ABSTRACT
The dinoflagellate family Warnowiaceae has often been considered to include some of the most complex cells among the protists. The number of described species is around 40, but both the species and generic concepts are in need of revision. Warnowiaceans are often fragile and readily change morphology under the light microscope, and they are usually regarded to be rare. They are particularly famous for the eye-like structures, termed ocelloids. Studies on warnowiaceans are hampered by lack of cultures, and our studies are therefore based on cells obtained directly from field samples. We provide a description of Nematodinium parvum comb. nov. (syn. Pouchetia parva, Warnowia parva), based on light, scanning and transmission electron microscopy, combined with phylogenetic analyses of ribosomal genes. It was described in 1908 by Lohmann from Kieler Bay, but is often common in Danish waters, allowing observations on distribution and behaviour. Crucial conditions for finding high cell abundances were periods of warm temperatures and a calm sea. Cells were yellowish, photosynthetic, and contained a net-like chloroplast, in addition to an ocelloid, trichocysts and harpoon-like nematocysts. They divided asexually but planozygotes were also seen. Following the demonstration of nematocysts, the species, which has been known as Warnowia parva since 1928, is transferred to Nematodinium. The finding of a peduncle indicates mixotrophy but all feeding experiments failed to identify a suitable prey. The phylogenetic analyses based on single-cell PCR and sequence determinations of small and large subunit rDNA confirmed that the systematics of Nematodinium and Warnowia is in a state of flux.
INTRODUCTION
Species of the dinoflagellate family Warnowiaceae (Gymnodiniales; Gómez et al. Citation2009) are some of the most complex unicellular organisms known. Both photosynthetic and phagocytotic species have been described, and species of all four main genera, Warnowia Er. Lindemann, Nematodinium Kofoid & Swezy, Erythropsidinium P.C. Silva and Proterythropsis Kofoid & Swezy, possess eye-like structures known as ocelloids, while the genera Nematodinium and Proterythropsis further contain nematocysts, and Erythropsidinium a so-called piston. The origin and function of ocelloids in the Warnowiaceae has formed the basis of much speculation since they were first observed (see details below). However, recent molecular studies have resolved their chimaeric nature and shown that chloroplast-encoded protein genes from the melanosome (retinal body) in a concatenated phylogenetic analysis grouped with dinoflagellates having the peridinin-type chloroplast. The chloroplasts in the peridinin-containing dinoflagellates originated from an endosymbiosis with a red alga (Gavelis et al. Citation2015). However, the genetic history of the ocelloid seems even more complicated, as Hayakawa et al. (Citation2015) showed that mRNA of rhodopsin pigment in Erythropsidinium originated from bacteria, and may have been acquired from either diatoms or haptophytes.
The first warnowiacean species described in some detail was Erythropsis agilis Hertwig, found by Richard Hertwig (Citation1884) in the Gulf of Naples, in the Mediterranean. The Ukrainian Metschnikoff had previously studied in some detail a single cell near Funchal at Madeira as early as 1872 (Metschnikoff Citation1874, Citation1885) but did not publish any illustrations nor a formal description. However, Metschnikoff was the first to interpret the ocellus as an eye, and he described both the lens and the pigment cup. Hertwig’s find was immediately repudiated by Vogt (Citation1885a) in Geneva, and the cell claimed to represent a ciliate that had engulfed a dead medusa, the eye coming from the medusa. This resulted in a brief but intense dispute (Hertwig Citation1885), but Vogt never admitted defeat, and one of his papers, which includes a poem on scientific errors and the use of osmium tetroxide (Vogt Citation1885b), makes for strange reading (‘Humans make errors as long as they live’). However, Metschnikoff (Citation1885) supported Hertwig, and an additional species was independently found at the same time in France by Pouchet (Citation1885a, Citation1885b, Citation1887) and described as Gymnodinium polyphemus C.H.G. Pouchet. The subsequent 135 years have resulted in the description of many species, 37 of which are mentioned in Kofoid & Swezy (Citation1921). The latter authors described many new species, often based on a single or a few individuals and using morphological features such as colour, and the structure and location of the ocelloid.
Later, Greuet (Citation1972) showed that many of the morphological features used to characterize the species vary during the cell life cycle. However, he was unable to construct an alternative taxonomic system, and went as far as saying that it would be more prudent at the time to consider the genera monospecific. A few years later, Elbrächter (Citation1979) studied over 50 specimens of Erythropsidinium during a cruise on the ‘Meteor’ and came to the conclusion that the six additional species of Erythropsidinium described by Kofoid & Swezy (Citation1921) may perhaps all be identical to Hertwig’s E. agilis, making this genus monospecific.
The number of species in the Warnowiaceae described is presently around 40, but since both the species and the generic concepts are uncertain, taxonomy of the group must start almost from scratch, taking care to use populations whose identity can be determined with confidence rather than one or a few cells, preferably from or near the type localities.
We have over a number of years studied warnowiaceans from Danish and Greenlandic waters. Here we present a description of Danish material characteristic of a species that agrees well with Lohmann’s Pouchetia parva (Lohmann Citation1908). This species was subsequently renamed Warnowia parva by Lindemann (Citation1928), as the generic name Pouchetia F. Schütt (Citation1895) is illegitimate in ‘botanical’ nomenclature, being a homonym of the Rubiaceae Pouchetia A. Richard ex de Candolle (Citation1830). Our study was based on detailed light, scanning and transmission electron microscopy combined with molecular data based on single-cell PCR. The aim was to seek a basis for a more reliable taxonomy for warnowiaceans.
Lohmann described Warnowia parva from Kieler Bay in the Baltic Sea, albeit in an incomplete way. The description is nevertheless sufficient to recognize it as the commonest warnowiacean species in the Danish parts of the Baltic. Below we transfer the species formally to Nematodinium as Nematodinium parvum comb. nov., and this name will be used in the following.
MATERIAL AND METHODS
Sampling
Surface water was sampled with a 30-litre bucket on numerous occasions. The material used here was sampled at (1) Rønbjerg in the Limfjord (May 2015, April 2019, May and June 2021); (2) Risø at Roskilde Fjord (May 2019 and September 2019) and (3) Skovshoved Harbour at the Sound (Øresund) (July 2021) (). For all locations the temperature range was 15–27°C and the salinity range 16–27. The bucket was subsequently transported as quickly as possible to a darkened culture room, and light from a Leica CLS 150X microscope light source (Leica, Wetzlar, Germany) was mounted on the edge of the bucket near the water surface. Cells accumulated at the water surface near the light source, and the highest cell number was noted between 30 min and 2 h of exposure.
Light microscopy of live and fixed cells
Live cells from freshly collected samples, or samples kept in the laboratory for up to two days, were concentrated as described above and observed using a Carl Zeiss Axio Imager.M2 (Zeiss, Oberkochen, Germany) equipped with Nomarski interference contrast and a 63× oil-immersion lens. Colour micrographs were taken with a Zeiss AxioCam HRc digital camera (Zeiss, Oberkochen, Germany). Images of live cells were used for morphometric measurements of cell length and width using the Zen image acquisition software (Zeiss). For 3D-microscopy of chloroplast and nucleus, cells were fixed in glutaraldehyde (final concentration 2%), filtered onto a filter with a pore size of 0.22 µm (Whatman, GE Healthcare, Chicago, Illinois, USA) and placed on a slide to which 20 µl of Vectashield had already been added. A second drop of Vectashield was placed on the upper side of the filter, before placing a coverslip on top. Vectashield mounting medium (Vector Laboratories, Burlingame, California, USA) contains 4ʹ-6-diamidino-2-phenylindole (DAPI), which stains DNA. Nuclear and chloroplast DNA was observed using the Zeiss filter set 49 (excitation band pass 365 nm and emission 445 nm). Chlorophyll autofluorescence was recorded using a Zeiss Axiocam 506 mono digital camera and a Zeiss filter set 09 (excitation BP450–490, emission LP515). Recorded z-stacks comprising 85–91 single images were imported into Imaris 9.2.1 (Bitplane Scientific Software, Zürich, Switzerland) for three-dimensional reconstructions. The snapshot option was applied for exporting combined stacks.
Electron microscopy
For scanning electron microscopy, a 800-µl suspension of swimming cells from Roskilde Fjord collected 17 June 2019 was fixed for 55 min at room temperature in a cocktail of 960 µl 4% OsO4 in seawater, 960 µl seawater and 480 µl saturated HgCl2. The cells were subsequently concentrated on a 5-µm Isopore Membrane filter (Millipore, Tullagreen, Cork, Ireland) in a Swinnex filter holder (Millipore), carefully rinsed for 15 min in distilled water, and dehydrated in an ethanol series, 12–20 min in each step, before staying overnight in absolute ethanol with molecular sieves to complete dehydration. It was critical point dried through liquid carbon dioxide in a BAL-TEC 030 Critical Point Drying apparatus (Balzers, Liechtenstein). The cells were coated with a thin layer of gold-palladium before being examined and photographed in a JEOL 6335F field emission Scanning Electron Microscope (Jeol, Tokyo, Japan).
For transmission electron microscopy, water samples from Rønbjerg, 21 May 2015, and Roskilde Fjord, 13 May 2019, were transported to University of Copenhagen, and cells concentrated as described above. Cells were fixed the day after collecting, having spent the night in a 15°C culture room. They were fixed in a cold osmium tetroxide-glutaraldehyde mixture in water (final concentration in the fixation cocktail 1.7% glutaraldehyde and 0.4% osmium tetroxide) for c. 1 h in a fridge, and subsequently concentrated into a pellet by centrifugation. After washing in cold (4°C) culture medium for 30–60 min, cells were post-fixed in 2% osmium tetroxide in distilled water for 1½ h or overnight in the fridge. This was followed by a brief rinse in culture medium, and dehydration in a series of cold ethanol, c. 20 min in each step of 30%, 50%, 70% and 96% ethanol. Dehydration was completed at room temperature in absolute ethanol (several changes over c. 1½ h) and propylene oxide (two changes over 10 min), followed by a 1:1 mixture of propylene oxide and Epon 812. The propylene oxide evaporated overnight in a fume hood, and the Epon was then replaced with another batch for c. 1½ h before transfer to Epon in a flat embedding mould and careful spreading of the cells on the bottom of the mould. The resin was subsequently cured at 65°C overnight or longer.
Portions of the resin containing the target cells were cut out, remounted on a stub, and sectioned with a diamond knife on an LKB-8800 ultramicrotome (LKB, Bromma, Sweden). Sections were collected on slot grids, stained in uranyl acetate and lead citrate, and examined in a JEM-1010 transmission electron microscope operating at 80 kV (JEOL, Tokyo, Japan).
Molecular sequencing
Material from all three locations was transferred with glass pipettes onto glass slides for single-cell isolations under a dissecting microscope. Individual cells were selected with a drawn-out glass pipette with a cotton plug. They were washed in three drops of filtered sea water on the glass slide and transferred to 0.2-ml PCR tubes containing 100 µl of water and 10% (w/v) Chelex 100 (Sigma-Aldrich #C7901). A total of 10 cells from each location were isolated for single-cell PCR.
For DNA extraction, PCR tubes were vortexed for 5s, spun down in a microcentrifuge for 10s, and subsequently incubated at 95°C for 20 min (Richlen & Barber Citation2005). After incubation, the tubes were centrifuged for 10s and stored at 4°C until used for PCR reactions. Two microlitres of the DNA extract were used as template in the subsequent PCR reactions, which involved the primer pairs: SSUF-SR7; SR4-SR9p; SR6-SSUR; 25F1-LSUr (see Table S1). PCR reactions were 25-µl reactions containing 2 mM MgCl2, 0.4 mM dNTPs, 0.625 units polymerase (Thermo Scientific #K0171), 0.8 µM primers, and using 40 cycles and an annealing temperature of 56°C. PCR products were sent to Macrogen (Macrogen Europe, Amsterdam, Netherlands) for purification and sequencing. For SSU rDNA, six cells (out of 10) provided high quality sequences (one from Roskilde, three from Rønbjerg and two from Skovshoved). For LSU rDNA, three cells (out of 10) resulted in high quality sequences (one from Rønbjerg and two from Skovshoved).
Alignment and phylogenetic analyses
For sequence trimming and assembly as well as BLAST we used Geneious Prime v2022.1.1. Three data matrices were constructured and the first included SSU rDNA sequences (1,755 base pairs) from a diverse assemblage of athecate dinoflagellates (warnowiaceans, and species of Gymnodinium F. Stein, Polykrikos Buetschli, Pheopolykrikos Chatton, Lepidodinium M.M. Watanabe, S. Suda, I. Inouye, Sawaguchi & Chihara, Karenia Gert Hansen & Moestrup, Karlodinium J. Larsen and a few more). Two thecate species of Peridinium Ehrenberg formed the outgroup (Fig. S1). This first phylogenetic analysis was included to further explore the phylogenetic status of warnowiaceans. Confirming that the Warnowiaceae formed a monophyletic group (Fig. S1) two additional (i.e. second and third) data matrices were compiled. The second SSU rDNA data matrix (1,749 base pairs) included a subset of the first data matrix but only comprised warnowiaceans (20 sequences) and Gymnodinium and Lepidodinium as the outgroup taxa. The third data matrix comprised 10 LSU rDNA sequences (720 base pairs) of warnowiaceans and had the same species of Gymnodinium and Lepidodinium as the outgroup. Concatenation was not attempted as only five warnowiacean taxa have had both their SSU and LSU rDNA sequences determined.
All sequences were retrieved from GenBank and aligned using MAFT, and all data matrices were used to build trees using MrBayes with the GTR+G model and four MCMC (Huelsenbeck & Ronquist Citation2001). The first alignment was run for 1 million generations (burn-in of the first 25% of the trees), the second and third alignments were run for 5 million generations (burn-in of the first 20% of the trees). All Bayesian analyses sampled a tree every 1,000 generation. For phylogentic analyses based on RAxML v8.2.11 (Stamatakis Citation2014) we started with complete random trees followed by 1,000 bootstrap replicates for the first data matrix and 10,000 bootstrap replicates for the second and third data matrix. All analyses were conducted using Geneious Prime (v2022.1.1 and v2023.0.4). The LSU and SSU rDNA gene sequences obtained have been submitted to GenBank, and the accession numbers are ON923865, OP442509 (LSU rDNA) and ON924314 (SSU rDNA).
Culturing attempts
Several attempts were made to identify the prey potentially used for food by Nematodinium parvum. Cells of N. parvum were isolated from different field locations in Denmark and tested against different species of prey (n ≤ 4 per test) and at culture conditions detailed in Table S2. Briefly, the tested prey included several species of dinoflagellates, ciliates, diatoms, green algae (served as either a mix or monospecific) and copepods (whole organisms or eggs). The light:dark cycle was 16:8 h, temperature was set at 15°C and the salinity was adjusted to the measured values from nature (15–20, depending on the sampling location).
RESULTS
Light microscopy
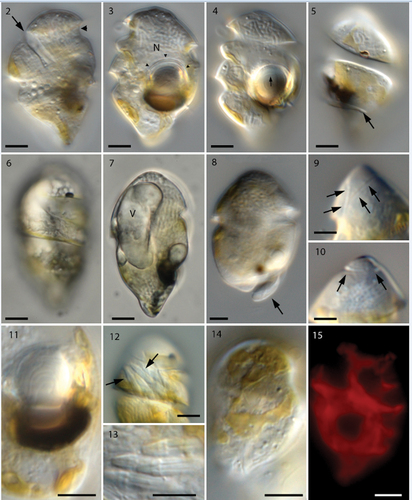
Live cells of Nematodinium parvum were observed on numerous occasions over a period of several years, but the images included here () are based on samples collected from Rønbjerg Harbour, the Limfjord, on 11 April 2019 and 24 May 2021 (). Cells measured 32.3 ± 2.95 µm (25.8–38.6 µm) in length and 20.3 ± 2.61 µm (16–25.1 µm) in width (n = 17). The cell outline was ovoidal with a widely rounded apex and a characteristic conical antapex (). However, some cells had a more pointed apex (). The cingulum started near the apical end, close to the uppermost part of the sulcus (arrow in ), and ended at the antapical end after three complete turns. A nose-like appendage of the antapical end delimited the last cingular turn (). The anterior rim of the cingulum continued as a flagellum-like extension to encircle the lower part of the cell (, arrow). A small, dark-coloured, elongate granule was present close to the anterior rim of the anteriormost turn of the cingulum ().
Figs 2–15. Longitudinal view of live cells of Nematodinium parvum in Nomarski interference contrast (Figs 2–14) and epifluorescence (Fig. 15).
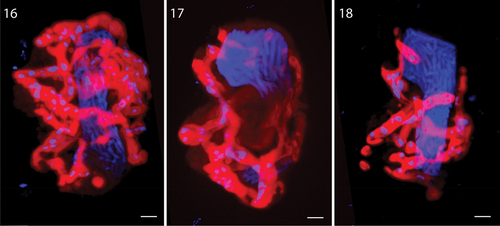
Figs 16–18. Glutaraldehyde-fixed cells of Nematodinium parvum from stacked series of images, used to construct the 3-dimensional structure of chloroplast and nucleus. The nucleus fills up a major part of the cell, reaching from the apex to the antapex. The chloroplast is highly reticulated but the morphology varies markedly between cells. The blue dots in the chloroplast lobes represent chloroplast-DNA.
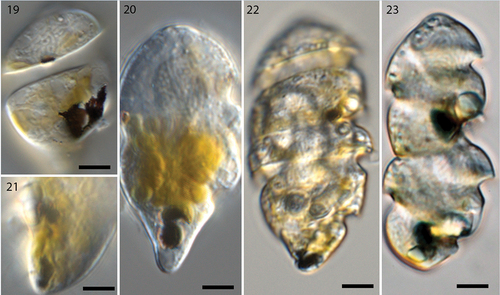
Figs 19–23. Live cells of Nematodinium parvum in different stages of mitotic division (Nomarski interference contrast).
The position and orientation of the ocelloid was always on the lower left side of the cell (). When fully mature, the dark-brown melanosome measured c. 10 µm in width and was 3 µm thick (). The concave part adjoined the hyalosome, which was c. 7 µm thick (). Higher magnification showed the hyalosome to contain concentric proximal ring-like structures ( cornea-like external envelope (, arrowheads). The apical furrow was U-shaped, and one branch started near the proximal part of the cingulum. The right branch was slightly longer than the left one (, arrows) and took a full turn on the dorsal side (, arrows). The nucleus made up a major portion of the cell, reaching from the apex to the antapex (). Parts of the golden-brown, permanent, chloroplast could be discerned in all cells (), always positioned along the perimeter as thin () or wide () lobes.
Epifluorescence microscopy showed the chloroplast lobes to be highly reticulated (). On a few occasions, cells contained a large elongate vacuole (19 µm long and 8 µm wide; ). Its contents remain obscure and it was not observed in sectioned material. When bundles of nematocysts were present (they were sometimes lacking), they were always located in the area between the first and second cingular curve () and measured 3.9 µm in length and 1.1 µm in width ().
Swimming behaviour
A few mililitres of illuminated surface water from the large buckets were transferred to Utermöhl sedimentation chambers or Petri dishes. The swimming behaviour was observed using an inverted microscope and showed that cells were always active and moving rapidly across the chamber or dish. When reaching the side of the chamber they stopped briefly, swam rapidly downwards, and later upwards again.
Three-dimensional configuration of nucleus and chloroplast
The 3D-outline of the nucleus and chloroplast were examined in a few glutaraldehyde-fixed cells to illustrate the morphological variation of the reticulate chloroplast (). Small perforations in the chloroplast lobes were filled with chloroplast-DNA as witnessed by the DAPI staining. The host nucleus was confirmed to take up a large proportion of the cell cytoplasm ().
Asexual cell division
Our observations over several years have resulted in some understanding of ocelloid transformation during asexual division (). The first sign of asexual division was observed early in the morning, when the melanosome began disintegrating by splitting into two parts (). At this stage, the hyalosome had apparently broken down. Next, a clear separation of the melanosome was observed, with one part located at the very antapex and the other in the position typical for the non-dividing stage (). Subsequently, when two daughter cells had started forming but had not yet separated, the ocelloid was in the process of maturing, one still in the same position (upper cell in ), the other in the antapex. In the final stage shown here, close to cytokinesis, the ocelloids in both daughter cells had matured and taken up their final position, the hyalosome and melanosome having reached the size seen in single cells. Cell fusion (sexual reproduction) or cyst stages were never observed under the light microscope (but see below).
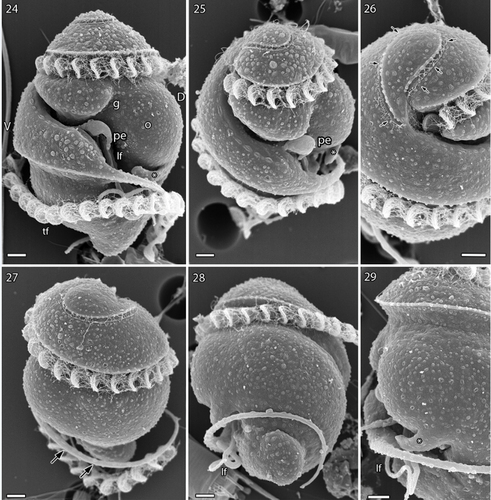
Scanning electron microscopy
The preparation contained both vegetative cells () and cells with double sets of both flagella, which we interpret as planozygotes (). The apical furrow was seen just below the cell apex (), and in a cell has been tilted to show the furrow from start to end (arrows). The apical furrow was horseshoe- or U-shaped, its two ends separated by a distance of just over 2 µm. A row of small projections was seen on the inner margin of the U (compare with ), and very thin fibrillary structures (apparently mucoid) were present on the outer margin of the furrow.
Figs 24–29. Scanning electron microscopy of Nematodinium parvum.
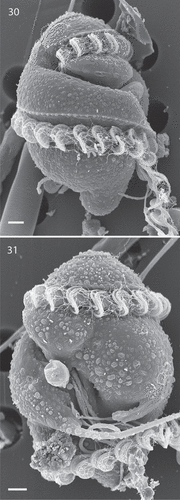
Figs 30, 31. Scanning electron microscopy of planozygotes of Nematodinium parvum.
The cingulum and the sulcus emerged close to the gap between the two ends of the apical furrow (). The cingulum took two whole turns around the cell but at the antapex the anterior, thickened rim of the cingulum continued () as a separate flagellum-like structure for another, third turn (). It delimited the nose-like antapical end of the cell (). The sulcus proceeded as a deep furrow along the ventral and left-hand side of the cell (), but the longitudinal flagellum was not visible until half-way down the cell ( and better in the planozygote in ). At the same level, a peduncle (pe) emerged as a short projection from the sulcus (). A relatively deep groove extended in an oblique anterior direction from this area to the anterior part of the cingulum (). The ocelloid was seen as a swelling on the cell surface, on the right side of the groove in . The proximal part of the longitudinal flagellum was in the upper half of the cell, but was located in a tube inside the cell (see below). An additional, finger-like projection emerged from the cell in the area just below the ocellus and extended in a direction opposite to the cingulum (, asterisk).
Sexual reproduction is illustrated in , which show two parallel longitudinal flagella () emerging from the bottom of the mid-region of the sulcus. Two parallel transverse flagella were also present, the curvatures of each flagellum very precisely aligned ().
Transmission electron microscopy
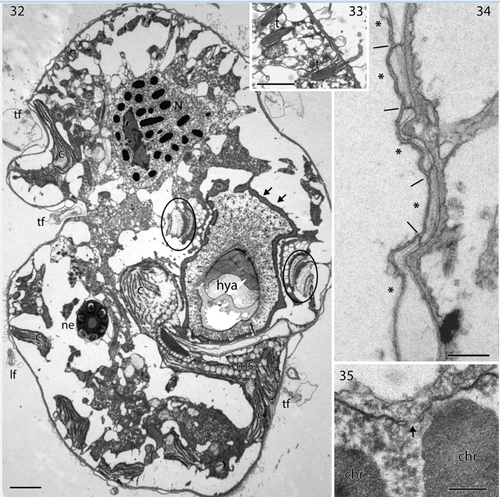
A detailed study of the ultrastructure of selected organelles will be published separately but a brief, general description of the complete cell is provided here. The main organelles and the two main turns of the cingulum are visible in . The nucleus was visible anteriorly and nuclear pores belong to the typical eukaryote type, without nuclear chambers (). Numerous trichocysts (t) are present in . The ocellus is visible in the right part of , showing the hyalosome (hya) and the melanosome (ms), and part of a canal from the exterior, which separates the hyalosome and melanosome. The hyalosome was surrounded by a layer of mitochondria (arrows), and two profiles of a horseshoe-shaped structure that surrounds the upper-middle part of the hyalosome were also visible (ovals). They represent the ring-like structure seen in the light microscope (). A nematocyst (ne) is visible on the left-hand side of and illustrated at higher magnification in transverse () and longitudinal sections (). Each nematocyst contained a ring of six elongate structures, perhaps acting as six dischargeable units.
Figs 32–35. Transmission electron microscopy of Nematodinium parvum.
Figs 36–41. Transmission electron microscopy showing details of Nematodinium parvum.
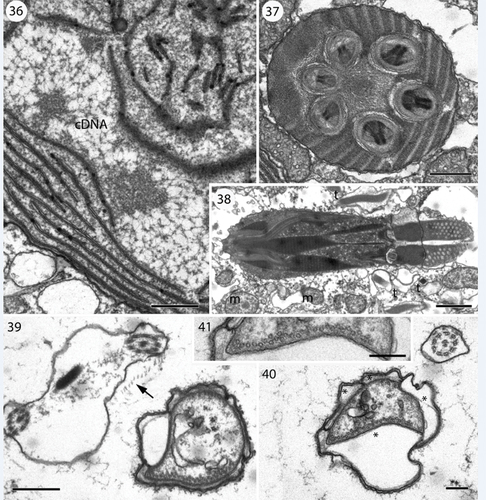
Chloroplast branches (c) were visible in several places, and a larger part of the chloroplast is visible to the left of the hyalosome in . In the chloroplast, extensive amounts of DNA fibres were visible at higher magnification (). The apical furrow was composed of three parallel rows of amphiesmal vesicles (). Vesicles of one row were restricted to the bottom part of the furrow, while the other two rows formed the sides of the furrow, extending over the furrow rims. The amphiesmal vesicles contained two plate-like structures, the lowermost structure well defined, the uppermost more diffuse in transverse section. The peduncle is illustrated in . It is lined here by four amphiesmal vesicles (asterisks in ), all with contents as the vesicles elsewhere in the cell. One side of the peduncle contained an irregular row of microtubules (), and a few separate microtubules were visible elsewhere in the peduncle lumen.
Distribution and behaviour of Nematodinium parvum in Danish waters
Cells of Nematodinium parvum were observed in water samples from Skovshoved, Svanemøllen (both in Øresund), Risø at Roskilde Fjord, and from Rønbjerg, Skive, Hjarbæk and Virksund harbours (all in the Limfjord). High abundance was often related to high temperatures (June–October, 20–27°C), a salinity range of 16–27 and absence of wind (calm surface water conditions). They were never observed in water samples from winter (February-March), when windy conditions, heavy rain and low temperatures often prevail.
Field observations indicated that N. parvum performed diel vertical migrations (DVM) as high abundances were found between 12:00 AM and 6:00 PM (even earlier when the sun rose at 4:00 AM), always at the surface (<1 m depth). In early autumn 2021 (October), when temperatures were still 15–20°C and there was no wind, cell concentrations were very high at 7:00 AM in Rønbjerg harbour ().
Nematodinium parvum most often co-occurred with ciliates (Mesodinium sp.), other dinoflagellates and larger zooplankton. Prolonged periods of high temperatures and a calm sea appear to be crucial conditions when searching for high concentrations of N. parvum in Danish waters.
Culturing experiments
Food uptake of Nematodinium parvum was never observed in the laboratory. The longest survival periods (13–14 days) were achieved when cells were provided with a ‘prey’ mixture of Eutreptiella and Mesodinium in TL30 medium with or without ammonium (modified from Erd-Schreiber medium; Throndsen Citation1969, Citation1978), and microturbulence generated by plankton wheel rotation (1 rpm). Similar results were achieved with Brachiomonas submarina in L1-Si+NH4 (modified from Guillard & Hargraves Citation1993), when N. parvum cells (n = 4) were observed to be alive after 13 days. However, they did not divide.
Molecular analyses
The phylogenetic tree based on the first data matrix revealed that the warnowiaceans formed a monophyletic lineage highly supported by posterior probability (0.99) but less so by RAxML bootstrap (70%) (Fig. S1). The sister group to the warnowiaceans comprised a diverse assemblage of other non-ocelloid-bearing athecate dinoflagellates including Gymnodinium catenatum H.W. Graham and Lepidodininium chlorophorum Elbrächter & Schnepf. Having received support for the monophyly of the warnowiaceans, the group of interest here, two additional phylogenetic analyses were completed. The first of these was based on SSU rDNA () and it revealed Nematodinium to appear in two and Warnowia in three of the five major lineages. Hence, both of these genera were considered polyphyetic as presently circumscribed. Two of the five lineages comprised sequences of presumably uncultured warnowiaceans, one representing cold-water species (Svalbard, Ross Sea and the Arctic Ocean), and the other comprised of taxa from the Sargasso Sea related to Erythropsidinium agile (Hertwig) P.C. Silva and an unidentied species of Warnowia from Florida (USA). The SSU rDNA sequence of Nematodinium parvum from Rønbjerg (Denmark) was closely related to two unidentified species of Warnowia, one from Canada and another from Spain, and an uncultured warnowiacean from China ().
Fig. 42. Phylogeny of 13 ocelloid-bearing dinoflagellates identified to genus level (one to species level) and seven uncultured eukaryotes anticipated to possess ocelloids analysed using Bayesian analysis and inferred from SSU rDNA sequences. Lepidodinium and Gymnodinium formed the outgroup taxa. Numbers at internodes represent posterior probabilities from Bayesian analysis and bootstrap values from RAxML (10,000 replicates). Filled circles indicate maximal support (1.0 in posterior probability and 100% in RAxML). Branch lengths are proportional to the number of character changes, see scale bar. Newly sequenced cell isolate is written in bold.
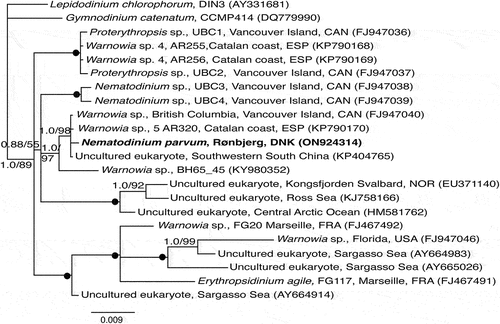
The phylogenetic analysis based on LSU rDNA comprised fewer warnowiaceans (10 sequences) but the polyphyletic nature of Warnowia and Nematodinium remained (). The two almost identical sequences from single-cell determinations of Nematodinium parvum from Denmark (see below) were closely related to two unidentified species of Warnowia from Spain (KP790242) and Canada (FJ947042).
Fig. 43. Phylogeny of 10 ocelloid-bearing dinoflagellates identified to genus level (one to species level) analysed using Bayesian analysis and inferred from partial LSU rDNA sequences. Lepidodinium and Gymnodinium formed the outgroup taxa. Numbers at internodes represent posterior probabilities from Bayesian analysis and bootstrap values from RAxML (10,000 replicates). Filled circles indicate maximal support (1.0 in posterior probability and 100% in RAxML). Branch lengths are proportional to the number of character changes, see scale bar. Newly sequenced cell isolates are written in bold.

Both analyses based on ribosomal genes showed species currently assigned to Nematodinium and Warnowia to be scattered within the warnowiaceans, allowing no clear taxonomic resolution ().
Sequence divergence
All SSU rDNA sequences determined of material from Rønbjerg, Skovshoved and Roskilde Fjord were 100% identical. The Danish material was 100% identical to Warnowia sp. from Spain (KP790170) and had a one base pair difference to Warnowia sp. from Canada (FJ947040) and an uncultured eukaryote from China (KP404765). The LSU sequences of cells from Skovshoved were all identical and differed by a single base pair compared to the sequence determined from the single cell from Rønbjerg. This difference was due to a transition between the purines guanine and adenine (G ↔ A). The LSU rDNA sequences of Nematodinium parvum determined here showed a 99.7% pairwise sequence similarity to Warnowia sp. from Spain (KP790242) and Warnowia sp. from Vancouver Island, Canada (FJ947042). Such low sequence divergence between these cells suggests that they are conspecific.
DISCUSSION
The organelles of Nematodinium parvum
Nematodinium parvum has a complex, net-like, golden-brown chloroplast with typical stacked thylakoids (and DNA) and it is therefore undoubtedly photosynthetic. Chloroplasts have been observed also in Nematodinium armatum (Dogiel) Kofoid & Swezy (Dogiel Citation1906; Hovasse Citation1951; Mornin & Francis Citation1967) and Nematodinium sp. (Hoppenrath et al. Citation2009; Gavelis et al. Citation2015). A chloroplast has not been illustrated in other warnowiaceans and it is clearly absent in Erythropsidinium (e.g. Greuet Citation1969; Gómez Citation2017) and Proterythropsis (Marshall Citation1925; Moestrup et al., unpublished observations). Future studies may reveal whether chloroplasts constitute a generic character that can be used to distinguish Nematodinium and Warnowia (present in Nematodinium and absent in Warnowia).
Compared to other warnowiaceans, Nematodinium parvum is unusual in possessing not only nematocysts but also trichocysts. The nematocysts will be described in more detail in a separate publication. We only found nematocysts with six dischargeable units (as in Proterythropsis vigilans; Moestrup et al., unpublished observations), whereas Gavelis et al. (2017) illustrated a variable number of 10 (11?)–15 in Nematodinium sp. and Hovasse (Citation1951) illustrated eight in Nematodinium armatum. Trichocysts are widespread in dinoflagellates, and they have also been observed in N. armatum (Mornin & Francis Citation1967). However, they seem to be uncommon in other species of the Warnowiaceae.
Feeding-related structures in Nematodinium parvum
Although all our attempts to establish cultures of Nematodinium parvum were unsuccessful, there can be little doubt that N. parvum is mixotrophic. In Proterythropsis vigilans, a food vacuole seen by Marshall (Citation1925) consisted of a greenish or yellowish food mass, probably from Thalassiosira or Phaeocystis cells. We also tested Thalassiosira weissflogii (Van Heurck) G.A. Fryxell & Hasle as food for N. parvum, but it did not feed on them. Gómez (Citation2017) illustrated Erythropsidinium scarlatinum (Kofoid et Swezy) P.C. Silva with ingested copepod eggs, but our feeding experiments with eggs from Acartia tonsa and Calanus sp. proved negative.
Cells of N. parvum possessed a peduncle, documented in both SEM and TEM. The peduncle was an extension of the cytoplasm supported internally by microtubules and emerged from the inner surface of the sulcus. A peduncle has been described in many dinoflagellates, starting with Gyrodinium lebouriae (Lee Citation1977), and is thought to act as a feeding-tube, which pierces the prey surface and sucks up the contents of the cell. Within the warnowiaceans, a peduncle has apparently not been found before, but it differs from the peduncle of other dinoflagellates by being bounded by both the cell membrane and amphiesmal vesicles. The peduncle has been studied in numerous other dinoflagellates and is always bounded by the plasma membrane only (e.g. Kubai & Ris Citation1969; Calado et al. Citation1998; Pandeirada et al. Citation2022). Due to the presence of amphiesmal vesicles in the peduncle it resembles the piston, although the long piston of Erythropsidinium at first sight is very different. The piston is believed to serve at least two different functions: food capture, and the sudden, jumpy movements of the usually motionless cell (Gómez Citation2008, Citation2017). Greuet (Citation1969) provided drawings of the piston, which he named stomopode. In addition to being surrounded by amphiesmal vesicles it also contained an internal row of microtubules. The drawings also show several vacuoles, and this recalls the vacuoles observed in the peduncle in . This raises the question of whether the piston in Erythropsidinium may have evolved as a modified peduncle. A piston was also observed by Greuet (Citation1968) in the related genus Leucopsis Greuet, nom. illeg. (subsequently renamed Greuetodinium A.R. Loeblich), which apparently has not been refound since the first description, but it has to our knowledge not been found in other dinoflagellates.
On the taxonomic identity of the Danish specimens
As stated above, the taxonomy of warnowiacean dinoflagellates is presently in a state of confusion, at the species as well as the genus level. The Danish material described agrees very precisely with Pouchetia parva Lohmann (Citation1908, p. 264), now known as Warnowia parva (Lohmann) Er. Lindemann. Lohmann’s description is brief, but the Danish material agrees in cell size, cell shape, including the characteristically rounded-conical antapex, and the yellow colour of the cytoplasm. Lohmann’s single illustration (fig. 23 in pl. XVII) is reproduced here as .
Fig. 44. Reproduction from Lohmann (Citation1908, pl. XVII, fig. 23). Lohmann drew the cell with the apical end downwards.
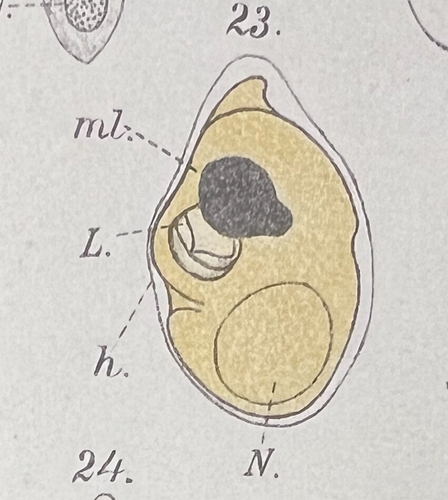
Lohmann (Citation1908) gave the length of Pouchetia parva as 33 µm, compared to our 26–39 µm and average of 32.2 µm. The ecology of the Danish material also agrees with Lohmann’s findings. Lohmann observed the species at the entrance to Kieler Bay (‘Ostsee’) in April-November, not far from our locations. Cells were not particularly numerous and mainly occurred in the upper layers of the water, a finding that Lohmann explains by its dependence on light. On 23 sampling days, cells appeared 15 times at the surface, twice at 5 m depth, five times at 10 m and only three times at 15 m. Lohmann gave the maximum concentration of cells near the surface as 2,300 cells l–1 on 16 June 1906.
Hulburt (Citation1957) reported Warnowia parva from Massachusetts and gave the cell length as 23–30 µm. He mentioned the presence of yellow chloroplasts but not the yellow colour of the cytoplasm. Lohmann’s and Hulburt’s descriptions differ from our cells in one respect: neither author mentions the presence of nematocysts. The many described species of Warnowia are believed to lack nematocysts, while the few known species of Nematodinium possess such organelles, and this is thought to be a main difference between the two genera.
Is Warnowia parva a species of Nematodinium?
Kofoid & Swezy (Citation1921), when describing their new genus Nematodinium, stated: “The diagnostic characteristic of this genus is the possession of nematocysts clustered about the nucleus” (p. 421). The accompanying figure of the type species (Kofoid & Swezy Citation1921, fig. NN, 4) shows some nematocysts near the nucleus and others along the right side of the cell; nematocysts do not appear as a cluster about the nucleus in this figure. In the description of the type species, Nematodinium partitum Kofoid & SwezyCitation1921, p. 425 wrote, “Scattered through the cytoplasm are numerous (15–20) nematocysts”, and “They are arranged in roughly three groups: one in the anterior part, one near the central region, and the third posteriorly”.
The position of the nematocysts in the type species therefore disagrees with the statement in the generic diagnosis. One explanation for this may be that the position of the nematocysts in the cell varies, and our experience has confirmed this.
As mentioned above, cells from Denmark possess nematocysts, but they are easily overlooked, and careful examination at high magnification is needed to see them. It appears possible that limitations in the magnification available to Lohmann would have caused him to miss the nematocysts in the material he observed. However, the shape and size of the cell and the yellow colour are identical to our material, and we have on numerous occasions observed cells in which nematocysts were not visible, perhaps for having been discharged. The phylogenetic analyses () revealed cells identified as Warnowia sp. with a morphology similar to our cells, but apparently lacking nematocysts (Hoppenrath et al. Citation2009, ; Reñé et al. Citation2015, ), clustering together with our cells in the molecular tree, supporting the difficulty in observing the nematocysts. Lebour (Citation1925) had the same experience with material from Plymouth, and Hulburt (Citation1957, p. 214) mentions that in Nematodinium armatum nematocysts may be present or absent, as reported also by Hoppenrath et al. (Citation2009) for Nematodinium sp.
Following the demonstration of nematocysts, we transfer Warnowia parva to Nematodinium under the International Code of Nomenclature for algae, fungi, and plants (Turland et al. Citation2018):
Nematodinium parvum (Lohmann) Moestrup comb. nov.
basionym
Pouchetia parva Lohmann Citation1908, Wissenschaftliche Meeresuntersuchungen, Abteilung Kiel 10, p. 264, pl. XVII, fig. 23.
homotypic synonym
Warnowia parva (Lohmann) Er. Lindemann (Citation1928, p. 52).
heterotypic synonym
Nematodinium pseudoarmatum Hovasse (Citation1951, p. 151).
Whether the two genera, Nematodinium and Warnowia, are separate genera must await examination of the type species of Nematodinium, N. partitum, originally described from La Jolla, California, and the type species of Warnowia, W. fusus (F. Schütt) Er. Lindemann, of uncertain origin but probably from the Bay of Naples or the Atlantic. It will cause some confusion if they have to be merged, as the name Nematodinium is older than Warnowia and the family name is Warnowiaceae.
Are Nematodinium armatum and Nematodinium parvum different species?
The two species are very similar, but an important difference appears to be the length of the cell and the typically conical shape of the antapex in N. parvum. Dogiel (Citation1906) gave the cell length of N. armatum as c. 50 µm, while Lohmann (Citation1908), as mentioned above, described N. parvum to be 33 µm long. Neither author gave information on cell width. Hulburt (Citation1957) provided a size range of his material from Massachusetts, in N. armatum 33–53 µm long and 20–33 µm wide, and in Warnowia parva 22.5–30 µm long and 15–18 µm wide. If these differences hold, the two species are different, although Lebour (Citation1925) stated the size of N. armatum as 28–50 µm and suggested that the cells described by Lohmann may have been encysted N. armatum. Considering that our material was very constant in size and never exceeded 39 µm, we currently regard N. armatum and N. parvum as different species.
In the phylogenetic tree based on LSU rDNA (), the sister taxon to N. parvum was a sequence (FJ947041) of Nematodinium sp. from Hoppenrath et al. (Citation2009), which is illustrated in their . The images show a yellowish cell with a cell length and shape more or less similar to N. armatum. This appears to support the conclusion above and to show the close relationship between N. parvum and N. armatum.
Phylogenetic inferences
The phylogenetic analyses conducted here, based on SSU and LSU rDNA, highlight the same challenge: very few warnowiaceans have been identified to the level of species. This makes the available molecular sequences of little use for phylogenetic inferences clarifying the taxonomy of the ocelloid-bearing dinoflagellates. Additionally, the phylogenetic trees also confirm the present state of confusion at the generic level: both Nematodinium and Warnowia taxa clustered within multiple lineages, as shown in and to some extent in . Since so few single-cell isolates have been determined for both ribosomal genes and morphologically characterized at the same time, concatenation would not have improved the outcome.
The numerous uncultured eukaryotes from Sargasso and polar seas clustering with the warnowiaceans () reveal the widespread biogeography of the Warnowiaceae. However, understanding their distribution pattern and ecological importance will require precise taxonomic identification.
CONCLUSION
The current study has illustrated that the taxonomy of species and genera in the Warnowiaceae is chaotic and no longer serves its purpose. We have with this article taken a modest step towards a new taxonomy for the group. By combining detailed light and electron microscopic analyses with molecular data we have characterized the species Nematodinium parvum. Yet, numerous questions remain unanswered within the Warnowiaceae such as the characters used to circumscribe genera, the number of species, and not the least how they feed. A major step forward towards solving some of these questions would be to resolve how to maintain the species in culture. They appear to be distributed in marine environments worldwide, but the number of species may be much lower than presently described.
Supplemental Material
Download MS Word (6.2 MB)ACKNOWLEDGEMENTS
We thank Anders Garm and Per Juel Hansen (Department of Biology, University of Copenhagen) for discussions on ocelloid dinoflagellates, and António Calado for important comments on the manuscript.
DISCLOSURE STATEMENT
No potential conflict of interest was reported by the authors.
Supplementary Information
Supplemental data for this article can be accessed online at https://doi.org/10.1080/00318884.2023.2244810
Additional information
Funding
REFERENCES
- Calado A.J., Craveiro S.C. & Moestrup Ø. 1998. Taxonomy and ultrastructure of a freshwater, heterotrophic Amphidinium (Dinophyceae) that feeds on unicellular protists. Journal of Phycology 34: 536–554.
- de Candolle A.P. [Ed.] 1830. Prodromus systematis naturalis regni vegetabilis …, vol. 4. Treuttel and Würtz, Paris, France. 683 pp.
- Dogiel V. 1906. Beiträge zur Kenntnis der Peridineen. Mittheilungen aus der Zoologischen Station zu Neapel 18: 1–45.
- Elbrächter M. 1979. On the taxonomy of unarmoured dinophytes (Dinophyta) from the Northwest African upwelling region. “Meteor” Forschungsergebnisse, D, Biologie 30: 1–22.
- Gavelis G.S., Hayakawa S., White III R.A., Gojobori T., Suttle C.A., Keeling P.J. & Leander B.S. 2015. Eye-like ocelloids are built from different endosymbiotically acquired components. Nature 523: 204–207.
- Gómez F. 2008. Erythropsidinium (Gymnodiniales, Dinophyceae) in the Pacific Ocean, a unique dinoflagellate with an ocelloid and a piston. European Journal of Protistology 44: 291–298.
- Gómez F. 2009. Molecular phylogeny of the ocelloid-bearing dinoflagellates Erythropsidinium and Warnowia (Warnowiaceae, Dinophyceae). Journal of Eukaryotic Microbiology 56: 440–445.
- Gómez F. 2017. The function of the ocelloid and piston in the dinoflagellate Erythropsidinium (Gymnodiniales, Dinophyceae). Journal of Phycology 53: 629–641.
- Greuet C. 1968. Leucopsis cylindrica nov. gen., nov. sp., péridinien Warnowiidae Lindemann. Considérations phylogénétiques sur les Warnowiidae. Protistologica 4: 419–422.
- Greuet C. 1969. Étude morphologique et ultrastructurale du trophonte d’Erythropsis pavillardi Kofoid et Swezy. Protistologica 5: 481–503.
- Greuet C. 1972. Les critères de détermination chez les péridiniens Warnowiidae Lindemann. Protistologica 8: 461–469.
- Guillard R.R.L. & Hargraves P.E. 1993. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32: 234–236.
- Hayakawa S., Takaku Y., Hwang J.S., Horiguchi T., Suga H., Gehring W., Ikeo K. & Gojobori T. 2015. Function and evolutionary origin of unicellular camera-type eye structure. PLOS One 10: Article e0118415.
- Hertwig R. 1884. Erythropsis agilis. Eine neue Protozoe. Morphologisches Jahrbuch 10: 204–213, pl. VI.
- Hertwig R. 1885. Ist Erythropsis agilis eine losgerissene Spastostyla sertulariarum? Zoologischer Anzeiger 8: 108–112.
- Hoppenrath M., Bachvaroff T.R., Handy S.M., Delwiche C.F. & Leander B.S. 2009. Molecular phylogeny of ocelloid-bearing dinoflagellates (Warnowiaceae) as inferred from SSU and LSU rDNA sequences. BMC Evolutionary Biology 9: 1–15.
- Hovasse R. 1951. Contribution à l’étude de la cnidogénèse chez les Péridiniens. Deuxième partie. Cnidogénèse cyclique chez Nematodinium armatum Dogiel. Archives de Zoologie Expérimentale et Générale 88 (Notes et Revue 3): 149–158.
- Huelsenbeck J.P. & Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
- Hulburt E.M. 1957. The taxonomy of unarmored Dinophyceae of shallow embayments on Cape Cod, Massachusetts. The Biological Bulletin 112: 196–219.
- Kofoid C.A. & Swezy O. 1921. The free-living unarmored Dinoflagellata. Memoirs of the University of California 5: 1–564.
- Kubai D.F. & Ris H. 1969. Division in the dinoflagellate Gyrodinium cohnii (Schiller). A new type of nuclear reproduction. Journal of Cell Biology 40: 508–528.
- Lebour M.V. 1925. The dinoflagellates of northern seas. Marine Biological Association of the United Kingdom, Plymouth, UK. 250 pp.
- Lee R.E. 1977. Saprophytic and phagocytic isolates of the colourless heterotrophic dinoflagellate Gyrodinium lebouriae Herdman. Journal of the Marine Biological Association of the UK 57: 303–315.
- Lindeman E. 1928. Abteilung Peridineae (Dinoflagellatae). In: Die Natürlichen Pflanzenfamilien, vol. 2, Peridineae, Diatomeae, Myxomycetes (Ed. by E. Jahn), pp 3–104. W. Engelmann, Leipzig, Germany.
- Lohmann H.V. 1908. Untersuchungen zur Feststellung des vollständigen Gehaltes des Meeres an Plankton. Wissenschaftliche Meeresuntersuchungen, Abteilung Kiel 10: 129–337.
- Marshall S.M. 1925. On Proterythropsis vigilans, n. sp. Quarterly Journal of Microscopical Science, ser. 2 69: 177–184, pl. 9.
- Metschnikoff E. 1874. Mittheilungen über eine Reise nach Madeira. Protokolle der Sitzungen der Kaiserlichen Gesellschaft der Liebhaber der Naturlehre, Anthropologie und Ethnographie in Moskau 10 (2): 6–9 [in Russian].
- Metschnikoff E. 1885. Zur Streitfrage über Erythropsis agilis. Zoologischer Anzeiger 8: 433–434.
- Mornin L. & Francis D. 1967. Fine structure of Nematodinium armatum, a naked dinoflagellate. Journal de Microscopie 6: 759–772.
- Pandeirada M.S., Craveiro S.C., Daugbjerg N., Moestrup Ø. & Calado A.J. 2022. Ultrastructure and phylogeny of Parvodinium cunningtonii comb. nov. (syn. Peridiniopsis cunningtonii) and description of P. cunningtonii var. inerme var. nov. (Peridiniopsidaceae, Dinophyceae). European Journal of Protistology 86: Article 125930.
- Pouchet G. 1885a. Nouvelle contribution à l’histoire des Péridiniens marins. Journal de l’Anatomie et de la Physiologie Normales et Pathologiques de l’Homme et des Animaux 21: 28–88, pls II–IV.
- Pouchet G. 1885b. Troisième contribution à l’histoire des Péridiniens. Journal de l’Anatomie et de la Physiologie Normales et Pathologiques de l’Homme et des Animaux 21: 525–534, pl. XXVI.
- Pouchet G. 1887. Quatrième contribution à l’histoire des Péridiniens. Journal de l’Anatomie et de la Physiologie Normales et Pathologiques de l’Homme et des Animaux 23: 87–112, pls IX, X.
- Reñé A., Camp J. & Garcés E. 2015. Diversity and phylogeny of Gymnodiniales (Dinophyceae) from the NW Mediterranean Sea revealed by a morphological and molecular approach. Protist 166: 234–263.
- Richlen M.L. & Barber P.H. 2005. A technique for the rapid extraction of microalgal DNA from single live and preserved cells. Molecular Ecology Notes 5: 688–691.
- Schütt F. 1895. Peridineen der Plankton-Expedition der Humboldt-Stiftung. Ergebnisse der Plankton-Expedition 4: 1–170.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313.
- Throndsen J. 1969. Flagellates of Norwegian coastal waters. Nytt Magasin for Botanikk 16: 161–216.
- Throndsen J. 1978. The dilution culture method. In: Phytoplankton manual (Ed. by A. Sournia), pp 218–224. IOC-UNESCO, Paris, France [Monographs on Oceanographic Methodology 6].
- Turland N.J., Wiersema J.H., Barrie F.R., Greuter W., Hawksworth D.L., Herendeen P.S., Knapp S., Kusber W.-H., Li D.-Z., Marhold K. et al. [Eds] 2018. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Koeltz Botanical Books, Glashütten, Germany. xxxviii +++ 254 pp. [ Regnum Vegetabile 159] DOI: https://doi.org/10.12705/Code.2018.
- Vogt C. 1885a. Über Erythropsis agilis Rich. Hertwig. Zoologischer Anzeiger 8: 53.
- Vogt C. 1885b. Ein wissenschaftlicher Irrthum. Die Natur 34: 183–187.