ABSTRACT
The activities of iron-oxidizing and reducing microorganisms impact the fate of arsenic in groundwater. Phylogenetic information cannot exclusively be used to infer the potential for iron oxidation or reduction in aquifers. Therefore, we complemented a previous cultivation-independent microbial community survey covering 22 arsenic contaminated drinking water wells in Bangladesh, with the characterization of enrichments of microaerophilic iron oxidizers and anaerobic iron reducers, conducted on the same water samples. All investigated samples revealed a potential for microbial iron oxidation and reduction. Microbial communities were phylogenetically diverse within and between enrichments as was also observed in the previous cultivation-independent analysis of the water samples from which these enrichments were derived. Enrichment uncovered a larger diversity in iron-cycling microorganisms than previously indicated. The iron-reducing enrichments revealed the presence of several 16S ribosomal RNA (16S rRNA) gene sequences most closely related to Acetobacterium, Clostridium, Bacillus, Rhizobiales, Desulfovibrio, Bacteroides, and Spirochaetes, in addition to well-known dissimilatory iron-reducing Geobacter and Geothrix species. Although a large diversity of Geobacteraceae was observed, they comprised only a small part of the iron-reducing consortia. Iron-oxidizing gradient tube enrichments were dominated by Comamonadaceae and Rhodocyclaceae instead of Gallionellaceae. Forty-five percent of these enrichments also revealed the presence of the gene encoding arsenite oxidase, which converts arsenite to less toxic and less mobile arsenate. Their potential for ferric (oxyhydr)oxides precipitation and arsenic immobilization makes these iron-oxidizing enrichments of interest for rational bioaugmentation of arsenite contaminated groundwater.
Introduction
Iron exists in groundwater systems predominantly in ferrous [Fe(II)] and ferric [Fe(III)] states (Chapelle Citation2001). At or above circumneutral pH, ferric iron primarily occurs as insoluble iron (oxyhydr)oxide minerals. In contrast, below pH 4.0 or under microaerophilic to anaerobic conditions, ferrous iron is relatively soluble in water and is mobile (Weber et al. Citation2006a). Microorganisms play a role in the cycling between the two redox states. Oxidation of organic carbon by heterotrophic iron-reducing bacteria is the dominant mechanism for ferric iron reduction in anaerobic groundwater systems (Lovley and Anderson Citation2000; Weber et al. Citation2006b). In contrast, chemolithotrophic microorganisms can obtain energy through the oxidation of ferrous iron in aerobic acidic environments (Clarke et al. Citation1997; Kozubal et al. Citation2008), at neutral pH under microaerophilic conditions (Emerson and Weiss Citation2004; Hallberg and Ferris Citation2004) or under nitrate reducing conditions (Weber et al. Citation2006a).
Iron can create, directly or indirectly, a number of nuisances in the extraction and human use of groundwater. Its oxidation can cause significant clogging problems in drinking water extraction (Ghiorse Citation1984; Emerson and De Vet Citation2015). Although iron is not considered to cause health problems in humans, its presence in potable water is rather unpleasant due to its rusty taste, the bad odors it spreads, and its tendency to stain clothing red. The emergence of red water in drinking water distribution systems might be caused by the activity of microaerophilic iron oxidizers, leading to extensive precipitation of iron (oxyhydr)oxides (Li et al. Citation2010).
Furthermore, the microbial mediated redox cycling of iron significantly influences the mobilization of toxic elements such as arsenic (Zobrist et al. Citation2000; Islam et al. Citation2004; Oremland and Stolz Citation2005). Several studies have documented a moderate to strong correlation between ferrous iron and arsenic in groundwater (e.g., Nickson et al. Citation2000; McArthur et al. Citation2001; Höhn et al. Citation2006). Arsenic is highly toxic to humans (Lloyd and Oremland Citation2006). Natural arsenic contamination in drinking water is a major concern for public health in Bangladesh and other countries in South and Southeast Asia, where people drink arsenic contaminated groundwater (Yu et al. Citation2003). These iron and arsenic are released into the groundwater due to the reductive dissolution of iron minerals containing arsenic (Islam et al. Citation2004). In contrast, aqueous arsenic contamination can be removed through biological oxidation of ferrous iron, as the precipitating ferric (oxyhydr)oxides bind arsenic (Katsoyiannis and Zouboulis Citation2004).
Thus, the activity of iron-reducing and oxidizing microorganisms indirectly affects arsenic concentrations in groundwater, in addition to the activity of specific microorganisms directly involved in the biogeochemical cycling of arsenic: arsenate-reducing bacteria releasing arsenite [As(III)] and arsenite-oxidizing bacteria producing arsenate [As(V)], which is less toxic, less mobile, and binds better to ferric (oxyhydr)oxides than arsenite (Cavalca et al. Citation2013). Knowledge on the microbial ecology of the iron cycle in relation to the generation and remediation of arsenic contaminated drinking water in South and Southeast Asia is still limited, in particular with respect to iron oxidation potential. Previously, we sampled 24 drinking water wells from four districts in Bangladesh and conducted a cultivation-independent 16S rRNA gene-based survey to obtain insight into the occurrence of microorganisms with potential for iron-cycling (Hassan et al. Citation2015). However, 16S rRNA gene-based molecular techniques do not necessarily inform on the physiological traits of the identified microorganisms. The capability to reduce iron is spread over many bacterial and archaeal genera, and within several of these genera some members do reduce iron while others do not (Lovley et al. Citation2004). Furthermore, it has become evident that the capability to oxidize iron is present in more genera than previously thought (Hedrich et al. Citation2011).
Therefore, to understand the role of iron-cycling in arsenic contaminated groundwater, it appeared essential to follow up our previous cultivation-independent work on iron-cycling with cultivation studies. Accordingly, this study aims to survey the abundance, distribution, and diversity of cultivatable iron-oxidizing and iron-reducing microorganisms in arsenic contaminated drinking water wells in Bangladesh. We hypothesized that a diverse range of cultivatable iron-cycling microorganisms is present in arsenic contaminated groundwaters in Bangladesh. We also hypothesized that enrichment would reveal iron-cycling microorganisms that had not been identified on the basis of our previous cultivation-independent analysis (Hassan et al. Citation2015) of the very same 22 samples investigated here. Furthermore, we investigated metabolic flexibility by determining the presence of arsenite oxidase (aioA) and arsenate reductase (arrA) genes, which would indicate the potential of the enrichments to oxidize arsenite or reduce arsenate, respectively.
Materials and methods
Field sampling
Between August 2011 and March 2012, a total of 22 groundwater samples were collected from shallow and deep tube wells from the Satkhira, Jessore, and Comilla districts in Bangladesh () (Hassan et al. Citation2015). Anaerobic groundwater samples were collected in sterile serum glass bottles by letting the bottles overflow, after three volumes of standing water in each tube well had been removed by hand pumping. Bottles with groundwater samples for culturing were capped with as little headspace as possible and transferred to the laboratory, where they were stored for less than 24 h at 4°C. Details on the hydrochemistry of these samples were reported in our previous study (Hassan et al. Citation2015), with key features indicated in . All samples were anaerobic, with pH values between 6.1 and 8.0.
Table 1. Enrichment of iron cycling microorganisms initiated from 22 drinking water wells in Bangladesh (indicated by sample code or ID, name of the village in which the well is located, district, well depth, and physicochemical parameters).
Enrichment of microaerophilic iron-oxidizing microorganisms
A gradient tube cultivation approach with opposing gradients of oxygen and ferrous iron was employed to enrich and maintain microaerophilic iron oxidizers (Emerson and Floyd Citation2005). Modified Wolfe's Mineral Medium (MWMM) consisted of the following ingredients (in g l−1 distilled water): NH4Cl, 1.0; MgSO4.7H2O, 0.2; CaCl2.2H2O, 0.1; K2HPO4, 0.05; and NaHCO3, 4.2. The system consisted of two layers of agarose in 16-mm screw cap glass tubes (Wang et al. Citation2009). The bottom layer contained 1.0 ml of FeS or FeCO3 solution and MWMM at a ratio of 1:1, amended with 1% (wt/vol) agarose. FeCO3 and FeS stock solutions were prepared according to Hallbeck et al. (Citation1993) and Emerson and Floyd (Citation2005), respectively. The top layer consisted of 5.0 ml of MWMM supplemented with 5 µl of vitamin solution (medium 141, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH [DSMZ]) and trace element solution each (medium 141, DSMZ), stabilized with 0.15% (wt/vol) agarose. Before autoclaving, the solutions for the upper layer were mixed with 0.5 ml of 50 mM NaHCO3 and flushed with CO2 for 2–3 min to attain a final pH of 6.4. The gradient tubes were inoculated ∼24 h after preparation to facilitate the diffusion of ferrous iron into the top layer where bacterial growth occurs. Tenfold serial dilutions (up to 10−3) of groundwater samples in sterile 0.85% NaCl solution were inoculated into gradient tubes. The tubes were inoculated just above the iron plug by pulling a micropipette vertically through the medium while the inoculum (10 µl) was added slowly from the micropipette. Abiotic controls were prepared without bacteria (an uninoculated control and a control inoculated with 10 µl sterile 0.85% NaCl) to distinguish between abiotic and biotic iron oxidation. Tubes were incubated in the dark at 28°C for 2–3 weeks. Microbial growth in the gradient tube was identified by the formation of a discrete brownish iron oxide band, after visual comparison to the more diffused bands that typically develop due to chemical oxidation of ferrous iron in abiotic control tubes (Supplementary Figure S1a). Three to four serial transfers were performed to obtain the dominant iron oxidizers; a sterile inoculating needle was used to extract cell material from the discrete brownish band and to transfer this material into a fresh tube. A Pasteur pipette was used to obtain bacterial cell material (∼0.5–1.0 ml) for molecular analysis. Cell material was centrifuged and the pellets were stored at−20°C until DNA extraction.
Enrichment of anaerobic iron-reducing microorganisms
Strictly anaerobic techniques were used to enrich iron-reducing microorganisms. The anoxic basal medium was based on Lin et al. (Citation2007) and contained (in g l−1 distilled water) KCl, 0.1; NH4Cl, 1.5; NaH2PO4, 0.6; NaHCO3, 2.5; Na2WO4.2H2O, 0.00025; 10 ml of trace element solution; and 10 ml of vitamin solution (medium 141, DSMZ), amended with 2.0 mM Na lactate and 3.0 mM Na acetate. A 100 mM stock solution of Fe(III)–nitrilotriacetic acid (NTA) was prepared by dissolving 1.64 g of NaHCO3, 0.256 g of trisodium NTA, and 0.27 g of FeCl3.6H2O in water to a final volume of 100 ml. The solution was made anoxic by purging with a O2-free mix of N2 and CO2 (80/20%), filter sterilized, and stored in an anaerobic, sterile serum bottle (Liu et al. Citation2002). We used Fe(III)–NTA (final conc. 5.0 mM) as an electron acceptor, resazurin dye (0.0005 g l−1) as an indicator for the presence of oxygen, and 0.1 mM cysteine as an oxygen scavenger. Anaerobic medium was prepared in serum vials sealed with butyl rubber septa and crimped with aluminum caps. The basal medium was flushed with a mix of N2 and CO2 (80/20%). Iron-reducing enrichments were performed on nine groundwater samples (). One milliliter of sample was inoculated into an anaerobic serum vial containing 9 ml of medium. These cultures were tenfold serial diluted up to 10−3 in the same medium and incubated in the dark at 28°C for 2–3 weeks. Positive iron reduction (medium turning colorless) was inferred by comparison with an uninoculated control (no color changes) (Supplementary Figure S1b). The highest dilution revealing iron reduction was used to inoculate fresh medium (2% vol/vol) and incubated again. This procedure was repeated three to four times.
For molecular analysis, iron-reducing cultures were vacuum-filtered over 45-mm-diameter, 0.2-µm pore size nitrocellulose membrane filters (Millipore, Billerica, MA, USA) and frozen at −20°C until DNA isolation.
DNA extraction
DNA was extracted using the soil DNA extraction kit (Mo BIO Laboratories Inc., Carlsbad, CA, USA) according to manufacturer's instructions. DNA was stored at −20°C.
16S rRNA gene-based profiling of bacterial, Gallionellaceae and Desulfuromonadales communities
Partial 16S rRNA gene sequences of bacteria, iron-oxidizing Gallionellaceae, and iron-reducing Desulfuromonadales, were amplified for denaturing gradient gel electrophoresis (DGGE) analysis, using the same primers and amplification conditions as in our previous study (Hassan et al. Citation2015; Supplementary Table S1). For bacteria, primer set 357F-GC clamp and 907r was used. The 16S rRNA gene fragments of Gallionellaceae were obtained with a nested PCR approach (Wang et al. Citation2009) using primer set 122F and 998R in the first round of amplification, followed by primer set 357F-GC and 907r in the second round, after diluting (1:100) the first round PCR products. A Desulfuromonadales-specific primer set (8f and 825r) was employed to amplify a 0.8-kb 16S rRNA gene fragment (Snoeyenbos-West et al. Citation2000). The order Desulfuromonadales comprises the iron-reducing families Geobacteraceae and Desulfuromonadaceae (Röling Citation2014). However, when Snoeyenbos-West et al. (Citation2000) developed their primers, members of current Desulfuromonadaceae were still included in the family Geobacteraceae. PCR products were then diluted (1:100) and used for the second round of amplification using bacteria specific primers 357F-GC and 518r. Each PCR reaction was carried out using 25 µl (total volume) of mixture containing 12 µl of GoTaq (Promega, Madison, WI, USA) ready Master Mix, 1 µl of each primer (0.4 µM final concentration), 8 µl of nuclease free water (Promega), and 3 µl of undiluted DNA as template.
DGGE was carried out using a Dcode™ Universal Mutation Detection System (Bio Rad Laboratories, CA, USA). PCR product was loaded onto a 1-mm-thick 8% (wt/vol) polyacrylamide (ratio of acrylamide to bisacrylamide, 37.5:1) gel containing a linear gradient of 30–55% of urea–formamide. The running conditions were 200 V at a constant temperature of 60°C in 1 × TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM Na-Ethylenediaminetetraacetic acid (EDTA), pH 8.0) for 4 h. The gels were stained in 1 × (Tris base, acetic acid and EDTA) (TAE) buffer containing 1 µg/ml of ethidium bromide and visualized using a UV transilluminator. To aid in the normalization of and comparison between gels, a DGGE marker (M12) with 12 bands at various positions was added to the external lanes of the gels, as well as to lanes in between every four samples. All gels to fingerprint a particular group of bacteria were run on the same day. The average between-gel similarity of the marker was 92%, with 3% standard deviation.
Bands were excised using sterile wide mouth blunt tips. Excised DNA bands were suspended in 1 × TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 7.5) and stored overnight at 4°C. One microliter of suspension was used as a template in the aforementioned PCR, using primers without GC clamp. Products were checked on 1.5% agarose gels and sequenced (Macrogen, Amstelveen, The Netherlands).
Analysis of potential for arsenic metabolism in enrichments
A degenerate oligonucleotide primer set was used to amplify the gene encoding arsenite oxidase, aioA (Supplementary Table S1). Partial arsenate respiratory reductase (arrA) genes were amplified using primers ArrAfwd and ArrArev (Supplementary Table S1). Amplification often yielded faint arrA products, consisting of multiple bands. Bands with the expected size (∼160–200 bp) were cut from 2% low melting point agarose gels, reamplified, and rechecked by agarose gel electrophoresis. aioA and arrA products were subjected to sequencing (Macrogen).
Phylogenetic analysis
Sequences were aligned with ClustalW using default settings and were manually edited. Phylogenetic analyses were performed with MEGA 4 (Tamura et al. Citation2007). Nucleotide distance and amino acid Poisson correction analyses were performed through maximum composite likelihood computation and trees were constructed using the neighbor-joining method with a bootstrap value of 1000 replicates. Gene sequences have been deposited in GenBank under accession numbers: KR095355 to KR095423 (bacterial 16S rRNA genes), KR095424 to KR095433 (aioA), and in the DNA Data Bank of Japan (DDBJ) database under accession numbers: LC043403 to LC043406 (arrA) and LC043407 to LC043433 (Desulfuromonadales-like 16S rRNA gene sequences).
Statistical analysis
Quantitative analysis of DGGE profiles was performed with GelCompar II (Applied Maths, Belgium) (van Verseveld and Röling Citation2004). Similarities between profiles were calculated using the Pearson correlation coefficient and visualized by the unweighted paired group clustering method with arithmetic means (UPGMA). The observed clusters of enrichments were related to hydrochemical characteristics of the groundwater from which these enrichments were derived, using nonparametric analysis of variance (Kruskal–Wallis) as described in our previous study (Hassan et al. Citation2015).
Results
Characterization of microaerophilic iron-oxidizing enrichments
All microaerophilic iron-oxidizing enrichments initiated with groundwater samples from 22 drinking water wells revealed iron oxidation within 7 days of incubation, most often at the highest dilution (10−3) tested (, Supplementary Figure S1a). DGGE analysis after four serial transfers of these enrichments revealed that fingerprints were generally dominated by a few brightly stained bands and several less intense bands (a: one to four bright bands per profile). Considerable variation between community profiles was observed: the fingerprints grouped into six clusters, at a 60% cutoff value (a). No significant correlation was detected between these clusters and the hydrochemical characteristics of the groundwater from which the enrichments were derived (all p > 0.05; Kruskal–Wallis).
Figure 1. (a) UPGMA cluster analysis of bacterial 16S rRNA gene-based DGGE profiles (30–55% denaturant gradient) of 22 microaerophilic iron-oxidizing enrichments, using Pearson correlation analysis as a measure of similarity. The enrichment ID refers to the location of the drinking water well (see ). Enrichments were assigned to clusters on the basis of >60% similarity. Names of enrichments belonging to a particular cluster received the same color, and this color is used in b to indicate the sequences of their excised bands. Numbers refer to the position of excised bands.
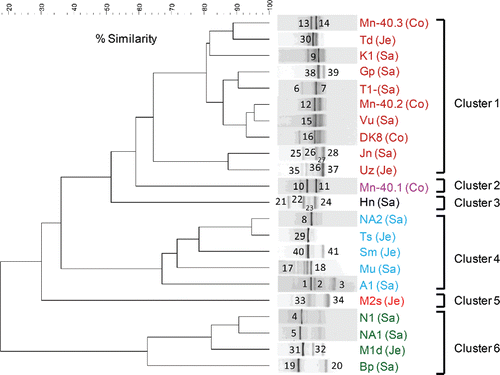
Figure 1. (b) Phylogenetic analysis of 16S rRNA gene sequences (525 unambiguously aligned nucleic acid positions) retrieved from the excised DGGE bands. Sequences are indicated by enrichment ID and the number of the excised band, as shown in (a). The three districts from which enrichments were obtained are indicated by distinct symbols: closed red circles, Satkhira; green triangles, Jessore; and blue squares, Comilla. The tree was constructed with the neighbor-joining method and bootstrap values (1000 replications) are indicated at the interior branches. The scale bar represents 5% sequence divergence.
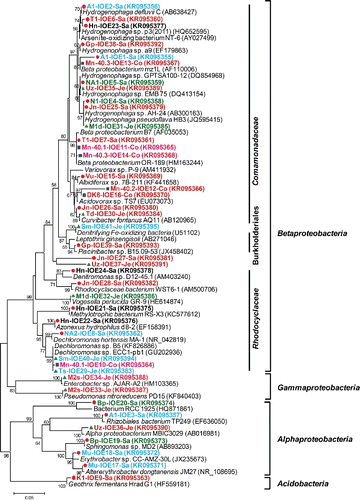
To gain insight into the identities of the major bacterial populations in the iron-oxidizing enrichments, 41 prominent DGGE bands were sequenced (see a for banding positions and b for phylogeny). The Betaproteobacteria constituted the dominant group (78% of the sequenced bands) followed by Alphaproteobacteria (15%) and Gammaproteobacteria (5%). Acidobacteria (Geothrix fermentans) contributed the remaining 2%. Betaproteobacteria sequences were most closely related to a variety of genera in especially the families Comamonadaceae and Rhodocyclaceae. Within the Comamonadaceae family, Hydrogenophaga sp. (11 bands; 27% of total), Curvibacter sp. (two bands; 5% of total), and Acidovorax sp. (two bands; 5% of total) were identified. Hydrogenophaga related bands were frequently detected in profiles belonging to clusters 1, 4, and 6 (). Within the Rhodocyclaceae family, Dechloromonas sp. (four bands; 10% of total) and Azonexus (one band; 2% of total) were observed. Dechloromonas appeared to be confined to cluster 4, with bands labeled 8 in enrichment NA2, 10 in Mn-40.1, 29 in Ts, and 40 in Sm (a). Furthermore, two bands relating to denitrifying iron-oxidizing bacteria were detected (5% of total; bands numbered 37 in sample Uz and 41 in Sm), while 14 genera of Betaproteobacteria were observed once, including a sequence with 97% similarity to the heterotrophic iron oxidizer Leptothrix (band 39 in Gp). Within the Alphaproteobacteria, sequences were most closely related to six different genera, corresponding to one band each (). A Gallionellaceae-specific PCR revealed that these microaerophilic, chemolithotrophic iron oxidizers were only detectable in two enrichments (; Hn and Ts).
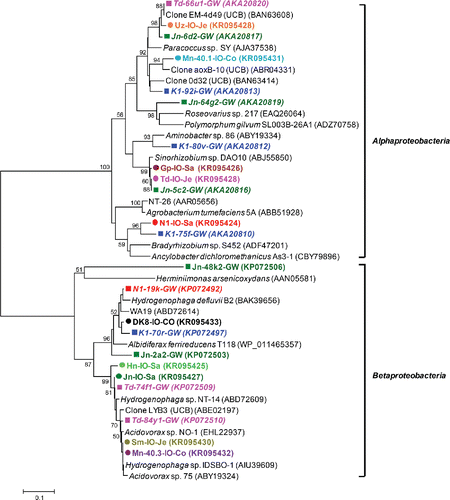
All iron-oxidizing enrichments were tested for the presence of the arsenite oxidase gene (aioA). Forty-five percent (10 out of 22 enrichments) was positive (). Amino acid sequences were all most closely related to the AioA sequences of known arsenite-oxidizing bacteria (; 86–99% amino acid similarity). Among them, AioA sequences most closely related to those encountered in Hydrogenophaga and Acidovorax species accounted for 50% of the total. These sequences were found in enrichments initiated with Hn, Jn, Sm, Mn-40.3, and DK8 groundwaters, and also the DGGE fingerprints of these enrichments revealed dominant bands with 16S rRNA gene sequences most closely related to Hydrogenophaga and Acidovorax, with the exception of Sm (bands labeled 25 in Jn, 13 in Mn-40.3, 23 in Hn, and 16 in DK8 in a). We also detected AioA sequences most closely related to AioA found in Paracoccus, Sinorhizobium, Bradyrhizobium, and Ancylobacter (). However, none of the 16S rRNA gene sequences of excised DGGE bands was closely related to any of these genera. The AioA sequences retrieved clustered well with AioA sequences detected in our previous cultivation-independent analysis (Hassan et al. Citation2015) ().
Figure 2. Unrooted neighbor-joining tree of amino acid sequences (162 unambiguously aligned positions) of the bacterial arsenite oxidase gene retrieved from iron-oxidizing enrichments. Bootstrap values (1000 replications) are indicated at the interior branches (bar = 0.1 substitutions/sequence position). The colored circles indicate the enrichments, with different colors referring to the various drinking water wells from which the enrichments were initiated. IDs in italics with colored squares indicate sequences derived directly from groundwater samples, without intermediate culturing (Hassan et al. Citation2015).
Characterization of iron-reducing enrichments
A total of nine groundwater samples were used to initiate iron-reducing enrichments. Iron was reduced in all enrichments, as indicated by the formation of ferrous iron after 3 weeks of incubation (). In five out of nine cases, iron reduction was observed at the highest dilution tested, that is, 10−3. Enrichments maintained their iron reduction capability during three serial transfers (Supplementary Figure S1b). Subsequent DGGE analysis revealed several dominant bands for each enrichment (two to five bands per profile), but considerable variation between the enrichments (18 different banding positions in nine profiles) was observed (a).
Figure 3. (a) UPGMA cluster analysis of bacterial 16S rRNA gene-based DGGE profiles (30–55% denaturant gradient) of nine iron reducing enrichments using Pearson correlation analysis to assess similarity. The enrichment ID refers to the location of the drinking water well (see ). Numbers refer to the position of excised bands.
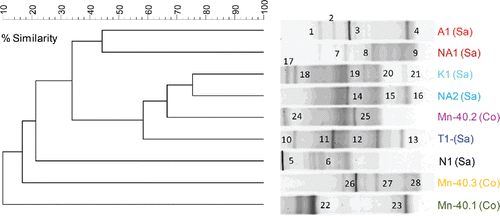
Figure 3. (b) Phylogenetic analysis of 16S rRNA gene sequences (525 unambiguously aligned nucleic acid positions) retrieved from the excised DGGE bands. Sequences are indicated by enrichment ID and the number of the excised band as shown in (a). Sequences are accompanied by a colored symbol, specific for each of the nine enrichments. The tree was constructed with the neighbor-joining method and bootstrap values (1000 replications) are indicated at the interior branches. The scale bar represents 2% sequence divergence.
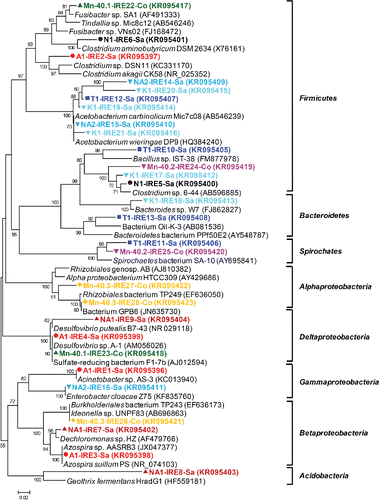
A total of 28 predominant bands were excised for sequence analysis. Phylogenetic diversity was high, besides members of several proteobacterial classes (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria), Acidobacteria, Bacteroidetes, Firmicutes, and Spirochaetes were observed) (b). Many bands (46%, 13 bands observed in seven out of nine enrichments) were most closely related to various Firmicutes genera, mostly Acetobacterium (21%, six bands in three enrichments—bands numbered 12 in T1; 14, 15 in NA2; and 19, 20, and 21 in K1 in a) and Clostridium (18%, five bands in four enrichments; bands labeled 5 and 6 in N1, 24 in Mn-40.2, 17 in K1, and 2 in A1). Betaproteobacteria and Deltaproteobacteria each contributed 11% of the sequenced bands (three bands each; b). The Betaproteobacteria sequences (Mn-40.3, NA1, and A1) belonged to various genera: Burkholderia and two genera in the Rhodocyclaceae, Dechloromonas and Azospira (b). All Deltaproteobacteria sequences were most closely related to sulfate reducing Desulfovibrio (b: bands labeled 4 in A1, 9 in NA1, and 23 in Mn-40.1; 90–99% nucleotide similarity). Other phyla contributed 4–7% of the sequenced bands, including an Acidobacterium sequence most closely related to the known iron reducer Geothrix fermentans (band 8 in NA1 in ; 79% similarity).
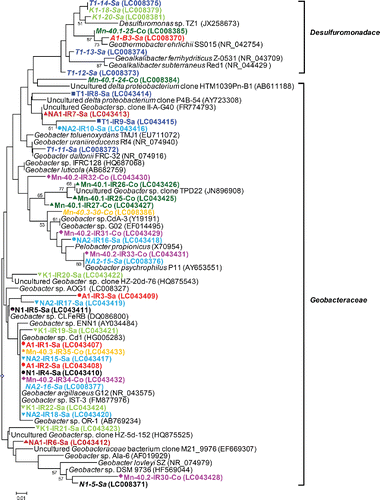
DGGE analysis after a PCR specific for iron-reducing Geobacteraceae and Desulfuromonadaceae indicated their presence and diversity within enrichments (a: one to eight bands per profile), as well as variation between the enrichments (20 different banding positions in nine profiles). As the employed primer set is not 100% specific for Desulfuromonadales (Snoeyenbos-West et al. Citation2000, Lin et al. Citation2005), a total of 35 DGGE bands were cut out and sequenced. Phylogenetic analysis revealed that most sequences were closely related to Geobacteraceae (77%, 27 bands in nine enrichments; b). The sequencing confirmed the diversity of the Geobacteraceae within and between enrichments.
Figure 4. (a) UPGMA cluster analysis of Desulfuromonadales 16S rRNA gene-based DGGE profiles (30–55% denaturant gradient) of nine iron-reducing enrichments using Pearson correlation to assess similarity. The enrichment IDs refer to the location of the drinking water well (see for details). Numbers refer to the position of excised bands subjected to sequencing. Sequences of bands with underlined, bold numbers in italics did not belong to Desulfuromonadales.
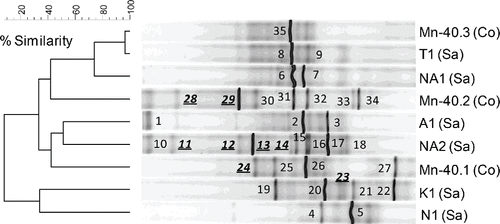
Figure 4. (b) Phylogenetic analysis of 16S rRNA gene sequences (122 unambiguously aligned nucleic acid positions) retrieved from the excised DGGE bands. Sequences are indicated by enrichment ID, the number of the excised band as shown in (a), and the district in which the well is located. Sequences are also accompanied by a colored symbol, specific for each of the nine enrichments and IDs in italics refer to water samples. The unrooted tree was constructed with the neighbor-joining method and bootstrap values (1000 replications) are indicated at the interior branches. The scale bar represents 1% sequence divergence.
Testing for the presence of the arsenate reductase gene (arrA) in the iron-reducing enrichments revealed that some 44% (four out of nine; ) was positive and contained arrA genes most closely related (80–99% nucleotide similarity) to those of Sulfurospirillum, Geobacter, or uncultured bacteria (). We did not observe any obvious correspondence between identities based on arrA sequences and identities based on bacterial 16S rRNA sequences retrieved from the same iron-reducing enrichments.
Table 2. Identities of amplified fragments of the arsenate respiratory reductase (arrA) gene (partial nucleotide sequences of 151 bp) detected in iron-reducing enrichments.
Discussion
Microbial diversity in iron-oxidizing enrichments
We initiated iron-oxidizing gradient cultures with the same groundwater samples that we used previously for direct molecular detection of iron-oxidizing Gallionellaceae (Hassan et al. Citation2015). We hypothesized a diverse range of cultivatable iron-cycling microorganisms to reside in arsenic contaminated groundwaters in Bangladesh. Indeed, all samples showed iron oxidation, and substantial diversity was found in these enrichments. As also hypothesized, we enriched microorganisms that had not been identified as iron oxidizers on the basis of our previous cultivation-independent 16S rRNA gene-based study (Hassan et al. Citation2015). However, as we did not isolate and characterize individual strains, it might be that not all identified microorganisms are capable of iron oxidation but live on the organic matter that is excreted by chemolithotrophic iron oxidizers (Ghiorse Citation1984). Nevertheless, since Fe(II) minerals were the sole source of electron donors in the enrichments and we targeted the dominant DGGE bands for phylogenetic analysis, we would expect to primarily encounter iron oxidizers.
Our culturing approach using opposing gradients of ferrous iron and oxygen is widely used to enrich and isolate Gallionella species (Emerson and Floyd Citation2005). Thereby, a striking finding was that the cultivation approach of the present study showed very limited persistence of Gallionella, whereas our prior study revealed that 77% of the 22 samples investigated contained Gallionellaceae (Hassan et al. Citation2015). The 16S rRNA gene of Gallionella could only be detected in 2 out of the 22 enrichment tubes, and only after using a Gallionellaceae specific PCR. Instead, a significant number of Comamonadaceae-related 16S rRNA gene sequences were detected. Similar findings were obtained by Blöthe and Roden (Blöthe and Roden Citation2009) in a cultivation-based study on groundwater seep material at circumneutral pH. Their 16S rRNA gene-based molecular analysis revealed that even though groundwater was dominated by Gallionella and Leptothrix spp., they also could not isolate Gallionella and Leptothrix spp., using the same opposing gradient cultivation method commonly used to enrich Gallionella (Emerson and Floyd Citation2005). Also several other studies have documented iron-oxidizing enrichments dominated by Comamonadaceae and Rhodocyclaceae rather than Gallionella spp. (Yu et al. Citation2010; Gülay et al. Citation2013). A possible explanation for the low encounter frequency of Gallionella spp. in our study might be a relatively lower abundance of Gallionella compared to other taxa in the groundwater samples, not allowing them to be recoverable in our dilution to extinction culturing approach. Another explanation might be that Gallionella was outcompeted by other microorganisms under the imposed culturing conditions, for example, because of a lower specific growth rate.
Based on our DGGE fingerprinting analysis, members of the Comamonadaceae were most frequently observed (11 out of 41 bands; 27% of total), especially Hydrogenophaga but also Curvibacter and Acidovorax species. Recently, Chan et al. (Citation2014) isolated iron-oxidizing Hydrogenophaga sp. P101 and Curvibacter sp. CD03 retrieved from alluvial sediment, Rifle aquifer in Colorado (USA) by microaerophilic iron-oxidizing gradient tube enrichment. Besides Hydrogenophaga, we also detected a number of sequences most closely related to the genera Leptothrix, Pseudomonas, and members of the Rhodocyclaceae (Dechloromonas), which contain iron-oxidizing members (Hedrich et al. Citation2011), and to anaerobic denitrifying iron-oxidizing Acidovorax (Carlson et al. Citation2013). Similar sequences were also reported in our previous cultivation-independent study (Hassan et al. Citation2015). Several microorganisms are able to oxidize ferrous iron using nitrate or oxygen as electron acceptors (Yu et al. Citation2010). Many denitrifying bacteria are capable of switching to microaerophilic iron oxidation (Benz et al. Citation1998; Edwards et al. Citation2003; Melton et al. Citation2012), which may explain the occurrence of 16S rRNA genes most closely related to anaerobic denitrifying iron-oxidizing bacteria in the gradient tubes of the present study.
Forty-five percent of the iron-oxidizing enrichments harbored genes encoding aioA. This gene is either used by heterotrophs to detoxify arsenite or by chemolithoautotrophs to yield energy from arsenite oxidation (Santini et al. Citation2002; Inskeep et al. Citation2007; Rhine et al. Citation2007). We found only four AioA sequences in our iron-oxidizing gradient tubes that were most closely related to the corresponding sequences of chemolithoautotrophic strains, that is, Ancylobacter dichloromethanicus As3-1b (Andreoni et al. Citation2012), the nitrate reducing Paracoccus sp. SY (Zhang et al. 2015) and Sinorhizobium sp. DAO10 (Rhine et al. Citation2006). The other six sequences were most closely related to heterotrophs. Congruence was observed between aioA and 16S rRNA gene data, both indicating the presence of Hydrogenophaga and Acidovorax species. Some Hydrogenophaga species are known to be capable of aerobic arsenite oxidation (vanden Hoven and Santini Citation2004; Salmassi et al. Citation2006), while several Acidovorax strains perform nitrate-dependent iron (Klueglein and Kappler Citation2013) and arsenite oxidation (Quéméneur et al. Citation2008). Also, some strains of Dechloromonas sp. oxidize iron (Weber et al. Citation2006b; Coby et al. Citation2011; Chakraborty and Picardal Citation2013) and arsenite under nitrate- or perchlorate-reducing conditions (Sun et al. Citation2009). However, to our knowledge prior studies did not investigate the presence of arsenite oxidase genes in iron-oxidizing enrichments, and overall our results indicate metabolic flexibility in several enrichments. The AioA sequences we identified were most closely related (>94% amino acid identity) to those identified on the basis of cultivation-independent analysis of the same water samples from which these enrichments were derived (Hassan et al. Citation2015).
Microbial diversity in iron-reducing enrichments
A substantial diversity was also found in the iron-reducing enrichments, including microorganisms that had not been identified as iron reducers on the basis of our previous cultivation-independent 16S rRNA gene-based study (Hassan et al. Citation2015). Geobacteraceae have often been demonstrated to constitute the most abundant iron reducers in iron-reducing subsurface environments (Lovley et al. Citation2011), including arsenic contaminated sediments in West Bengal, India (Islam et al. Citation2004; Héry et al. Citation2010). We encountered Geobacteraceae in all our enrichments. However, Geobacter spp. appeared to constitute a minor fraction of the communities observed in this study, in line with our previous cultivation-independent analysis of the same samples (Hassan et al. Citation2015). Geobacteraceae could only be revealed after specifically targeting their 16S rRNA genes. Enrichment recovered other and also more Geobacter phylotypes compared to our prior cultivation-independent analysis (Hassan et al. Citation2015).
Besides Geobacter spp., other microorganisms can substantially contribute to iron reduction in iron-reducing environments (Lin et al. Citation2007; Li et al., 2011), and therewith affect the release of arsenic into groundwater (Rowland et al. Citation2009). Dominant phylotypes present in our iron-reducing enrichments belonged to the Firmicutes, a phylum also identified in our previous cultivation-independent study (Hassan et al. Citation2015). Several Acetobacterium sequences were observed, in addition to Clostridia and a sequence most closely related to Bacillus sp. strain IST-38. Acetobacterium species have been identified as anaerobic hydrogen consuming acetogens in iron corrosive settings (Mori et al. Citation2010), but iron reduction by Acetobacterium isolates has not yet been reported, to our knowledge. Clostridium 16S rRNA gene sequences were also retrieved during a cultivation-independent analysis of incubated arsenic contaminated West Bengal sediments, India (Islam et al. Citation2004). The capability of clostridia to reduce iron is well known. Subsurface iron-reducing Clostridium strains have for instance been isolated from a landfill leachate polluted aquifer in The Netherlands (Lin et al. Citation2007). They use ferric iron as an electron sink generating additional ATP via substrate level phosphorylation during acetate production (Lovley et al. Citation2011). Also Bacillus sp. strain IST-38, isolated from a groundwater seep in Alabama (USA), is capable of such fermentative iron reduction (Blöthe and Roden Citation2009).
Other sequences were also closely related to sequences of iron reducers, or sequences retrieved from iron-reducing environments. Geothrix is a dissimilatory iron-reducing bacterium, previously encountered in iron-reducing zones of hydrocarbon polluted aquifers and redox dynamic hydrocarbon polluted surface sediments (Coates et al. Citation1999; Klueglein et al. Citation2013). Sulfate reducing Desulfovibrio species were encountered in three iron-reducing culture enrichments (NA1, A1, and Mn-40.1). This is surprising perhaps; several Desulfovibrio species can enzymatically reduce iron, but do not conserve sufficient energy to support growth (Lovley et al. Citation1993). Active Desulfovibrio were recently identified in arsenic mobilizing Cambodian aquifer sediments (Héry et al. Citation2015). We identified a sequence most closely related to the fermentative iron-reducing Bacteroides strain W7 isolated from the anode suspension of a microbial electrolysis cell (Wang et al. Citation2010). Several sequences fell in the Rhizobiales order, a group not known to reduce iron. However, members of this group can reduce uranium (Vishnivetskaya et al. Citation2010). Rhizobiales were also abundant in anode biofilm communities that had been enriched for electricity generation (Ishii et al. Citation2008; Kaku et al. Citation2008). Interestingly, we encountered in the iron-reducing enrichments Azospira and Dechloromonas, which are known denitrifying iron-oxidizing Betaproteobacteria (Weber et al. Citation2006a). Dechloromonas species have previously been isolated from an arsenic contaminated dimictic lake in Arlington, MA (Gibney and Nüsslein Citation2007) and nitrate reducing Wisconsin River sediment (Chakraborty and Picardal Citation2013), and Azospira from an animal waste lagoon (Byrne-Bailey and Coates Citation2012). These species were also encountered in anaerobic microbial iron-cycling reactors (Coby et al. Citation2011). We identified sequences of both Dechloromonas sp. and Clostridium in our iron-reducing enrichments that were >97% similar to those detected in the previous cultivation-independent 16S rRNA gene survey of potentially iron-cycling microbial communities (Hassan et al. Citation2015). Overall, the observations complement our previous findings (Hassan et al. Citation2015), confirming the abundance and diversity of microorganisms with potential for iron reduction in the investigated Bangladeshi aquifers. However, we cannot be completely certain that all microorganisms encountered in our enrichments are indeed capable of iron reduction, as we did not perform isolation and characterization of strains.
Some iron reducers can sometimes also reduce arsenate and possess the respiratory arsenate reductase gene arrA (Ohtsuka et al. Citation2013; Kudo et al. Citation2014; Osborne et al. Citation2015). arrA genes closely related to those of Geobacter species have been detected frequently in arsenic rich sediments (Lear et al. Citation2007; Héry et al. Citation2015). While some 44% of our iron-reducing enrichments held arrA genes, their sequences were more closely related to uncultivated species or Sulfurospirillum than to Geobacter species.
Implications for the (im)mobilization of arsenic
The mobilization of arsenic in subsurface environments has enormous toxic consequences for millions of people in Bangladesh who are vulnerable through their use of groundwater as drinking water and through arsenic entering the food chain (Huq et al. Citation2006). Currently available remediation technologies have major disadvantages such as they are often expensive and result in secondary exposure to and environmental pollution with arsenic, for example, through inadequate handling and disposal of arsenic binding water filters (Gonzaga et al. Citation2006). The nature of the minerals formed during microbial iron oxidation may have important implications for arsenic mobility. Biological ferrous oxidation can effectively sequester arsenic via the precipitation of ferric oxyhydroxide minerals (Hohmann et al. Citation2009). Several microbial iron oxidizers produce poorly soluble crystalline iron oxides (Miot et al. Citation2009; Liu et al. Citation2013; Li et al. 2015). Biogenic amorphous iron oxides play a major role in removing arsenite due to their strong capacity to adsorb or coprecipitate arsenite (Omoregie et al. Citation2013). With respect to microaerophilic iron oxidation, most attention has been focused on Gallionella (Hedrich et al. Citation2011). Its role and effectiveness in arsenic removal from water by producing arsenic adsorbing ferric (oxyhydr)oxides has been established (Katsoyiannis and Zouboulis Citation2004; Zouboulis and Katsoyiannis Citation2005). Recently, it was shown that Citrobacter freundii strain PXL1 could remove arsenite from water in a sewage plant, in a process associated with iron oxidation (Li et al. Citation2015). The Comamonadaceae and Rhodocyclaceae identified here in many gradient tube enrichments may also be of interest with regard to biological iron and arsenic removal, in particular since consortia containing these bacteria revealed the presence of aioA genes. Arsenite oxidases oxidize arsenite to the less toxic and less mobile arsenate. This potential for ferric (oxyhydr)oxides precipitation and arsenic immobilization makes these iron-oxidizing enrichments of interest for bioaugmentation of reactors treating arsenite contaminated groundwater retrieved from drinking water wells. They may also be of interest for in situ removal of arsenic and iron by subsurface arsenic removal (SAR) technology (van Halem et al. Citation2010). SAR was recently introduced in Bangladesh. It comprises the injection of oxygenated water into aquifers so as to oxidize ferrous iron abiotically and therewith precipitate iron and adsorb and coprecipitate arsenic (van Halem et al. Citation2010). The activity of iron-oxidizing microorganisms that also oxidize arsenite could potentially enhance SAR efficiency down to lower residual concentrations of arsenic.
Whether biological arsenic removal from groundwater in Bangladesh would be conducted in situ or ex situ, care should be taken to avoid anaerobic conditions. Groundwater in Bangladesh also contains a wide range of anaerobic iron reducers (Hassan et al. Citation2015, this study), which may then become active and release arsenic as a result of the reduction of precipitated and arsenic adsorbing ferric (oxyhydr)oxides. Specialized arsenate reducers and iron reducers possessing arrA (Hassan et al. Citation2015, this study) may enhance the release of arsenic by reducing arsenate to more mobile and more toxic arsenite.
Supplementary_Information.docx
Download MS Word (1.9 MB)Funding
This work was financially supported by The Netherlands Organization for Scientific Research in the integrated program of WOTRO Science for Global Development (W01.65.324.00/project 4), as well as by EU-FP7 (KBBE2012.3.4-02; # 311815; SYNPOL), BBSRC, EU-H2020 (Corbel; INFRADEV-4-2014-2015; #654248), and BBSRC (DTC grant).
References
- Andreoni V, Zanchi R, Cavalca L, Corsini A, Romagnoli C, Canzi E. 2012. Arsenite oxidation in Ancylobacter dichloromethanicus As3-1b strain: detection of genes involved in arsenite oxidation and CO2 fixation. Curr Microbiol 65:212–218.
- Benz M, Brune A, Schink B. 1998. Anaerobic and aerobic oxidation of ferrous iron at neutral pH by chemoheterotrophic nitrate-reducing bacteria. Arch Microbiol 169:159–165.
- Blöthe M, Roden EE. 2009. Microbial iron redox cycling in a circumneutral-pH groundwater seep. Appl Environ Microbiol 75:468–473.
- Byrne-Bailey KG, Coates JD. 2012. Complete genome sequence of the anaerobic perchlorate-reducing bacterium Azospira suillum strain PS. J Bacteriol 194:2767–2768.
- Carlson HK, Clark IC, Blazewicz SJ, Iavarone AT, Coates JD. 2013. Fe(II) oxidation is an innate capability of nitrate-reducing bacteria that involves abiotic and biotic reactions. J Bacteriol 195:3260–3268.
- Cavalca L, Corsini A, Zaccheo P, Andreoni V, Muyzer G. 2013. Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Future Microbiol 8:753–768.
- Chakraborty A, Picardal F. 2013. Induction of nitrate-dependent Fe(II) oxidation by Fe(II) in Dechloromonas sp. strain UWNR4 and Acidovorax sp. strain 2AN. Appl Environ Microbiol 79:748–52.
- Chan C, Cabaniss K, Williams K, Moore M, Michael H, Caplan J, Lin C. 2014. Fe-oxidizing microorganisms in microscopic model aquifer systems: feedbacks between flow and biomineralization. In: Proceedings of the ninth international symposium on subsurface microbiology. Pacific Grove, California USA, October 5-10, pp 22.
- Chapelle FH. 2001. Ground-Water Microbiology and Geochemistry. New York: John Wiley & sons, pp 272–275.
- Clarke WA, Konhauser KO, Thomas JC, Bottrell SH. 1997. Ferric hydroxide and ferric hydroxysulfate precipitation by bacteria in an acid mine drainage lagoon. FEMS Microbiol Rev 20:351–361.
- Coates JD, Ellis DJ, Gaw CV, Lovley DR. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int J Syst Bacteriol 49:1615–1622.
- Coby AJ, Picardal F, Shelobolina E, Xu H, Roden EE. 2011. Repeated anaerobic microbial redox cycling of iron. Appl Environ Microbiol 77:6036–6042.
- Edwards KJ, Rogers DR, Wirsen CO, McCollom TM. 2003. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α-and γ-Proteobacteria from the deep sea. Appl Environ Microbiol 69:2906–2913.
- Emerson D, Weiss JV. 2004. Bacterial iron oxidation in circumneutral freshwater habitats: findings from the field and the laboratory. Geomicrobiol J 21:405–414.
- Emerson D, Floyd MM. 2005. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol 397:112–123.
- Emerson D, De Vet W. 2015. The role of FeOB in engineered water ecosystems: A Review. J Am Water Works Assoc 107:e47–57. http://dx.doi.org/10.5942/jawwa.2015.107.0004.
- Ghiorse W. 1984. Biology of iron-and manganese-depositing bacteria. Annu Rev Microbiol 38:515–550.
- Gibney BP, Nüsslein K. 2007. Arsenic sequestration by nitrate respiring microbial communities in urban lake sediments. Chemosphere 70:329–336.
- Gonzaga MIS, Santos JAG, Ma LQ. 2006. Arsenic phytoextraction and hyperaccumulation by fern species. Sci Agric 63:90–101.
- Gülay A, Musovic S, Albrechtsen HJ, Smets BF. 2013. Neutrophilic iron-oxidizing bacteria: occurrence and relevance in biological drinking water treatment. Water Sci Technol Water Supply 13:1295–1301.
- Hallbeck L, Ståhl F, Pedersen K. 1993. Phylogeny and phenotypic characterization of the stalk-forming and iron-oxidizing bacterium Gallionella ferruginea. J Gen Microbiol 139:1531–1535.
- Hallberg R, Ferris FG. 2004. Biomineralization by Gallionella. Geomicrobiol J 21:325–330.
- Hassan Z, Sultana M, van Breukelen BM, Khan SI, Röling WFM. 2015. Diverse arsenic-and iron-cycling microbial communities in arsenic-contaminated aquifers used for drinking water in Bangladesh. FEMS Microbiol Ecol doi:10.1093/femsec/fiv026.
- Hedrich S, Schlömann M, Johnson DB. 2011. The iron-oxidizing proteobacteria. Microbiology 157:1551–1564.
- Héry M, van Dongen BE, Gill F, Mondal D, Vaughan DJ, Pancost RD, Polya DA, Lloyd JR. 2010. Arsenic release and attenuation in low organic carbon aquifer sediments from West Bengal. Geobiology 8:155–168.
- Héry M, Rizoulis A, Sanguin H, Cooke DA, Pancost RD, Polya DA, Lloyd JR. 2015. Microbial ecology of arsenic-mobilizing Cambodian sediments: lithological controls uncovered by stable-isotope probing. Environ Microbiol 17:1857–1869.
- Hohmann C, Winkler E, Morin G, Kappler A. 2009. Anaerobic Fe(II)-oxidizing bacteria show As resistance and immobilize As during Fe(III) mineral precipitation. Environ Sci Technol 44:94–101.
- Höhn R, Isenbeck-Schröter M, Kent D, Davis J, Jakobsen R, Jann S, Niedan V, Scholz C, Stadler S, Tretner A. 2006. Tracer test with As(V) under variable redox conditions controlling arsenic transport in the presence of elevated ferrous iron concentrations. J Contam Hydrol 88:36–54.
- Huq SMI, Joardar J, Parvin S, Correll R, Naidu R. 2006. Arsenic contamination in food-chain: transfer of arsenic into food materials through groundwater irrigation. J Health Popul Nutr 24:305.
- Inskeep WP, Macur RE, Hamamura N, Warelow TP, Ward SA, Santini JM. 2007. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ Microbiol 9:934–43.
- Ishii S, Shimoyama T, Hotta Y, Watanabe K. 2008. Characterization of a filamentous biofilm community established in a cellulose-fed microbial fuel cell. BMC Microbiol 8:6.
- Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, Lloyd JR. 2004. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68–71.
- Kaku N, Yonezawa N, Kodama Y, Watanabe K. 2008. Plant/microbe cooperation for electricity generation in a rice paddy field. Appl Microbiol Biotechnol 79:43–49.
- Katsoyiannis IA, Zouboulis AI. 2004. Application of biological processes for the removal of arsenic from groundwaters. Water Res 38:17–26.
- Klueglein N, Kappler A. 2013. Abiotic oxidation of Fe(II) by reactive nitrogen species in cultures of the nitrate‐reducing Fe(II) oxidizer Acidovorax sp. BoFeN1–questioning the existence of enzymatic Fe(II) oxidation. Geobiology 11:180–190.
- Klueglein N, Lösekann-Behrens T, Obst M, Behrens S, Appel E, Kappler A. 2013. Magnetite formation by the novel Fe(III)-reducing Geothrix fermentans strain HradG1 isolated from a hydrocarbon-contaminated sediment with increased magnetic susceptibility. Geomicrobiol J 30:863–873.
- Kozubal M, Macur R, Korf S, Taylor W, Ackerman G, Nagy A, Inskeep W. 2008. Isolation and distribution of a novel iron-oxidizing crenarchaeon from acidic geothermal springs in Yellowstone National Park. Appl Environ Microbiol 74:942–949.
- Kudo K, Yamaguchi N, Makino T, Ohtsuka T, Kimura K, Dong DT, Amachi S. 2014. Release of arsenic from soil by a novel dissimilatory arsenate-reducing bacterium, Anaeromyxobacter sp. Strain PSR-1. Appl Environ Microbiol 79:4635–4642.
- Lear G, Song B, Gault AG, Polya DA, Lloyd JR. 2007. Molecular analysis of arsenate-reducing bacteria within Cambodian sediments following amendment with acetate. Appl Environ Microbiol 73:1041–1048.
- Li B, Pan X, Zhang D, Lee DJ, Al-Misned FA, Mortuza MG. 2015. Anaerobic nitrate reduction with oxidation of Fe(II) by Citrobacter Freundii strain PXL1–a potential candidate for simultaneous removal of As and nitrate from groundwater. Ecol Eng 77:196–201.
- Li D, Li Z, Yu J, Cao N, Liu R, Yang M. 2010. Characterization of bacterial community structure in a drinking water distribution system during an occurrence of red water. Appl Environ Microbiol 76:7171–7180.
- Li H, Peng J, Weber KA, Zhu Y. 2011. Phylogenetic diversity of Fe(III)-reducing microorganisms in rice paddy soil: enrichment cultures with different short-chain fatty acids as electron donors. J Soils Sedim 11:1234–1242.
- Lin B, Braster M, van Breukelen BM, van Verseveld HW, Westerhoff HV, Röling WFM. 2005. Geobacteraceae community composition is related to hydrochemistry and biodegradation in an iron-reducing aquifer polluted by a neighboring landfill. Appl Environ Microbiol 71:5983–5991.
- Lin B, Braster M, Röling WFM, van Breukelen BM. 2007. Iron-reducing microorganisms in a landfill leachate-polluted aquifer: complementing culture-independent information with enrichments and isolations. Geomicrobiol J 24:283–294.
- Liu C, Gorby YA, Zachara JM, Fredrickson JK, Brown CF. 2002. Reduction kinetics of Fe(III), Co (III), U (VI), Cr (VI), and Tc (VII) in cultures of dissimilatory metal‐reducing bacteria. Biotechnol Bioeng 80:637–649.
- Liu Q, Guo H, Li Y, Xiang H. 2013. Acclimation of arsenic-resistant Fe(II)-oxidizing bacteria in aqueous environment. Inter Biodeter Biodegrad 76:86–91.
- Lloyd JR, Oremland RS. 2006. Microbial transformations of arsenic in the environment: from soda lakes to aquifers. Elements 2:85–90.
- Lovley DR, Roden EE, Phillips E, Woodward J. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar Geol 113:41–53.
- Lovley DR, Anderson RT. 2000. Influence of dissimilatory metal reduction on fate of organic and metal contaminants in the subsurface. Hydrogeol J 8:77–88.
- Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn (IV) reduction. Adv Microb Physiol 49:219–286.
- Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru A, et al. 2011. Geobacter: the microbe Electric's physiology, ecology, and practical applications. Adv Microb Physiol 59:1–100.
- McArthur JM, Ravenscroft P, Safiulla S, Thirlwall MF. 2001. Arsenic in groundwater: testing pollution mechanisms for sedimentary aquifers in Bangladesh. Water Resour Res 37:109–117.
- Melton ED, Schmidt C, Kappler A. 2012. Microbial iron (II) oxidation in littoral freshwater lake sediment: the potential for competition between phototrophic vs. nitrate-reducing iron (II)-oxidizers. Front Microbiol 3:197.
- Miot J, Benzerara K, Morin G, Kappler A, Bernard S, Obst M, Férard C, Skouri-Panet F, Guigner JM, Posth N. 2009. Iron biomineralization by anaerobic neutrophilic iron-oxidizing bacteria. Geochim Cosmochim Acta 73:696–711.
- Mori K, Tsurumaru H, Harayama S. 2010. Iron corrosion activity of anaerobic hydrogen-consuming microorganisms isolated from oil facilities. J Biosci Bioeng 110:426–430.
- Nickson RT, McArthur JM, Ravenscroft P, Burgess WG, Ahmed KM. 2000. Mechanism of arsenic release to groundwater, Bangladesh and West Bengal. Appl Geochem 15:403–413.
- Ohtsuka T, Yamaguchi N, Makino T, Sakurai K, Kimura K, Kudo K, Homma E, Dong DT, Amachi S. 2013. Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate-reducing bacterium Geobacter sp. OR-1. Environ Sci Technol 47:6263–6271.
- Omoregie EO, Couture RM, van Cappellen P, Corkhill CL, Charnock JM, Polya DA, Vaughan D, Vanbroekhoven K, Lloyd JR. 2013. Arsenic bioremediation by biogenic iron oxides and sulfides. Appl Environ Microbiol 79:4325–4335.
- Oremland RS, Stolz JF. 2005. Arsenic, microbes and contaminated aquifers. Trends Microbiol 13:45–49.
- Osborne TH, McArthur JM, Sikdar PK, Santini JM. 2015. Isolation of an arsenate-respiring bacterium from a redox front in an arsenic-polluted aquifer in West Bengal, Bengal Basin. Environ Sci Technol. doi:10.1021/es504707x
- Quéméneur M, Heinrich-Salmeron A, Muller D, Lièvremont D, Jauzein M, Bertin PN, Garrido F, Joulian C. 2008. Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl Environ Microbiol 74:4567–4573.
- Rhine ED, Phelps CD, Young LY. 2006. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ Microbiol 8:899–908.
- Rhine ED, Ni Chadhain SM, Zylstra GJ, Young LY. 2007. The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem Biophys Res Commun 354:662–667.
- Röling WFM. 2014. The Family Geobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, (eds). The Prokaryotes. Berlin Heidelberg:Springer, 157–172.
- Rowland HAL, Boothman C, Pancost R, Gault AG, Polya DA, Lloyd JR. 2009. The role of indigenous microorganisms in the biodegradation of naturally occurring petroleum, the reduction of iron, and the mobilization of arsenite from West Bengal aquifer sediments. J Environ Qual 38:1598–1607.
- Salmassi TM, Walker JJ, Newman DK, Leadbetter JR, Pace NR, Hering JG. 2006. Community and cultivation analysis of arsenite oxidizing biofilms at Hot Creek. Environ Microbiol 8:50–59.
- Santini JM, Sly LI, Wen A, Comrie D, Wulf-Durand PD, Macy JM. 2002. New Arsenite-Oxidizing Bacteria Isolated from Australian Gold Mining Environments—Phylogenetic Relationships. Geomicrobiol J 19:67–76.
- Snoeyenbos-West OL, Nevin KP, Anderson RT, Lovley DR. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb Ecol 39:153–167.
- Sun W, Sierra-Alvarez R, Milner L, Oremland RS, Field JA. 2009. Arsenite and ferrous iron oxidation linked to chemolithotrophic denitrification for the immobilization of arsenic in anoxic environments. Environ Sci Technol 43:6585–6591.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599.
- van Halem D, Olivero S, de Vet WW, Verberk JQ, Amy GL, van Dijk JC. 2010. Subsurface iron and arsenic removal for shallow tube well drinking water supply in rural Bangladesh. Water Res 44:5761–5769.
- van Verseveld HW, Röling WFM. 2004. Cluster analysis and statistical comparison of molecular community profile data. In:Kowalchuk GA, de Bruijn FJ, Head IM, et al., editors. Molecular Microbial Ecology Manual. 2nd edn. New York, NY: Springer, pp 1373–1397.
- vanden Hoven RN, Santini JM. 2004. Arsenite oxidation by the heterotroph Hydrogenophaga sp. str. NT-14: the arsenite oxidase and its physiological electron acceptor. Biochim Biophys Acta 1656:148–155.
- Vishnivetskaya TA, Brandt CC, Madden AS, Drake MM, Kostka JE, Akob DM, Küsel K, Palumbo AV. 2010. Microbial community changes in response to ethanol or methanol amendments for U (VI) reduction. Appl Environ Microbiol 76:5728–5735.
- Wang A, Liu L, Sun D, Ren N, Lee DJ. 2010. Isolation of Fe(III)-reducing fermentative bacterium Bacteroides sp. W7 in the anode suspension of a microbial electrolysis cell (MEC). Int J Hydrogen Energy. 35:3178–3182.
- Wang J, Muyzer G, Bodelier PL, Laanbroek HJ. 2009. Diversity of iron oxidizers in wetland soils revealed by novel 16S rRNA primers targeting Gallionella-related bacteria. ISME J 3:715–725.
- Weber KA, Achenbach LA, Coates JD. 2006a. Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol 4:752–764.
- Weber KA, Pollock J, Cole KA, O'Connor SM, Achenbach LA, Coates JD. 2006b. Anaerobic nitrate-dependent iron (II) bio-oxidation by a novel lithoautotrophic betaproteobacterium, strain 2002. Appl Environ Microbiol 72:686–694.
- Yu R, Gan P, MacKay AA, Zhang S, Smets BF. 2010. Presence, distribution, and diversity of iron-oxidizing bacteria at a landfill leachate-impacted groundwater surface water interface. FEMS Microbiol Ecol 71:260–271.
- Yu WH, Harvey CM, Harvey CF. 2003. Arsenic in groundwater in Bangladesh: A geostatistical and epidemiological framework for evaluating health effects and potential remedies. Water Resour Res 39:1146–1163.
- Zhang J, Zhou W, Liu B, He J, Shen Q, Zhao F-J. 2015. Anaerobic arsenite oxidation by an autotrophic arsenite-oxidizing bacterium from an arsenic-contaminated paddy soil. Environ Sci Technol. 49:5956–5964.
- Zobrist J, Dowdle PR, Davis JA, Oremland RS. 2000. Mobilization of arsenite by dissimilatory reduction of adsorbed arsenate. Environ Sci Technol. 34:4747–4753.
- Zouboulis AI, Katsoyiannis IA. 2005. Recent advances in the bioremediation of arsenic-contaminated groundwaters. Environ Int. 31:213–219.
