM, VIC or NED for detection using an ABI 3700 capillary machine. These primers were used to genotype a set of 95 plants from the Collection and wild-collected plants. Details of the plants are given in .
Table 1 Samples genotyped in this study
PCR conditions for SSR genotyping reactions were an initial denaturation at 95°C for 2 min, followed by 30 cycles of 94°C for 30 s, primer-specific annealing temperature for 30 s and 72°C for 45 s. A final 30-min incubation at 72°C was conducted to promote 3'-A nucleotide addition. Primer sequences for novel markers and the annealing temperatures used are shown in . Amplification of the chloroplast SSRs used here was described in Smissen et al. (Citation2008). One sample was genotyped in duplicate for all loci as an internal control.
Table 2 Details of nuclear simple sequence repeat (SSR) markers used in this study
A UPGMA tree and polymorphic information content (PIC: Botstein et al. Citation1980) statistics for nuclear SSR data were generated using Powermarker v. 3.25 (Liu & Muse Citation2005) with pairwise genetic distance between samples estimated by 1 – (number of shared alleles/total number of alleles).
Results
SSR enrichment and characterization
SSR enrichment was only modestly successful, with ~ 20% of clones containing a repeat region of more than six repeats. However, it was deemed more efficient to sequence a larger number of clones than to attempt improved enrichment. Forty primer pairs were designed and screened against a panel of twelve samples, and eight of these were chosen for subsequent genotyping, as they were both polymorphic and interpretable as diploid loci.
Genotyping
In some cases the majority of detected allelic variation was interpretable solely as the result of changes in the number of repeat units present, whereas in other cases variation appeared to be complex and probably included nucleotide substitutions and size variation in the flanking regions. Discrimination of distinct size classes of alleles was most problematic for locus PhSSR84 because of almost continuous variation in fragment size data for some samples. However, this locus displayed a high number of alleles (18) and was thus highly informative despite this minor ambiguity around allele calling. The interpretation of fragment size variation as allelic variation for this marker is specified in . Variation detected in each locus is described in . The genotype for one sample (Ruawai) was generated in duplicate for all markers as an internal control, with no variation between the duplicates detected.
Table 3 Interpretation of fragment size variation as allelic variation for the PhSSR84 marker
Table 4 Polymorphism in markers used in this study
Five alleles were detected for the trnL intron SSR and four for the trnL–trnF intergenic spacer SSR. Taken together, these two markers distinguish 13 distinct chloroplast haplotypes (designated A–M in ) in our sampling.
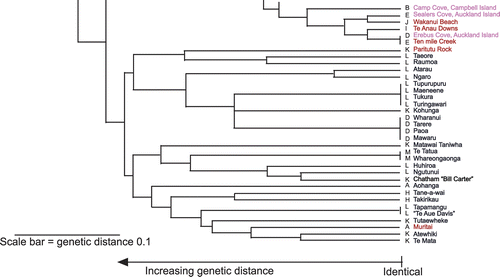
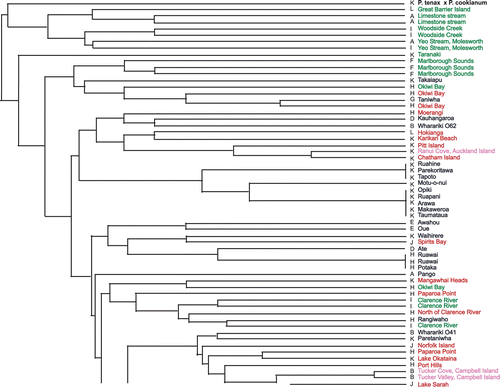
Fig. 1 UPGMA dendrogram generated from single sequence repeat (SSR) genotypes. Green labels, Phormium cookianum; blue labels, weaving cultivars; red labels, wild-collected P. tenax; pink labels, subantarctic samples; black labels, other samples. Letters (A-M) before names indicate cpDNA haplotypes.
Genotypes were obtained for 49 weaving cultivars (all from the North Island), six New Zealand subantarctic islands plants (Auckland and Campbell Islands), a plant cultivated by Mr Bill Carter on Chatham Island, and from 38 wild provenances from throughout New Zealand also growing at Landcare Research's Lincoln site.
Relationships among plants in the National New Zealand Flax Collection
All wild-sourced plants displayed unique genotypes in our dataset. Most of the 49 Maori weaving plants also displayed unique genotypes, but several groups of cultivars with identical genotypes were identified. These were:
Group 1 Parekoritawa, Ruahine and Tapoto;
Group 2 Taumataua, Makaweroa, Opiki, Ruapani and Arawa;
Group 3 Ruawai and Potaka;
Group 4 Te Tatua and Whareongaonga;
Group 5 Tapamangu and Unknown (‘Te Aue Davis’);
Group 6 Tupurupuru, Maeneene, Tukura and Turingawari; and
Group 7 Wharanui, Mawaru, Tarere and Paoa.
An UPGMA tree was constructed based on shared alleles for the nuclear loci (). Chloroplast haplotypes are also shown in for each plant, alongside its name. Noteworthy groupings (discussed below) appearing in this tree include the clustering of Taniwha close to wild Okiwi Bay (Marlborough, eastern coast) P. tenax; the clustering of the plant from Ranui Cove, Auckland Island, with Chatham and Pitt Island samples; and the clustering of other subantarctic island samples with South Island P. tenax samples. The alignment of Atarau with Ngaro and Atewhiki with Te Mata parallels particular morphological features shared between each pair of cultivars (described below).
Discussion
The individual markers used in this research differ in their information content and some caveats are necessary in interpreting the analysis of them. We interpret the two chloroplast SSR markers to be part of the same maternally inherited chloroplast DNA molecule. Although chloroplast DNA is most often maternally inherited in flowering plants, this is not always the case, and we have no direct evidence regarding its inheritance in Phormium. However, its haploid genetics appear to be confirmed by the fact that no heterozygotes were detected for either of these makers.
Some of the nuclear SSR loci are much more informative than others, depending both on the number of alleles present for each locus and their frequencies. The PIC statistic attempts to capture this variation and indicates that PhSSR105, PhSSR118 and PhSSR84 are the most informative loci and PhSSR108 is the least informative (Table 4).
The UPGMA clustering of samples () does not take into account the fact that some alleles are common in the Collection and some are rare. That two samples share a very common allele may be much less significant than two samples sharing a very rare one. For this and other reasons, we caution that most of the relationships suggested by the clusters in are not reliable indicators of relationships among the plants. However, the fact that none of the wild material share identical profiles, and that a moderate number of markers in total have been examined, gives us some confidence that those Maori cultivars sharing identical haplotypes are highly likely to be either close kin (originally from the same geographical area or wild population) or vegetative clones (scions) of the same original wild collection. This conclusion is well supported by both morphological and phenological observations over 20 years (Scheele unpublished data) and qualitative and quantitative assessments completed of their leaf and fibre properties for weaving (Harris et al. Citation2005a ,Citationb ,Citation2007 ,Citation2008; K Tawiri personal communication).
Of interest in Group 1 is the grouping of the variegated cultivar Parekoritawa with Tapoto and Ruahine. A cursory examination would suggest the former was a quite different plant; but on any bush, a number of the leaves revert from sulphur-yellow striping to wholly green, and these leaves look strikingly similar to Tapoto. Leaf and fibre qualities are also similar, as previously observed by Harris et al. (Citation2008). It is possible that these cultivars all originated as vegetative propagates of a single genetic individual and that their variable morphology (variegated or non-variegated) is the result of somatic mutation since they came into cultivation.
The cultivars in Group 2 are similar looking plants, and are classified as excellent muka varieties (Scheele Citation2005); that is, their fibres are strong, silky and easy to extract using the traditional technique of a mussel shell. Orchiston obtained these cultivars from the North Island's East Coast and the Bay of Plenty. The exception would seem to be Opiki, with the plant obtained near Foxton, Manawatu. It is described by Orchiston as an all-purpose flax, lacking the superior fibre qualities of the other cultivars in the group. This apparent anomaly, however, reinforces the long-held opinion of one of the authors (SMS) that the Opiki held in the Lincoln collection is, in fact, Arawa. Scheele was present when the divisions were first taken from bushes in the original collection in Gisborne in 1987. The large bushes were growing very closely together and not always easily distinguishable. Arawa and Opiki were growing next to each other. It is Scheele's opinion (formed once the clones matured 4 years later) that the division was taken from the wrong plant.
Orchiston described Potaka (Group 3) as similar to Ruawai, both in appearance and weaving qualities. Both were obtained on the North Island East Coast. Similarly, Te Tatua and Whareongaonga (Group 4) were acquired around Gisborne. Orchiston makes some distinctions between the latter two in her descriptions of the plants, but to our eye they look the same in form, leaf colour, inflorescence structure and capsules, with similar weaving qualities. We have long been convinced that Tapamangu (Group 5) and the unlabelled plant that Orchiston received from the weaver Te Aue Davis, were clones of the same stock, because of appearance and equivalent fibre qualities.
Phormium in the Collection were divided and replanted in autumn 2007 and the plants are now big enough for comparison of their immature forms. The cultivars listed in Group 6 are too similar in form, growth habit, leaf colour and shading to be distinguished. Old plants of these cultivars in the original reference collection grown at Havelock North also look just the same.
The cultivars in Group 7 are relatively soft leaved plants, yellow–green, having inclined scapes and capsules with a twist, suggesting an interspecific hybrid origin. All are among the first of the cultivars to flower, just as P. cookianum flowers earlier than P. tenax. It is difficult morphologically to separate Wharanui (Urewera district), Mawaru (Central North Island) and Tarere (East Coast, North Island); Paoa has thicker and firmer leaves. Again, genetic identity at our DNA fingerprint loci does not necessarily indicate that the plants concerned can be considered the same or interchangeable. It is possible that these cultivars too originated as vegetative propagates of a single genetic individual and that their variable morphology, such as Paoa's stronger leaves, is the result of somatic mutation since they came into cultivation. Alternatively, they may share a similar geographic origin but not be propagates of the same original genetic individual.
Similar profiles, such as those of Atewhiki and Te Mata, may also indicate kinship, but to what degree and whether as a result of breeding before or after entering cultivation is not clear. These two cultivars have distinctively coloured leaves that make them attractive as ornamentals, and may have been collected and redistributed as garden plants. Ngaro and Atarau also share similar profiles. Ngaro is a tall cultivar with stiff, bronzed leaves, and thick, strong fibre that shows through on the surface of the leaf. Ngaro was very popular with flax millers and was used in breeding research at Moutoa Estate in Manawatu Province in the first half of the 20th century (Orchiston obtained our clone from Moutoa). Atarau has a very similar DNA profile, and many of the same physical characteristics as Ngaro (thick, bronzed leaves, extractable fibre though not as strong). It was located by Orchiston in Whanganui and it is possible that it shares a similar geographic origin to Ngaro. Whanganui flaxes were well regarded by early Maori weavers and flax millers (Flax Commissioners Report 1870).
The clustering of Taniwha with wild P. tenax plants from Okiwi Bay likely reflects a similar genetic background in that fruit morphology suggests Taniwha may be of interspecific hybrid origin (in having terete and very narrow, twisted but upright seed pods, that become papery with age) while the Okiwi Bay population of P. tenax is also of interspecific hybrid origin (Smissen et al. Citation2008).
Origins of subantarctic island plants
Phormium does not grow naturally on the subantarctic islands and was taken there by humans (Walls Citation1998 ,Citation2009). On Campbell Island, Phormium stands are found in Tucker Cove, the site of a former farming homestead, Camp Cove (associated with a 19th century sod hut), and Tucker Valley, at the site of a World War II coastwatch station. The late George Poppleton, a leader of a New Zealand Meterological Service team stationed there in the 1950s, confirmed (G Poppleton personal communication) that he took divisions of Phormium from Tucker Cove and planted them in Tucker Valley. Phormium is evident in photos taken at Camp Cove in 1905 but does not appear in early photos of Tucker Cove; Oliver & Sorensen (cited in Walls Citation1998) report its presence at both sites — though growing poorly — in the early 1940s. Camp Cove, then, is the likely source of the Tucker Cove population. Walls (Citation1998) considered that the stands looked like Phormium growing in southern New Zealand.
On the Auckland Islands, Phormium grows in four places: Erebus Cove and at the mouth of Sealers Creek in Port Ross, Ranui Cove (opposite Ewing Island) and at Tandy Inlet on the south-east of the main island (Walls Citation2009). Erebus Cove and Tandy Inlet were both used as campsites by sealers, who were hunting around the southern and western coasts of the South Island and in the subantarctic islands in the early 19th century. They used Phormium for ropes, twine and matting. There is no known archaeological evidence of sealers camps at Sealers Creek (Prickett Citation2009), and Walls (Citation2009) considers that the small population may have become established from seeds blown or washed across the harbour from a clump at Beacon Point (along from Erebus Cove). The Ranui Cove flax is different from the other populations, droopy rather than erect, and with weak fibre. It has not been seen to flower at Ranui Cove, but divisions planted at Lincoln have produced short inflorescences with tight bunches of short, stout capsules, like the Chatham Island flax (Phormium aff. tenax). Ranui Cove was settled for a brief period between 1843 and 1856 by Maori and Moriori from the Chatham Islands (Shand Citation1893; King Citation1989) Another Phormium selection (not analysed here) grows at Crozier Point, east of Ranui Cove, and is shorter with yellow margins, like a Pitt Island flax.
In our UPGMA clustering analysis the subantarctic island samples divide into two groups. One includes most Auckland and Campbell Island plants and is associated with wild P. tenax from the South Island, including Phormium from the southern coast. The Ten Mile Creek (South Island West Coast, near Greymouth) and Erebvs Cove, Auckland Island, plants share identical nuclear genotypes but differ in their chloroplast haplotype. This may be a case of chance-sharing of genotypes because both are highly homozygous (seven of the eight loci) for high-frequency alleles. They may have independently derived their nuclear genotype following inbreeding from a similar set of ancestral South Island genotypes. Again, we know that sealers were active on the West Coast and that ‘every bay, creek, and river was examined by them in quest of these objects, and the fruit of their labour most amply recompensed them’ (Turnbull, quoted in McNab Citation1907).
The other group includes only the Ranui Cove (Auckland Island) sample, which is associated clearly in the UPGMA tree with the Chatham and Pitt Island samples. The hypothesis that this flax originated in the Chatham Islands and was brought to Auckland Island by Maori and Moriori who briefly settled there will be further addressed in future work.
Future application
Our characterization of highly polymorphic SSR loci from Phormium will potentially allow tests of relatedness among flax plants held by botanical institutions, marae and individuals, occurring at historical sites, and in the horticultural industry. Our markers also provide opportunities to study the process of speciation, evolution and interspecific hybridization in Phormium, particularly the role of ecological factors in effecting reproductive isolation between species when intrinsic reproductive isolating mechanisms are not present (Smissen et al. Citation2008). Phormium has also received research attention in relation to its breeding biology and ecological genetics (Craig & Stewart Citation1988; Craig Citation1989; Beccera & Lloyd Citation1992; Jesson et al. Citation2006; Houliston et al. Citation2008) and these markers are likely to be valuable tools for further work in these areas.
Acknowledgements
The authors acknowledge material, intellectual or technical contributions to this work by Te Roopu Raranga/Whatu o Aotearoa, Te Runanga o Ngai Tahu, PJ de Lange and Amanda Baird of the New Zealand Department of Conservation, Bill Carter, Rene Orchiston, Savannah River Ecology Lab, Nicola Bolstridge and Katarina Tawiri (Landcare Research), Canterbury Sequencing and Genotyping (University of Canterbury), and Geoff Walls. The manuscript benefited by input from G Houliston PJ Bellingham and C Bezar (Landcare Research) and two anonymous reviewers. This research was supported by funding from the Foundation for Research, Science and Technology through the Defining New Zealand's Land Biota OBI.
References
- Allan , HH and Zotov , VD . 1937 . An artificial cross between Phormium colensoi and P. tenax . The New Zealand Journal of Science and Technology , 18 : 831 – 838 .
- Beccera , JX and Lloyd , DG . 1992 . Competition-dependent abscission of self-pollinated flowers of Phormium tenax (Agavaceae): a second action of self-incompatibility at the whole flower level . Evolution , 46 : 458 – 469 .
- Best , E . 1899 . The art of the whare pora . Transactions and Proceedings of the New Zealand Institute , 31 : 625 – 658 .
- Best , E . 1942 . Forest lore of the Maori , Wellington : Dominion Museum .
- Botstein , D , White , RL , Skolnick , M and Davis , RW . 1980 . Construction of a genetic linkage map in man using restriction fragment length polymorphisms . American Journal of Human Genetics , 32 : 314 – 331 .
- Craig , JL . 1989 . Seed set in Phormium: interactive effects of pollinator behaviour, pollen carryover and pollen source . Oecologia , 81 : 1 – 5 .
- Craig , JL and Stewart , AM . 1988 . Reproductive biology of Phormium tenax: a honeyeater-pollinated species . New Zealand Journal of Botany , 26 : 453 – 463 .
- Cross BD 1912 . Investigations on Phormium with regard to the improvement of its economic importance . Unpublished MA thesis , Canterbury College , Christchurch, , New Zealand .
- Dawson , MI . 2000 . Index of chromosome numbers of indigenous New Zealand spermatophytes . New Zealand Journal of Botany , 38 : 47 – 150 .
- De Lange , PJ and Murray , BG . 2002 . Contributions to a chromosome atlas of the New Zealand flora – 37. Miscellaneous families . New Zealand Journal of Botany , 40 : 1 – 23 .
- Doyle , JJ and Dickson , EE . 1987 . Preservation of plant samples for DNA restriction endonuclease analysis . Taxon , 36 : 715 – 722 .
- Flax Commissioners 1870 . Report of the Flax Commissioners to the New Zealand House of Representatives D-14: 9 .
- Glenn , TC and Schable , NA . 2005 . Isolating microsatellite DNA loci . Methods in Enzymology , 395 : 202 – 222 .
- Harris , W , Scheele , SM , Brown , CE and Sedcole , JR . 2005a . Ethnobotanical study of growth of Phormium varieties used for traditional Maori weaving . New Zealand Journal of Botany , 43 : 83 – 118 .
- Harris , W , Scheele , SM and Forrester , GJ . 2005b . Varietal differences and environmental effects on leaves of Phormium harvested for traditional Maori weaving . New Zealand Journal of Botany , 43 : 791 – 816 .
- Harris , W , Scheele , SM , Forrester , GJ , Kanawa , KT , Murray , M and Pahewa , E . 2008 . Varietal differences and environmental effects on fibre extracted from Phormium leaves and prepared for traditional Maori weaving . New Zealand Journal of Botany , 46 : 401 – 423 .
- Harris , W , Scheele , SM , Forrester , GJ , Murray , M , Kanawa , KT and Pahewa , E . 2007 . Varietal differences and environmental effects on the characteristics of leaf strips of Phormium prepared for traditional Maori plaiting . New Zealand Journal of Botany , 45 : 111 – 137 .
- Hector , J . 1872 . Phormium tenax as a fibrous plant , Wellington : Government Printer .
- Houliston , GJ , Heenan , PB and Smissen , RD . 2008 . Experimental crosses in Phormium confirm the lack of intrinsic barriers to interspecific hybridization and the fertility of putative wild hybrids . New Zealand Journal of Botany , 46 : 393 – 399 .
- Jesson , LK , Milicich , LD and Newman , SC . 2006 . Does competition-dependent incompatibility provide reproductive assurance in Phormium tenax? . New Zealand Journal of Botany , 44 : 249 – 259 .
- King , M . 1989 . Moriori. A people rediscovered , Auckland : Viking Press .
- Liu , K and Muse , SV . 2005 . Powermarker: integrated analysis environment for genetic marker data . Bioinformatics , 21 : 2128 – 2129 .
- McBreen , K , Lockhart , PJ , McLenachan , PA , Scheele , S and Robertson , AW . 2003 . The use of molecular techniques to resolve relationships among traditional weaving cultivars of Phormium . New Zealand Journal of Botany , 41 : 301 – 310 .
- McNab , R . 1907 . Murihiku and the Southern Islands , Invercargill : William Smith .
- Meudt , HM and Clarke , AC . 2007 . Almost forgotten or latest practice? AFLP applications, analyses and advances . Trends in Plant Science , 12 : 106 – 117 .
- Moore , LB and Edgar , E . 1970 . Flora of New Zealand. Vol II , Wellington : Government Printer .
- Oliveira , EJ , Pádua , JG , Zucchi , MI , Vencovsky , R and Vieira , MLC . 2006 . Origin, evolution and genome distribution of microsatellites . Genetics and Molecular Biology , 29 : 294 – 307 .
- Prickett N 2009 . Sealing in the Auckland Islands . Dingwall PR , Jones K , Egerton R In care of the Southern Ocean. An archaeological and historical survey of the Auckland Islands . New Zealand Archaeological Association Monograph 27 .
- Scheele S 2005 . Harakeke. The Rene Orchiston Collection . Lincoln , Manaaki Whenua Press .
- Shand , A . 1893 . The occupation of the Chatham Islands by the Maoris in 1835. Part V: The residence at the Auckland Islands in 1843 . Journal of the Polynesian Society , 2 : 79 – 86 .
- Smissen , RD and Heenan , PB . 2007 . DNA fingerprinting supports hybridisation as a factor explaining complex natural variation in Phormium (Hemerocallidaceae) . New Zealand Journal of Botany , 45 : 419 – 432 .
- Smissen , RD , Heenan , PB and Houliston , GJ . 2008 . Genetic and morphological evidence for localised interspecific gene flow in Phormium (Hemerocallidaceae) . New Zealand Journal of Botany , 46 : 287 – 297 .
- Walls , GY . 1998 . Vegetation monitoring in the subantarctic islands. Conservation Advisory Science Notes 174 , Wellington : New Zealand Department of Conservation .
- Walls GY 2009 . Picking up the plant trail . In: Dingwall PR , Jones K , Egerton R In care of the Southern Ocean. An archaeological and historical survey of the Auckland Islands . New Zealand Archaeological Association Monograph 27 .
- Wehi , PM and Clarkson , BD . 2007 . Biological flora of New Zealand 10. Phormium tenax, harakeke, New Zealand flax . New Zealand Journal of Botany , 45 : 521 – 544 .