Abstract
We report here the genetic diversity of killer whales around New Zealand and compare samples collected in this region (n = 11) with larger geographic databases of mtDNA control region sequences to investigate the relationship of the New Zealand killer whales with more distant populations/ecotypes. Eight variable sites defined four haplotypes, revealing a low mtDNA genetic diversity when compared with other cetacean species and to that observed worldwide for killer whales. The geographic distribution and segregation of haplotypes within New Zealand suggested that this population could be geographically structured. Only one of the New Zealand haplotypes matched with those from other distant regions (the Eastern North Atlantic and Western South Atlantic populations).
Introduction
The killer whale Orcinus orca (Linnaeus, 1758) is reported around New Zealand coastal and offshore waters, off subAntarctic islands and the Ross Sea Dependency, Antarctica (Thomas et al. Citation1981; Baker Citation1999; Visser Citation2000; Department of Conservation Citation2012a,Citationb) (). It is believed that a population of around 100–200 killer whales frequent the main islands of New Zealand (Visser Citation2000). It has been proposed, although not confirmed, that this population is divided into several subpopulations (Visser Citation2000). Extensive movements around the main islands of New Zealand have been documented for a few individuals occurring regularly within coastal waters (Visser Citation1999a, Citation2000). This might be related with seasonality, as it has been observed in other areas around the world (Baird Citation2000). Less is known about killer whales around the New Zealand offshore and subAntarctic islands (Kermadec, Chatham, Antipodes, Snares, Auckland and Campbell; Baker Citation1999; Visser Citation2000); however, they have been regularly recorded on the Antarctic Ross Sea Dependency (Thomas et al. Citation1981; Andrews et al. Citation2008; Ainley et al. Citation2009; Lauriano et al. Citation2011).
Four morphological forms or ecotypes of killer whales have been described in the southern hemisphere (referred to here as Types A–D; Pitman & Ensor Citation2003; Pitman et al. Citation2011). Differences among the four forms is supported by historical (Mikhalev et al. Citation1981; Berzin & Vladimirov Citation1983) and contemporary (Pitman et al. Citation2007) morphological data, diet specialisation (Berzin & Vladimirov Citation1983) and genetic analysis (LeDuc et al. Citation2008; Morin et al. Citation2010; Foote et al. Citation2013). New Zealand is the only place in the southern hemisphere where three out of the four forms of killer whales have been reported (Visser Citation1999b; Pitman & Ensor Citation2003; Pitman et al. Citation2011; Foote et al. Citation2013). As these southern hemisphere forms have been described only recently, little is known about their distribution or movements.
It is believed that dependence on local prey contributes to non-random distribution and population differentiation among killer whales in other parts of the world, resulting in different foraging specialists. In the North Pacific, there is reduced gene flow among three different ecotypes: fish-eating ‘residents’; marine mammal-eating ‘transients’; and ‘offshore’ feeding on marine fish and, possibly, other offshore prey (Hoelzel et al. Citation1998; Ford et al. Citation2000; Hoelzel et al. Citation2007; Krahn et al. Citation2007; Dahlheim et al. Citation2008; Morin et al. Citation2010; Pilot et al. Citation2010). Benthic foraging of stingrays by killer whales has been reported to occur commonly in New Zealand, but seems uncommon elsewhere (Visser Citation1999a; Duignan et al. Citation2000).
Previous genetic analyses have included a small number (n = 5) of New Zealand killer whale samples for worldwide phylogeography studies, identification of putative forms/species and estimation of divergence times among types (Hoelzel et al. Citation2002b; Morin et al. Citation2010; Foote et al. Citation2013). Here, we report the genetic diversity of killer whales around New Zealand, and compare samples collected in this region with larger geographic databases (Hoelzel et al. Citation2002b; Morin et al. Citation2010; Foote et al. Citation2013) to investigate the relationship of the New Zealand killer whales with more distant populations/ecotypes.
Materials and methods
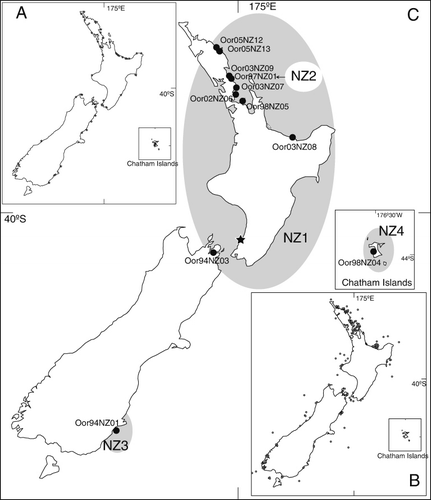
A total of 11 samples were available for analysis (, SF1). One sample was collected in the Chatham Islands, eight on the North Island and two on the South Island of New Zealand. Nine skin samples were collected from stranded whales. Four samples were collected from solitary live stranded animals, which were refloated and returned to the sea, and five were collected from dead stranded individuals. Two samples were collected from wild-ranging killer whales in the Bay of Islands, North Island, using a biopsy system (Krützen et al. Citation2002). A comparison of post ocular patches and dorsal fin photographs of three of the refloated stranded killer whales and the two wild-ranging killer whales indicated that they corresponded to different individuals. One sample included in our analysis was included previously in a published study (Hoelzel et al. Citation2002b). All samples were stored in 70% ethanol at −15 °C and archived in the New Zealand Cetacean Tissue Archive at the University of Auckland (Thompson et al. Citation2013).
Genomic DNA was extracted using a standard phenol/chloroform extraction protocol (Baker et al. Citation1994). Symmetrical amplification of the mtDNA control region was performed via the polymerase chain reaction (PCR Saiki et al. Citation1988) following standard protocols (Palumbi Citation1996). Two overlapping 800 base pair (bp) fragments of the mtDNA control region were amplified using the primers light-strand tPro-whale M13Dlp1.5 and heavy-strand Dlp8G (Garrigue et al. Citation2004), and the primers Dlp7 (5′-CCYCTTAAATAAGAC ATCVCAATGG-3′) and M13-t-phe (5′-TGTAAAACGACGGCCAGTTANNCATTTTCAGTGYWTTGCTTT-3′). Thermocycle profiles consisted of initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 40 s, and extension at 72 °C for 40 s. A final extension period at 72 °C for 10 min was included. PCR products were cleaned with ExoSAP-IT (USB) and sequenced in both directions with BigDye™ terminator chemistry on an ABI3100 DNA sequencer (Applied Biosystem), using the same amplification primers as sequencing primers. Sequencher™ (version 4.1.2, Genes Codes Co.) was utilised for the alignment and assembly of the overlapping sequences. The program MacClade version 4.0 (Maddison & Maddison Citation2000) was used to identify variable sites and unique haplotypes. Genetic diversity was estimated at haplotype (h) and nucleotide (π) levels using Arlequin version 3.5.2.1 (Excoffier et al. Citation2005). Molecular identification of sex was based on amplification of sry fragment following the protocol of Gilson et al. (Citation1998) and confirmed by field morphological observations of dorsal fin (Heyning & Dahlheim Citation1988).
Sequences were compared to the worldwide sample datasets reported by Hoelzel et al. (Citation2002b) and Morin et al. (Citation2010), including also the recent sequence of a Type D killer whale obtained by Foote et al. (Citation2013), to identify shared haplotypes. Morin et al. (Citation2010) and Foote et al. (Citation2013) complete mitochondrial genome data was trimmed to match the consensus region used in our analysis. In addition to the previous dataset, a sequence from Colombia (sample provided by S. Caballero), one from Chile (sample provided by J. Acevedo) and seven from the Japan and Korea meat markets were included in our analysis for comparisons (for more details on Japan and Korea markets surveys see Baker et al. Citation1996, Citation2000, Citation2006).
Results
The full length of the mtDNA control region (915 bp) was sequenced from seven of the individual New Zealand killer whales and nearly complete lengths from three other individuals (SF1). One of the samples could be sequenced for only the first 582 bp of the control region. The molecular sex identification confirmed the sex identification in the field of all the samples: nine males and two females. A total of eight variable sites defined four haplotypes ().
Table 1 Variable sites of New Zealand killer whales (Orcinus orca) haplotypes compared to haplotype ANTRS from Hoelzel et al. (2002b) representing a Type A killer whale and Morin et al. (Citation2010) southern hemisphere killer whales Types A, B and C haplotypes. Numbers indicate the position in relation to the beginning of the control region 5′ end.
Among the 11 individuals, the most common haplotyope (NZ1) was represented by eight individuals from the North Island and the northern tip of the South Island (). The other three haplotypes (NZ2, NZ3 and NZ4) were represented by only one individual each. Two of these were observed in the southern part of the South Island, the Chatham Islands and one in the North Island, within the range of the common NZ1 haplotype.
Given the finding of only four haplotypes and the relatively small sample size, the mtDNA diversity observed in the New Zealand population was low compared with other cetacean species (New Zealand killer whales: π = 0.0018 ± 0.0013 and h = 0.491 ± 0.175; see Pichler & Baker [Citation2000] and Hoelzel et al. [Citation2002a]) for values of genetic diversity on cetacean species) and to that observed at worldwide level for killer whales (π = 0.0052 ± 0.0031, Hoelzel et al. Citation2002b) and some regional populations of odontocetes.
The New Zealand killer whale mitochondrial sequences obtained here were compared to haplotypes from other geographic populations around the world. Two of the four New Zealand haplotypes (NZ3 and NZ4) did not match with other haplotypes from either datasets (sequences were deposited in GenBank accession no. KC465959 and KC465960, respectively). Our NZ1 haplotype corresponded to one of the haplotypes from New Zealand (WSPNZ1) previously reported by Hoelzel et al. (Citation2002b) and one (SWPUNZ) reported by Morin et al. (Citation2010). Our NZ2 haplotype matched with one haplotype from Hoelzel et al. (Citation2002b); WSPNZ2), for the reason that this individual (Oor97NZ01) was included in both studies. The only New Zealand haplotype shared with any other region around the world was NZ1. Based on the available sequence length this haplotype is shared with whales from the Eastern North Atlantic and Western South Atlantic populations. It is possible that this identity is the result of a weak phylogeographic signal in the killer whale mtDNA control region, or of recent common ancestry of these extensively separated populations, as has been recently shown by Morin et al. (Citation2010).
Discussion
The geographic distribution and segregation of the New Zealand mtDNA haplotypes obtained here suggest that this population could be geographically structured. The mostly non-overlapping distribution of the four haplotypes suggests some degree of segregation that may be the result of a reduced number of matrilineal groups and some degree of local philopatry. A more intensive and wide-ranging sampling is needed to assess this, together with the incorporation of demographic data into the analysis.
An alternative explanation for the observed geographic distribution of haplotypes could be that the different haplotypes represent the different morphological forms or ecotypes of killer whales reported in New Zealand (Visser Citation1999b, Citation2000; Pitman & Ensor Citation2003; Pitman et al. Citation2011). Photographic records indicate that five of our New Zealand samples (Oor97NZ01, Oor03NZ07, Oor03NZ08, Oor05NZ12, Oor05NZ13; SF ), which included both NZ1 and NZ2 haplotypes, were collected from the type of killer whales found worldwide (Pitman & Ensor Citation2003). It is not possible to rule out that the more geographically and genetically distant killer whales from Chatham Islands and South Island represent different forms as no photographic records were available. However, two specific fixed differences reported for Types B and C killer whales (in positions 476 [T] and 536 [G] of the mtDNA control region; Morin et al. Citation2010), were not present in New Zealand killer whales. This suggests that these individuals did not correspond to either of these forms (SF ). The comparison of New Zealand haplotypes with the recently reported sequence of a Type D killer whale (Foote et al. Citation2013) also confirmed that our samples did not represent this poorly described form.
Supplementary files
Supplementary file 1: Table 1. Details of sample collection from killer whales (Orcinus orca) in New Zealand waters.
Supplementary file 2: Figure 1. Photographs of four New Zealand killer whales (Orcinus orca) sampled for this genetic analysis (samples codes Oor03NZ07, Oor03NZ08, Oor05NZ12, Oor05NZ13). Photographs of individual associated to sample Oor97NZ01 can be found in Visser & Fertl (2000).
Photographs of individuals associated to sample Oor97NZ01 can be found in Visser & Fertl (Citation2000).
Figure 1. Photographs of four New Zealand killer whales (Orcinus orca) sampled for this genetic analysis.
Download MS Word (24.7 KB)Table 1. Details of sample collection from killer whales (Orcinus orca) in New Zealand waters.
Download MS Word (313.4 KB)Acknowledgements
We thank A. Hickey and D. Steel for laboratory analysis and G. Stone, K. Russell, N. Patenaude, E. Slooten, J. Berghan, T. Wilson, M. Tully and the Department of Conservation for providing tissue samples. S. Caballero and J. Acevedo kindly provided the Colombian and Chilean killer whale samples, respectively. S. Lavery and M. Dalebout provided the Japan and Korea markets killer whales sequences. A. Foote provided the sequence of Type D killer whale. We thank the Department of Conservation, (L. Boren and T. Beauchamp) and the Museum of New Zealand Te Papa Tongarewa (A. van Helden), co-administrators of the New Zealand Marine Mammal Stranding and Sightings Database for providing access to the database. Funding for fieldwork was provided by the Conservation Action Fund of the New England Aquarium, the Bermuda Underwater Exploration Institute, the National Geographic Society, Northland Marine Mammal Trust, Department of Conservation (Northland). Genetic analyses were supported by a New Zealand Marsden Fund and an International Fund for Animal Welfare (IFAW) grants to C.S. Baker, a University of Auckland International Doctoral Scholarship to C. Olavarría and a University of Auckland Postgraduate Tuition Fee Bursary to G. Tezanos-Pinto. Biopsy samples were collected under permit to CSB from the New Zealand Department of Conservation and animal ethics protocols AEC/02/2002/R9 and AEC/02/2005/R334 from the University of Auckland.
References
- Ainley DG, Ballard G, Olmastroni S 2009. An apparent decrease in the prevalence of ‘Ross Sea Killer Whales’ in the southern Ross Sea. Aquatic Mammals 35: 334–346. 10.1578/AM.35.3.2009.334
- Andrews RD, Pitman RL, Ballance LT 2008. Satellite tracking reveals distinct movement patterns for Type B and Type C killer whales in the southern Ross Sea, Antarctica. Polar Biology 31: 1461–1468. 10.1007/s00300-008-0487-z
- Baird RW 2000. The killer whale. Foraging specializations and group hunting. In: Mann J, Connor RC, Tyack PL, Whitehead H eds. Cetacean societies. Field studies of dolphins and whales. Chicago and London, The University of Chicago Press. Pp. 127–153.
- Baker AN 1999. Whales and dolphins of New Zealand & Australia. 3rd edition. Wellington, Victoria University Press. 131 p.
- Baker CS, Cipriano F, Palumbi SR 1996. Molecular genetic identification of whale and dolphin products from commercial markets in Korea and Japan. Molecular Ecology 5: 671–685. 10.1111/j.1365-294X.1996.tb00362.x
- Baker CS, Lento GM, Cipriano F, Dalebout ML, Palumbi SR 2000. Scientific whaling: source of illegal products for market? Science 290: 1695–1696. 10.1126/science.290.5497.1695b
- Baker CS, Lukoschek V, Lavery S, Dalebout ML, Yong-un M, Endo T et al. 2006. Incomplete reporting of whale, dolphin and porpoise ‘bycatch’ revealed by molecular monitoring of Korean markets. Animal Conservation 9: 474–482. 10.1111/j.1469-1795.2006.00062.x
- Baker CS, Slade RW, Bannister JL, Abernethy RB, Weinrich MT, Lien J et al. 1994. Hierarchical structure of mitochondrial DNA gene flow among humpback whales Megaptera novaeangliae, world-wide. Molecular Ecology 3: 313–327. 10.1111/j.1365-294X.1994.tb00071.x
- Berzin AA, Vladimirov VL 1983. Novyi vid kosatki (Cetacea, Delphinidae) iz vod Antarktiki [A new species of killer whale (Cetacea, Delphinidae) from Antarctic waters]. Zoologicheskiy Zhurnal 62: 287–295.
- Dahlheim ME, Schulman-Janiger A, Black N, Ternullo R, Ellifrit D, Balcomb KC 2008. Eastern temperate North Pacific offshore killer whales (Orcinus orca): occurrence, movements, and insights into feeding ecology. Marine Mammal Science 24: 719–729. 10.1111/j.1748-7692.2008.00206.x
- Department of Conservation 2012a. New Zealand marine mammal sightings database. Wellington, New Zealand, Department of Conservation. http://www.doc.govt.nz/conservation/native-animals/marine-mammals/marine-mammal-sightings/ (accessed 1 November 2013).
- Department of Conservation 2012b. New Zealand marine mammal stranding database. Wellington, New Zealand, Department of Conservation.
- Duignan PJ, Hunter JEB, Visser IN, Jones GW, Nutman A 2000. Stingray spines: a potential cause of killer whale mortality in New Zealand. Aquatic Mammals 26: 143–147.
- Excoffier L, Laval G, Schneider S 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50.
- Foote AD, Morin PA, Pitman RL, Ávila-Arcos MC, Durban JW, van Helden A et al. 2013. Mitogenomic insights into a recently described and rarely observed killer whale morphotype. Polar Biology 36: 1519–1523.
- Ford JKB, Ellis GM, Balcomb KC 2000. Killer whales: the natural history and genealogy of Orcinus orca in British Columbia and Washington State. Vancouver, UBC Press.
- Garrigue C, Dodemont R, Steel D, Baker CS 2004. Organismal and ‘gametic’ capture-recapture using microsatellite genotyping confirm low abundance and reproductive autonomy of humpback whales on the wintering grounds of New Caledonia. Marine Ecology Progress Series 274: 251–262. 10.3354/meps274251
- Gilson A, Syvanen M, Levine K, Banks J 1998. Deer gender determination by Polymerase Chain Reaction: validation study and application to tissues, bloodstains and hair forensic samples from California. California Fish and Game 84: 159–169.
- Heyning JE, Dahlheim ME 1988. Orcinus orca. Mammalian Species 304: 1–9. 10.2307/3504225
- Hoelzel AR, Dahlheim ME, Stern SJ 1998. Low genetic variation among killer whales (Orcinus orca) in the Eastern North Pacific and genetic differentiation between foraging specialists. The Journal of Heredity 89: 121–128. 10.1093/jhered/89.2.121
- Hoelzel AR, Goldsworthy SD, Fleischer RC 2002a. Population genetic structure. In: Hoelzel AR ed. Marine mammal biology. An evolutionary approach. Bodmin, Cornwall, UK, Blackwell Science. Pp. 325–352.
- Hoelzel AR, Natoli A, Dahlheim ME, Olavarría C, Baird RW, Black NA 2002b. Low worldwide genetic diversity in the killer whale (Orcinus orca): implications for demographic history. Proceedings of the Royal Society of London Series B-Biological Sciences 269: 1467–1473. 10.1098/rspb.2002.2033
- Hoelzel AR, Hey J, Dahlheim ME, Nicholson C, Burkanov V, Black N 2007. Evolution of population structure in a highly social top predator, the killer whale. Molecular Biology and Evolution 24: 1407–1415. 10.1093/molbev/msm063
- Krahn MM, Herman DP, Matkin CO, Durban JW, Barrett-Lennard L, Burrows DG et al. 2007. Use of chemical tracers in assessing the diet and foraging regions of eastern North Pacific killer whales. Marine Environmental Research 63: 91–114. 10.1016/j.marenvres.2006.07.002
- Krützen M, Barre LM, Moller LM, Heithaus MR, Simms C, Sherwin WB 2002. A biopsy system for small cetaceans: darting success and wound healing in Tursiops sp. Marine Mammal Science 18: 863–878. 10.1111/j.1748-7692.2002.tb01078.x
- Lauriano G, Fortuna CM, Vacchi M 2011. Occurrence of killer whales (Orcinus orca) and other cetaceans in Terra Nova Bay, Ross Sea, Antarctica. Antarctic Science 23: 139–143. 10.1017/S0954102010000908
- LeDuc RG, Robertson KM, Pitman RL 2008. Mitochondrial sequence divergence among Antarctic killer whale ecotypes is consistent with multiple species. Biology Letters 4: 426–429. 10.1098/rsbl.2008.0168
- Maddison DR, Maddison WP 2000. MacClade: analysis of phylogeny and character evolution. 4.0 ed. Sunderland, MA, Sinauer Associates.
- Mikhalev YA, Ivashin MV, Savusin VP, Zelenaya FE 1981. The distribution and biology of killer whales in the Southern Hemisphere. Report of the International Whaling Commission 31: 551–566.
- Morin PA, Archer FI, Foote AD, Vilstrup J, Allen EE, Wade P et al. 2010. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Research 20: 908–916. 10.1101/gr.102954.109
- Palumbi SR 1996. Nucleic acids II: the Polymerase Chain Reaction. In: Hillis D, Moritz C, Mable BK eds. Molecular systematics. 2nd edition. Sunderland, MA, Sinauer Associates. Pp. 205–248.
- Pichler FB, Baker CS 2000. Loss of genetic diversity in the endemic Hector's dolphin due to fisheries-related mortality. Proceedings of the Royal Society of London Series B-Biological Sciences 267: 97–102. 10.1098/rspb.2000.0972
- Pilot M, Dahlheim ME, Hoelzel AR 2010. Social cohesion among kin, gene flow without dispersal and the evolution of population genetic structure in the killer whale (Orcinus orca). Journal of evolutionary Biology 23: 20–31. 10.1111/j.1420-9101.2009.01887.x
- Pitman RL, Ensor P 2003. Three forms of killer whales (Orcinus orca) in Antarctic waters. Journal of Cetacean Research and Management 5: 131–139.
- Pitman RL, Perryman WL, Leroi D, Eilers E 2007. A dwarf form of killer whale in Antarctica. Journal of Mammalogy 88: 43–48. 10.1644/06-MAMM-A-118R1.1
- Pitman RL, Durban JW, Greenfelder M, Guinet C, Jorgensen M, Olson PA et al. 2011. Observations of a distinctive morphotype of killer whale (Orcinus orca), type D, from subantarctic waters. Polar Biology 34: 303–306. 10.1007/s00300-010-0871-3
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT et al. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239: 487–491. 10.1126/science.2448875
- Thomas JA, Leatherwood S, Evans WE, Jehl JR Jr, Awbrey FT 1981. Ross sea killer whale distribution, behavior, color pattern and vocalizations. Antarctic Journal 16: 157–158.
- Thompson KF, Millar CD, Baker CS, Dalebout M, Steel D, van Helden AL et al. 2013. A novel conservation approach provides insights into the management of rare cetaceans. Biological Conservation 157: 331–340. 10.1016/j.biocon.2012.07.017
- Visser IN 1999a. Benthic foraging on stingrays by killer whales (Orcinus orca) in New Zealand waters. Marine Mammal Science 15: 220–227. 10.1111/j.1748-7692.1999.tb00793.x
- Visser IN 1999b. Antarctic orca in New Zealand waters? New Zealand Journal of Marine and Freshwater Research 33: 515–520. 10.1080/00288330.1999.9516896
- Visser IN 1999c. Propeller scars on and known home range of two orca (Orcinus orca) in New Zealand waters. New Zealand Journal of Marine and Freshwater Research 33: 635–642. 10.1080/00288330.1999.9516906
- Visser IN 2000. Orca (Orcinus orca) in New Zealand waters. Unpublished PhD thesis. University of Auckland, Auckland, New Zealand. 199 p.
- Visser IN, Fertl D 2000. Stranding, resighting, and boat strike of a killer whale (Orcinus orca) off New Zealand. Aquatic Mammals 26: 232–240.
