 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
We investigated the microbial community composition in three different mine tunnels from Hualgayoc (Cajamarca, Peru) and the metabolic activity of acidophilic sulfate-reducing bacteria (aSRB) enriched from acidic sediments of the mining tunnels. Microbial community composition in environmental samples was analyzed by 16S rRNA amplicon sequencing. Metallibacterium and Acidiphilium were found as the most abundant bacteria in PM1 and PM2 sites, respectively, while Cyanobacteria was abundant in PM3 site. Isosphaera and Pseudomonas thrived in the acidic water of PM2 site as well. Sulfate reduction at low pH was then evaluated in microcosm experiments showing activity even at pH 3.4. Hydrogen was the most favorable electron donor in terms of sulfate reducing rates (0.5 mM day−1 at pH 5.1) and sulfide production (4.9 mM at the end of the experiment). Sequences affiliated to Desulfosporosinus and Desulfovibrio were abundant in the hydrogen microcosms (60.3 and 17.8%, respectively). These sequences were not detected in sediments, but their occurrence in the microcosms suggests their low abundance in the studied AMD systems. Our results expand the phyla detected in AMD environments and contribute to the understanding of aSRB for the possibility of applying these microorganisms in bioremediation.
Introduction
Acid mine drainage (AMD) causes harmful effects on the environment, being considered the second biggest problem after global warming according to the United Nations (Tuffnell Citation2017). AMD is formed by consecutive steps of chemical oxidation of metal sulfides, mainly pyrite (FeS2) and pyrrhotite (Fe(1-x)S) (Schippers et al. Citation2010), producing a solution with high concentrations of ferrous iron (Fe(II)), protons (H+) and sulfate (SO42−) (Dold Citation2014; Karimian et al. Citation2018; Nordstrom and Alpers Citation1999; Sánchez España et al. Citation2005; Sánchez-Andrea et al. Citation2014a). This process is accelerated by chemoautotrophic iron oxidizing microorganisms generating ferric iron which acts as a strong metal sulfide oxidant. Additionally, other metal sulfides can be also oxidized in a parallel way releasing additional metal(oid)s such as copper (Cu), zinc (Zn), lead (Pb), or arsenic (As) (Iakovleva et al. Citation2015).
Despite the unfavorable conditions, well-adapted microorganisms perform crucial biogeochemical processes. Bacteria, archaea and eukarya represented by fungi and algae, are present in different proportions in AMD environments such as water (Falagán et al. Citation2014; González-Toril et al. Citation2011; Rowe et al. Citation2007), sediments (Lucheta et al. Citation2013; Sánchez-Andrea et al. Citation2011) and biofilms (Mesa et al. Citation2017). Microbial community composition varies from one place to other due to changes in pH, temperature, metal concentrations, redox potential, and even climate (e.g., seasonal variations) (Auld et al. Citation2017; Méndez-García et al. Citation2015). Bacteria phyla such as Proteobacteria, Nitrospirae, Actinobacteria, Acidobacteria, and Firmicutes are usually reported in AMD surveys, and their metabolic capabilities related to the biogeochemical cycling of iron, sulfur, carbon, and nitrogen, have been well-studied (Méndez-García et al. Citation2015; Mesa et al. Citation2017; van der Graaf et al. Citation2020).
An interesting microbial metabolism found in AMD environments is the sulfidogenic activity performed by sulfate-reducing bacteria (SRB). SRB catalyzes the dissimilatory reduction of sulfate (or potentially other sulfur oxidized compounds) resulting in the production of sulfide with the subsequent acidity neutralization of its surrounding environment (e.g., Ayangbenro et al. Citation2018; Sánchez-Andrea et al. Citation2011, Citation2013). The majority of SRB are acidotolerant or neutrophilic microorganisms, but the existence of acidophilic SRB (aSRB) has been reported recently (Alazard et al. Citation2010; Mardanov et al. Citation2016; Sánchez-Andrea et al. Citation2015). aSRB may not only survive in aqueous environments with pH as low as 3.0 (e.g., acidic pit lakes) but also contribute to a great extent to the self-attenuation of toxic metal pollution (Diez-Ercilla et al. Citation2014; Falagán et al. Citation2014; Sánchez-Andrea et al. Citation2012; Sánchez-España et al. Citation2020; van der Graaf et al. Citation2020).
Hualgayoc Province is situated in Cajamarca Region on the Llaucano river basin in northern Peru (). The Hualgayoc district is a mining district of the central Peruvian Andes. Its geology is represented by calcareous sediments (Goyllarisquizga formation) and the occurrence of sphalerite-galena, pyrite-quartz and other minerals (e.g., chalcopyrite, pyrrhotite, arsenopyrite, and hematite as accessory minerals). Ore minerals have been extracted in this area by small and medium scale mining for several years (Canchaya Citation1990). Several mining companies in this district had activities related to the extraction of silver, zinc and lead. Some of these mining activities dated back to the Spanish colonial period and resulted in many environmental liabilities. The non-remediated liabilities are exposed to the environment releasing AMD and affecting the local ecosystems ([DIGESA] Dirección General de Salud ambiental Citation2012).
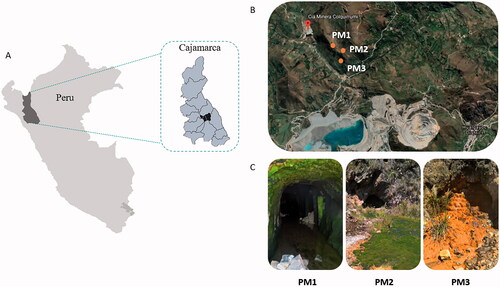
Figure 1. Map of Hualgayoc district in Cajamarca (Peru) (A) and the localization of the three mine tunnels (PM1, PM2, and PM3) (B). Pictures showing field features of PM1, PM2, and PM3 sampling sites (C).
In the current research we focus on three previously unexplored AMD-generating mine tunnels located in the Hualgayoc mine district (Cajamarca, Peru). This is the first study about the microbiology of such AMD systems. The main goal of our study was to investigate if the three mining sites host different microbial composition due to their geochemical differences. We also investigated whether the current extreme conditions would restrict the microbial diversity able to thrive in these environments down to a core microbiota of AMD. To accomplish it, the 16S rRNA gene amplicon sequencing analysis was carried out in both water and sediment samples. Furthermore, we investigated the metabolic activity of aSRB using an enrichment approach, with ionic and nonionic electron donors, and with sediments as inocula in low-pH microcosms. Our study revealed the occurrence of different microbial communities in each old Peruvian mining sites and key aSRB with high biotechnological potential in biomining and bioremediation processes.
Materials and methods
Site description and sample collection
Three mine tunnels were selected for their accessibility: PM1 (6°44′35.75″S, 78°38′28.54″O), PM2 (6°44′41.97″S, 78°38′17.81″O), and PM3 (6°44′47.24″S, 78°38′24.21″O) (); each site was separated from the others by approximately 100 m. While PM1 was easily accessible, both PM2 and PM3 sites were relatively closed with bricks and concrete (). From each sampling site, a stream of AMD constantly flows throughout the mining area forming a low-depth runoff. Some interesting features were observed during sampling such as the occurrence of benthic algae in PM1 site, higher levels of microbial mats in PM2, and chemical precipitates in sediments with yellowish/orange color in PM3 site (). Water and sediment samples (each one in triplicates) were collected at each sampling point in June 2017. On-site parameters like pH and redox potential (ORP) were measured with Oakton® instruments (model 110 series). Water samples were collected with sterile glass bottles (250 ml) which were filled up and sealed. Sediment samples were collected using 50 mL cut syringes which were introduced in the sediment until a depth of 10 cm. The sediments extracted were put down in falcon tubes of the same volume (50 mL) without air bubbles and then sealed with parafilm to reduce the entrance of oxygen. Water and sediment samples were collected at the tunnel entrance in PM1 site, and from the external part of the tunnel entrance in PM2 and PM3 sites. All samples were labeled and kept at 4 °C upon transportation to the Laboratory of Microbiology of the National University of Cajamarca (Peru). Once in the laboratory, sediment samples were divided for nucleic acid extraction and chemical analysis (stored at −20 °C) and for microbial cultures (stored at 4 °C). Water samples were filtered using 0.22 µm membrane filters (Membrane solutions, WA, US) and a vacuum pump (Welch, IL, US) by an external service (NKAP laboratory, Cajamarca). Filters were stored at −20 °C for DNA extraction. All samples were conditioned for transport to the Laboratory of Microbiology of the Wageningen University (The Netherlands) in which subsequent analysis were performed.
Chemical analyses of water and sediments
Total metal content in water was quantified in the Regional Water Laboratory-Regional Government of Cajamarca (Peru) using Inductively Coupled Plasma Optical-Emission Spectrometry (ICP-OES) in a Thermo Scientific iCAP 7000 series after previous acid digestion.
Major elements and trace metals in the sediments were quantified in the Geochemistry Laboratory of the Spanish Geological Survey (IGME-CSIC). All major elements (except Na) were measured by X-ray fluorescence (XRF) using a Zetium equipment from PANalitical. Na was measured by Atomic Absorption Spectrometry (AAS) in a VARIAN FS-220. Total sulfur was measured in a Eltra CS-800 elemental analyzer. Trace elements (As, Cu, Zn) were measured by inductively coupled plasma mass spectrometry (ICP-MS) in an Agilent 7500 ce after previous acid digestion.
DNA extraction from environmental samples
Water and sediment samples from each sampling point (PM1, PM2, and PM3) were used for DNA extraction. Sediment samples were pretreated following the method suggested by Fang et al. (Citation2015). Briefly, 0.5 g of each sediment was weighed and placed into 10 ml conical tubes with 3 ml of pretreatment solution composed by 0.1 M L−1 EDTA; 0.1 M L−1 Tris (pH 8); 1.5 M L−1 NaCl; 0.1 M L−1 Na2HPO4; 0.1 M L−1 Na2HPO4. Filters were placed with 10 ml of the same pretreatment solution into 15 ml conical tubes for 30 min and vortex for 15 min. In both cases (filters and sediments), tubes were placed into the incubator shaker (New BrunswickTM Innova®, Eppendorf, Germany) with the following setting up: 180 rpm × 15 min × 65 °C. The liquid phase was collected and centrifuged at 12000 × g for 5 min, the supernatant was discarded and pellets were suspended with the pretreatment solution and centrifuged again. The process was repeated until the supernatant was clear or colorless; all pellets obtained were stored at 5 °C. DNA extraction was performed using the FastDNA® SPIN Kit for Soil and the FastPrep® Instrument (MP Biomedicals, Santa Ana, CA, USA), following the instructions of the manufacturer. The dsDNA obtained was quantified with a Nanodrop spectrophotometer (DeNovix, USA). DNA purification was done using the DNA Clean and Concentrator TM-5.zymo Research, USA) and stored at − 20 °C until use.
Sulfate reduction microcosms assays
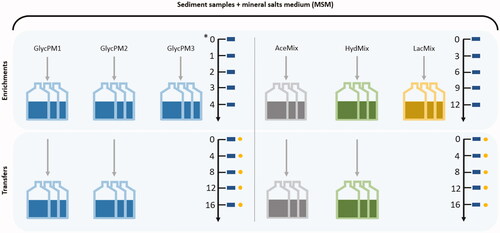
Microcosms promoting sulfate reduction at low pH were performed in triplicates and separated in two sets (enrichments and transfers) with different electron donors and incubation times (). Mineral Salts Medium (MSM) and solutions were used as previously described in Stams et al. (Citation1993) and later modified by Sánchez-Andrea et al. (Citation2015). In short, the media was supplemented with 0.1 g L−1 yeast extract and 1.42 g L−1 Na2SO4 (Sigma-Aldrich, St. Louis, MO, USA), and bicarbonate solution was eliminated from the original medium composition to allow pH modifications. Glycerol, acetate, and lactate were added from sterile 1 M stock solutions and used as electron donors and carbon sources (to a final concentration of 5 mM). Hydrogen was also tested as an electron donor with 1 mM of acetate as an additional carbon source (Widdel and Bak Citation1992). Anoxic conditions were achieved by sealing 100 mL serum bottles under N2/CO2 (80:20, v/v) or H2/CO2 (80:20, v/v) gas mixture at 1.5 atm with butyl rubber septa using the atmosphere exchanger device (GR instruments, The Netherlands). The pH of media was adjusted to a range varying between 3.4 and 4.8 in the enrichments and between 4.7 and 5.2 in the transfers with HCl or NaOH before autoclaving. Sample inoculation was done in an anaerobic chamber at 25 °C. Enrichments started with the inoculation of glycerol microcosms (GlycPM1, GlycPM2 and GlycPM3) with each sediment sample in a separate way (). The incubation time of these enrichments was 4 weeks and 16 days in the transfer. Acetate (AceMix), hydrogen (HydMix) and lactate (LacMix) microcosms () were subsequently inoculated using a mixed sediment sample (to enhance the chances of sulfate reduction regardless the origin of inocula). The incubation time of these enrichments and transfers was 12 days and 16 days, respectively. All microcosms were incubated statically in the dark at 30 °C and monitored periodically for pH and sulfide production. Sulfide was measured with the methylene blue assay (Cline Citation1969). GlycPM3 and LacMix microcosms were incubated but due to lack of sulfide activity transfers were not performed. Microcosms of each electron donor with the best sulfide activity were used to screening sulfate metabolism and acetate production in the transfers every 4 days using Dionex 2000 ion chromatograph and LKB high-performance liquid chromatography (HPLC) respectively.
Figure 2. Sulfate reduction at low pH in microcosms from both enrichments and transfers. Glycerol (GlycPM1, PM2, PM3), acetate (AceMix), hydrogen + acetate (HydMix) and lactate (LacMix) were used as electron donors. Arrows with numbers indicate the incubation time (in days, except for glycerol microcosms in the enrichment which were in weeks (*)) and the scale time of sampling to measure pH and sulfide (rectangles), as well as sulfate and acetate (circles). GlycPM1 microcosms were not selected for sulfate measuring. GlycPM3 and LacMix microcosms were not transferred.
DNA extraction from microcosms
High-activity microcosms from the transfers (GlycPM2, AceMix, and HydMix) were used for DNA extraction following the procedure for sediments explained above. Additionally, LacMix (from the enrichment) and GlycPM1 (from the transfer) microcosms were also included in this process due to their low pH at which they began.
High-throughput sequencing and analysis
The 16S rRNA genes were amplified from all sediment samples at each sampling point (PM1, PM2, PM3) in triplicates. Water samples were also amplified successfully except for samples from the PM1 site and one replicate from the PM3 site (DNA was not recovered). The aforementioned microcosms were amplified as well. DNA concentration of each sample was adjusted to 20 ng µL−1. Barcoded amplicons from the bacterial V4 region of 16S rRNA gene were generated through PCR using the primers 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR mastermix was prepared with a total volume of 50 µL per sample containing 10 µL 5x HF PCR buffer, 1 µL (200 µM) dNTPs, 0.5 µL (2 U µL−1) Phusion Hot start II DNA polymerase, 1 µL (10 µM) barcoded forward and reverse primers, 1 µL template DNA and nuclease-free water up to final volume. All samples were done by triplicates adding a negative control without the addition of a template. The PCR set up consisted of a pre-denaturation at 98 °C for 30 s, followed by 35 cycles of 10 s denaturation at 98 °C, 10 s annealing at 50 °C, 10 s elongation at 72 °C; and 10 min final extension step at 72 °C. After PCR, triplicates of each sample were mixed and 5 µL of the PCR product was loaded in 1% (w/v) agarose gel to check the size of the amplicons through electrophoresis. PCR products were purified using the Clean PCR − 96-well plate kit (CleanNA, The Netherlands) following the instruction of the manufacturer, and dsDNA concentration was determined by fluorescence using Denovix DS-11 FX (DeNovix, USA) throughout the Qubit dsDNA BR assay. Sequencing was carried out on an Illumina HiSeq 2000 system at GATC-Biotech (Konstanz, Germany).
16S rRNA gene amplicon sequences were analyzed using NGtax pipeline (Ramiro-Garcia et al. Citation2016). Reads were quality checked and assigned to samples based on nucleotide barcodes. Quality-controlled sequences were denoised. Chimeras were predicted and removed before downstream analyses. Chimera-free sequences were clustered into operational taxonomic units (OTUs) defined by a 97% sequence similarity. Taxonomic affiliation was performed through a comparison against the SILVA non-redundant database (v 128). Data analysis and plotting were carried out in R studio.
Results
Physicochemical features of AMD samples
The chemical composition as well as pH, redox potential and temperature of water and sediment samples are summarized in and . While water and sediment samples from the same site had similar redox potential (ORP), they differ in pH ( and ). Water samples from PM1 and PM2 were highly acidic (pH 2.3 and 2.4, respectively) while the corresponding pore waters from sediment samples presented pH values of 5.8 and 6 respectively, likely due to microbial anaerobic respiratory and acid-neutralizing processes such as sulfate reduction (Sánchez-Andrea et al. Citation2012). Water and sediment samples from PM3 were less acidic with pH of 5.4 and 6.5, respectively. Dissolved metals in the water samples were in decreasing order of abundance from iron (the most abundant trace metal) to cadmium (the least abundant trace metal): Fe > Zn > Mn > Al > Cu > As > Cd (). All sediment samples had abundance of silica, alumina, ferric iron oxides, phosphate and sulfur (). Metals and metalloids were present from moderate (Cu, As) to high (Zn) concentrations in the studied sediments ().
Table 1. Physico-chemical parameters -pH, ORP (mV), temperature (°C)- and chemical composition (metals and metalloid concentration in mM) measured in water samples.
Table 2. Physico-chemical parameters -pH, ORP (mV)- and major element concentration (including major oxides and trace metal(oid)s in mM units) measured in sediment samples.
Microbial diversity and composition of AMD samples
Water and sediment samples possessed low alpha diversity (Shannon index (H’) average of the three sites = 2.3 ± 0.3 in sediments and 2.0 ± 0.4 in waters). This diversity is typical of some AMD environments (García-Moyano et al. Citation2015; Mesa et al. Citation2017; Sajjad et al. Citation2018; Sánchez-Andrea et al. Citation2011), and differed from each other at the same sampling site and between sites. A total of 92 OTUs were obtained after analysis. Water samples from PM2 and PM3 had 30 and 21 OTUs, respectively. Sediment samples from PM1, PM2, and PM3 had 26, 33, and 24 OTUs, respectively. The beta-diversity analysis revealed three general clusters representing each of the three sites (). Per site, water samples clustered together and separate from the sediment samples.
Figure 3. Principal coordinate analysis (PCoA) plot of 16S rRNA sequences (A) and bar plot showing microbial composition at the phylum level in environmental samples (B). Consensus of replicates by sample type is showed in the bar plot. Taxonomic affiliation is described at right. Abbreviations: Wat PM2: water sample PM2; Wat PM3: water sample PM3; Sed PM1: sediment sample PM1; Sed PM2: sediment sample PM2; Sed PM3: sediment sample PM3.
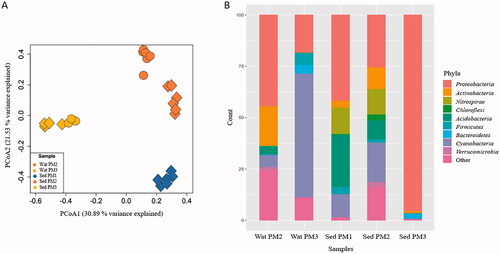
At the phylum level, and reflecting the observation of the beta diversity analysis, the microbial composition of water samples differed from that of sediment samples in each site and within sites (). The main phyla detected in both samples of each sampling site but at different relative abundances were Proteobacteria, Cyanobacteria, Actinobacteria, Nitrospirae, Chloroflexi, Acidobacteria, Firmicutes, Bacteroidetes and Verrucomicrobia. Proteobacteria was the most abundant phylum in sediments of PM1 (41.8%). Acidobacteria was more abundant in PM1 sediments (25.5%) in comparison to other sampling sites. Similar to PM1, both samples from PM2 were dominated by Proteobacteria (water: 44.4%, sediment: 25.6%). Actinobacteria, Acidobacteria and Chloroflexi phyla were also found in PM2. Nitrospirae was similarly abundant in sediment samples from PM1 and PM2. At PM3, Proteobacteria was highly abundant in sediment samples (95.5%), while Cyanobacteria was highly abundant in water samples (60.1%). Firmicutes and Bacteroidetes were less abundant at PM3.
At the genus level, each site and sample type (water or sediment) also reflected variable microbial composition (). Sediment samples from PM1 were characterized by sequences affiliated to Metallibacterium (31.2 ± 7.7%), followed by sequences related to an uncultured Acidobacteriaceae (Acidobacteria) (23.2 ± 4.1%), and Leptospirillum (12.7 ± 8.9%). The dominant genera identified in the sequences of water samples from PM2 were Acidiphilium (34.2 ± 36.8%), an uncultured Acidimicrobiales (Actinobacteria) (17.6 ± 18.3%), Isosphaera (Planctomycetes), (8.1 ± 9.5%), and Pseudomonas (5.7% ± 14.1%). Sediment samples from PM2, which had the major number of OTUs, were especially characterized by sequences related to an undetermined Cyanobacteria cluster (19.1 ± 14.6%) and Leptospirillum (12.3 ± 3.6%).
Table 3. Bacterial genera found in environmental samples of each sampling site (PM1, PM2 and PM3) and their potential metabolic profiles.
Water samples from PM3 were mostly sequences affiliated to one undetermined Cyanobacteria cluster (49.4 ± 4.9%), and Sideroxydans (15.0 ± 2.8%). In the same way, other sequences belonged to another undetermined Cyanobacteria cluster (6.7 ± 1.9%), Desulfosporosinus (5.5 ± 1.0%), and Microbacter (4.0 ± 3.7%). Abundant sequences in sediments from PM3 were affiliated to an uncultured Betaproteobacteria cluster (26.1 ± 19.0%), Ferritrophicum (20.5 ± 13.1%), Sideroxydans (17.2 ± 5.1%) and Rhodanobacter (12.5 ± 12.3%). Sequences belonging to rare taxa (0.1 − 1% relative abundance) were also detected in water and sediment samples.
Screening of sulfate reducing activity at a low pH
The increase of pH and sulfide production were tracked as a proxy for sulfate reduction activity. An increase of more than 2 pH units took place in all microcosms from the beginning of the enrichment set with the exception of GlycPM2 (pH increase of 0.9 units) and GlycPM3 (did not show activity). The microcosms incubated with hydrogen (HydMix) showed the most prominent increase reaching 2.7 pH units over the initial pH (Supplementary information 1). In contrast, pH did not show the same increase in the transfers (increase lower than 1 pH unit) despite the less acidic conditions of these cultures. HydMix microcosms did, however, show an important increase (2 pH units). Sulfide was detected in all microcosms though more sulfide was detected in the transfers than the enrichments, especially in HydMix microcosms (Supplementary information 2). Sulfide concentrations in GlycPM1/PM2 and AceMix microcosms of the transfers suggest that despite the presence of electron donors (5 mM), sulfide production was not highly promoted, conversely to HydMix microcosms which reached up to 4.9 ± 0.4 mM at the end of the experiments. The lowest pH tested at which sulfide was detected varied from 3.4 in the enrichment to 4.7 in the transfer of GlycPM1 microcosms. Latent phase for sulfide production also varied from 1 week (for GlycPM1/PM2 microcosms) to 3 days (for AceMix, and HydMix) in the enrichments, and up to 4 days (GlycPM1/PM2, AceMix, and HydMix) in the transfers.
Sulfate and acetate concentrations were measured in the high-activity replicates of GlycPM2, AceMix, and HydMix microcosms of the transfer set (Supplementary information 2). Sulfate concentration increased in the first part of the incubation (from the starting point up to day 8) and decreased up to the final part (from day 12 up to day 16) in GlycPM2 microcosm. Conversely, in AceMix microcosm, a slight sulfate uptake occurred in the first part of the incubation (from the starting point up to day 8), and a similar sulfate increase in the final part (from day 12 up to day 16). Sulfate was almost totally depleted in HydMix microcosm (8.1 mM). The sulfate reduction rate (mM day−1) in each microcosm was: 0.2 in GlycPM2 at pH 5, 0.03 in AceMix at pH 5.2, and 0.5 in HydMix at pH 5.1.
A substantial increase in acetate generation was detected uncoupled and coupled to sulfate reduction in GlycPM2, reaching up to 8.1 mM at the end of incubation. Similarly, this occurred in AceMix microcosm, reaching up to 10.1 mM at the end of incubation. Acetate was also generated in HydMix microcosm at a moderate extent (5.7 mM). Sulfate reduction processes were accompanied by the formation of black precipitates (likely, iron sulfides) at a different time and pH in the microcosms (Supplementary information 3).
Microbial diversity and composition of microcosms
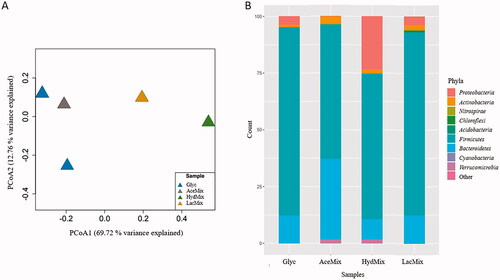
High-activity microcosms (GlycPM2, AceMix and HydMix) of the transfer were selected after an incubation time of 16 days for microbial community analysis. Additionally, one LacMix and one GlycPM1 microcosm (from the enrichment and transfer, respectively) were also included after an incubation time of 12 and 16 days, respectively. All analyzed microcosms possessed lower alpha diversity than environmental samples (H’ average of the three sites = 1.3 ± 0.4) and different beta diversity (). A total of 21 OTUs were obtained. The number of OTUs were 4, 7, 7, 9 and 12 for GlycPM1, GlycPM2, AceMix, HydMix, and LacMix respectively. In general, the most abundant phyla were Firmicutes (GlycPM1/PM2: 83.1%; AceMix: 59.2%; HydMix: 64%; LacMix: 80.6%), followed by Bacteroidetes (GlycPM1/PM2: 12.4%; AceMix: 35.6%; HydMix: 8.9%; LacMix: 12.7%), Proteobacteria (GlycPM1/PM2: 3.7%; HydMix: 23.4%; LacMix: 3.6%), and Actinobacteria (GlycPM1/PM2: 0.9%; AceMix: 3.4%; HydMix: 1.9%; LacMix: 2.4%). Verrucomicrobia (AceMix: 1.7%; HydMix: 1.9%) and Chloroflexi (LacMix: 0.8%) were also found at low abundance ().
Figure 4. Principal coordinate analysis (PCoA) plot of 16S rRNA sequences (A) and bar plot showing microbial composition at the phylum level in microcosms (B). Taxonomic affiliation is described at right in the bar plot. Abbreviations: Glyc: glycerol PM1/PM2 microcosms; AceMix: acetate microcosm; HydMix: hydrogen microcosm; LacMix: lactate microcosm.
At the genus level, GlycPM1/PM2 and AceMix microcosms were similar in their microbial community composition (). Clostridium sensu stricto 12 affiliated sequences were the most abundant (GlycPM1: 74.6%; GlycPM2: 49.6%; and AceMix: 45.1%). Other sequences affiliated to Microbacter (GlycPM1: 19.9%; GlycPM2: 4.8%; and AceMix: 35.6%), Desulfosporosinus (GlycPM1: 2.6%; GlycPM2: 14.4%; and AceMix: 8.2%), and Cellulomonas (GlycPM2: 1.9%; and AceMix: 3.4%) were coenriched in these microcosms. Sequences affiliated to the genera Thiomonas (2.9%), Caproiciproducens (23.7%), and Clostridium sensu stricto 10 (5.2%) were detected only in GlycPM1, GlycPM2, and AceMix microcosms respectively.
Figure 5. Heat map showing genera abundance in microcosms and cluster analysis dendrogram based on Bray–Curtis similarity. Abbreviations: GlycPM1: glycerol PM1 microcosm; GlycPM2: glycerol PM2 microcosm; AceMix: acetate microcosm; HydMix: hydrogen microcosm; LacMix: lactate microcosm.
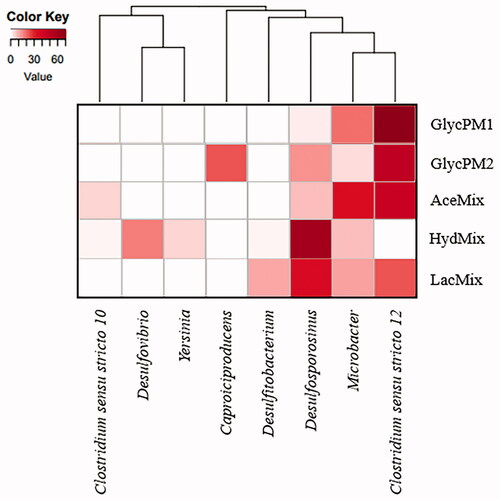
HydMix and LacMix microcosms shared some microbial phylotypes in common (). HydMix microcosm showed sequences identified as Desulfosporosinus (60.3%) as the most abundant followed by Desulfovibrio (17.8%), Microbacter (8.9%), Yersinia (5.6%), Cellulomonas (1.9%), Clostridium sensu stricto 10 (1.4%), and Desulfitobacterium (1.4%). In LacMix microcosm the sequences were affiliated to Desulfosporosinus (23.8%), and Clostridium sensu stricto 12 (23.8%), followed by the genera Microbacter (12.7%), Desulfitobacterium (11.5), Thermincola (3.6%), Acidocella (3.6%), Fonticella (3.2%), and Cellulomonas (1.6%).
Discussion
Physicochemical characteristics
The abundance of silica (SiO2) and alumina (Al2O3) and the marked scarcity of calcium (Ca2O) in the analyzed sediments is coherent with the dominant presence of aluminosilicate minerals and the lack of carbonates in the area (Canchaya Citation1990). These two geological features point to a low buffer capacity of the sediment substrate to neutralize the acidity of the leachates. The abundance of iron and sulfur in the three mine sites (0.7–2.9 mM Fe2O3, 0.7–1.9 mM S) relates to the presence of pyrite. PM3 showed much higher iron contents with respect to the rest, consistent with the abundance of iron oxyhydroxides (e.g., schwertmannite, jarosite, goethite, etc.) suggested by the typical orange color observed in these sediments (). Finally, the phosphate contents of most sediments (18–38.4 × 10−3 mM P2O5) may derive from the abundance of benthic green algae in the stream banks (). Arsenic (9.25–22.6 mM) is likely derived from the dissolution of the arsenopyrite present in the mine sites and could be an adsorbed constituent on the iron oxyhydroxides of the sediments (Islam et al. Citation2005). On the other hand, Cu (0.8–14.6 mM) and Zn (44.5–76.1 mM) would result from chalcopyrite and sphalerite dissolution in the mine sites. These metals could be associated with sulfates or oxyhydrosulfates, or with secondary (i.e., newly formed) metal sulfides (e.g., CuS, ZnS) resulting from SRB activity below the sediment surface (Rowe et al. Citation2007).
Microbial community composition and potential ecological function of the three studied sites
PM1 site
shows the geomicrobiological model of PM1 site. Metallibacterium and Leptospirillum were the two most abundant genera. High number of sequences belonging to Fe(III) reducing heterotrophic bacteria from the genus Metallibacterium are ubiquitous in acidic environments (Bomberg et al. Citation2019; Brantner et al. Citation2014; Ziegler et al. Citation2013). This genus has one reported species so far (M. scheffleri). It has been featured as a versatile facultative anaerobe able to reduce nitrate (Bartsch et al. Citation2017) to alkalinizing its aqueous medium through ammonium production. Alkalinization contributes to the development of microniches in acidic environments (Ziegler et al. Citation2013) which is a very convenient feature for the establishment of some other less acidophilic microbial populations (including SRB). Sequences belonging to the genus Leptospirillum (Nitrospirae) were also detected abundant in sediment samples. Members of this genus are chemolithoautotrophic iron oxidizers broadly known for its role in AMD generation (Baker and Banfield Citation2003). Interestingly, the pH found in these sediments (pH 5.8) was relatively higher than the optimum pH for growth of this acidophile (pH <3). We therefore hypothesize that Leptospirillum has colonized superficial layers of these sediments which are in contact with the acidic water (pH 2.3). Leptospirillum spp. is often found together with other iron oxidizers such as Acidithiobacillus spp. in AMD environments (Lu et al. Citation2010). However, we did not obtain any OTUs corresponding to Acidithiobacillus spp., in this study. The relatively higher abundance of Leptospirillum over Acidithiobacillus spp. was explained by its higher tolerance to higher metal content and lower pH (Dopson and Holmes Citation2014). Oxygen requirements of Leptospirillum can be supplied by benthic algae and Cyanobacteria (Portielje and Lijklema Citation1995). Presence of benthic algae was visible () signaling an important primary production.
Figure 6. Geomicrobiology of water and sediment samples of PM1 mine tunnel from Hualgayoc. pH and redox potential are mentioned in each environment. Bacterial taxa and chemical compounds (oxides and sulfides) are depicted by different shapes (see legend). Bacterial metabolisms and chemical speciation of carbon, oxygen, phosphate, iron, nitrogen and sulfur are represented by arrows. Dotted arrows indicate alternative processes that can support additional bacterial metabolisms. Dotted shapes indicate alkalization propitiated by bacterial metabolism. Question marks refer to unknown components like bacteria phylotypes and metabolism products.
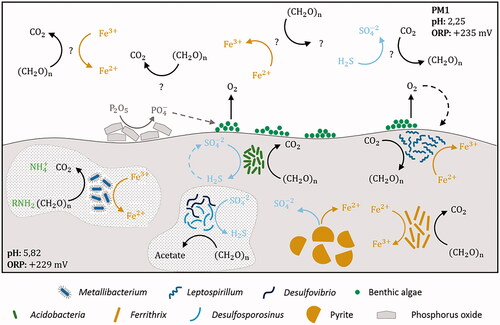
Non-common sulfur-cyclers were detected in PM1 sediments while common sulfate reducers could be occurring in low abundance. Sequences belonging to an uncultured Acidobacteriaceae (Acidobacteria) were abundant. A relatively low number of Acidobacteria phylum members have been cultured so far. Recently, Acidobacteria (subdivision 1 and 3) were reported to encode the complete canonical pathway for dissimilatory sulfite and sulfate reduction (Hausmann et al. Citation2018). Occurrence of dsrL genes (typically found in sulfur oxidizers) on dsr operons in proximity to dsrAB genes let to the hypothesis that some Acidobacteria may also operate the sulfate-reduction pathway in reverse to perform sulfide oxidation (Hausmann et al. Citation2018) (). We did not detect OTUs of the typical SRB (Desulfosporosinus and Desulfovibrio) despite having detected activity in the cultures enriched with this sediment as inoculum. Therefore, their abundance might be below the detection limits of the sequencing approach used. Although low abundance could mean low contribution to biogeochemical cycling, contribution to sulfur and carbon, and even iron cycling (Neal et al. Citation2001) by Desulfosporosinus spp. has recently been reported (Hausmann et al. Citation2019).
PM2 site
The geomicrobiological model of this site is showed in . The availability of direct solar light and the hydrology of PM2 acidic mine effluent (with low flow and low depth of the water course) allowed the presence of abundant microbial mats (). The production of organic compounds (an important carbon input in the AMD system) promotes heterotrophic populations such as the well-documented acidophile Acidiphilium (Alphaproteobacteria) (Prieto-Barajas et al. Citation2018; Rowe et al. Citation2007), whose OTUs were found abundant in these samples. Acidiphilium spp., which can perform heterotrophic oxygen or Fe(III) respiration (Baker and Banfield Citation2003), are often reported in sediments (García-Moyano et al. Citation2015; Lucheta et al. Citation2013) and shallow AMD environments (Mesa et al. Citation2017), but also in acidic pit lakes (Sánchez-España et al. Citation2020; van der Graaf et al. Citation2020). Other OTUs corresponded to uncultured Acidimicrobiales (Actinobacteria), which harbors iron-oxidizing and iron-reducing phylotypes depending on the presence or absence of oxygen (González-Toril et al. Citation2011), and even an additional role removing sulfate (Eisen et al. Citation2016), suggest an active cycling of iron and sulfur in this site ().
Figure 7. Geomicrobiology of water and sediment samples of PM2 mine tunnel from Hualgayoc. pH and redox potential are mentioned in each environment. Bacterial taxa and chemical compounds (oxides and sulfides) are depicted by different shapes (see legend). Bacterial metabolisms and chemical speciation of carbon, oxygen, phosphate, iron, nitrogen, sulfur and arsenic are represented by arrows. Dotted arrows indicate alternative processes that can support additional bacterial metabolisms. Dotted shape indicates alkalization propitiated by bacterial metabolism. Question marks refer to unknown components like metabolism products.
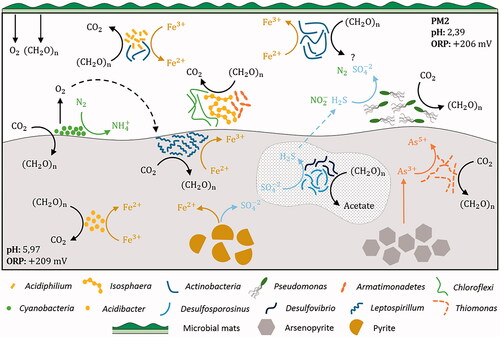
Isosphaera has not been reported under acidic conditions, and together with Pseudomonas, to the best of our knowledge, it is the first time both genera are found in extreme acidic water (pH 2.4). Pseudomonas (Gammaproteobacteria) has been shown to be involved in nitrite reduction and sulfide oxidation (Mahmood et al. Citation2009) and with detoxification abilities (e.g., biofilm formation and siderophores production) (Ferreira et al. Citation2018; Teitzel and Parsek Citation2003). Interestingly, surveys reporting Pseudomonas spp. range from pH 3.5–3.7 in water (Sato et al. Citation2019). Isosphaera (Isosphaeraceae family), whose sequence were relatively abundant, has only one reported species (I. pallida (Giovannoni et al. Citation1987) featured as an oligotrophic strictly aerobic and moderately thermophilic bacterium isolated from hot springs in Yellowstone National Park (Ward et al. Citation2006). Although it is not consistent with the temperature measured in the PM2 site (14.6°C), it could either be another mesophilic phylotype or still thermophilic since exothermic microbial oxidation of pyrite creates microenvironments with relatively high temperatures that could host thermophiles (Sánchez-Andrea et al. Citation2011; van der Graaf et al. Citation2020). Uncultured phylotypes conformed the microbial community in sediment samples from PM2. Some sequences affiliated to an uncultured Cyanobacteria were the most abundant in this sediment. These microorganisms are normally reported in surface and shallow AMD systems as primary producers (producing oxygen and fixing nitrogen and carbon) (Mesa et al. Citation2017), but also in deeper subsurface environments with hydrogen-based metabolisms (Puente-Sánchez et al. Citation2018). Some unexpected bacteria (undetermined) of the Armatimonadales (Armatimonadetes) and Ktedonobacteraceae (Chloroflexi) were detected in these samples. Although these phylotypes are not new in AMD environments and have been related to carbohydrate-based metabolism (Lee et al. Citation2014; García-Moyano et al. Citation2015; Mesa et al. Citation2017; Gavrilov et al. Citation2019), their roles in the microbial community are still not clear.
Microorganisms potentially active in the iron and arsenic cycles were present in sediment samples from PM2. Acidibacter (Gammaproteobacteria), is involved in iron metabolism. A. ferrirreducens, possesses high metal resistance with the minimal inhibitory concentration (MIC) for metals being 5 mM for Cu and 50 mM for Zn (Falagán and Johnson Citation2014). We found even higher concentration of heavy metals in the studied sediments (e.g., 14.6 mM Cu and 62.9 mM Zn). Thiomonas spp. (found in both PM2 sediment as rare taxa and in microcosms assays), is a chemolithotrophic As(III)-oxidizing Betaproteobacteria member. The oxidative dissolution of arsenopyrite (FeAsS) by Thiomonas releases As(III) and reduced sulfur into solution, being able to oxidize subsequently the As(III) to As(V) (Battaglia-Brunet et al. Citation2011; Kelly et al. Citation2007). The biological oxidation of As(III) to the less toxic form As(V) by Thiomonas species can mean a natural attenuation of toxic metals in AMD environments (Bryan et al. Citation2009).
PM3 site
shows the geomicrobiological model of PM3 site. Many uncultured bacteria were found in PM3, both in water and sediment samples. The main variables influencing this different microbial community include physicochemical characteristics such as the higher pH in this mine site, a relatively lower redox potential (in comparison with the other two mine sites), and the higher metal concentration. Sequences affiliated to an uncultured Cyanobacteria were found as the most abundant, confirming an important primary production in water. The presence of dissolved iron (15.9 × 10−1 mM Fe) in this site is again consistent with the occurrence of OTUs related with iron metabolism such as Sideroxydans, who belongs to Gallionellaceae family (Betaproteobacteria) which holds many neutrophilic Fe(II) oxidizers (Hallbeck and Pedersen Citation2014). Sideroxydans can fix CO2 (), an important strategy for adapting to oligotrophic environments like AMD settings and show a great metal tolerance (Sajjad et al. Citation2018).
Figure 8. Geomicrobiology of water and sediment samples of PM3 mine tunnel from Hualgayoc. pH and redox potential are mentioned in each environment. Bacterial taxa and chemical compounds (oxides and sulfides) are depicted by different shapes (see legend). Bacterial metabolisms and chemical speciation of carbon, oxygen, iron, nitrogen, hydrogen and sulfur are represented by arrows. Dotted arrows indicate alternative processes that can support additional bacterial metabolisms. Dotted shape indicates the probable occurrence of archaea. RISC: reduced inorganic sulfur compound.
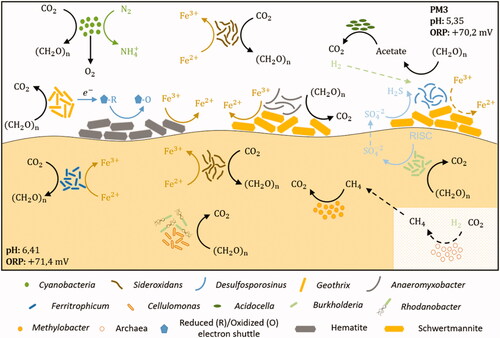
Sequences affiliated to Desulfosporosinus, Microbacter, and Acidocella-like bacteria were surprisingly found in the PM3 waters, when normally they coexist in sulfidogenic enrichments or reactors (Sánchez-Andrea et al. Citation2013, Citation2014b). Desulfosporosinus (Firmicutes), an ubiquitous and metabolically versatile SRB, is commonly found in acidic reductive environments (Chen et al. Citation2016; Johnson and Hallberg Citation2003; Méndez-García et al. Citation2015; Sánchez-Andrea et al. Citation2011). However, the finding of this genus in this AMD site (ORP: + 70 mV) could point toward a greater tolerance to oxidant conditions (Karnachuk et al. Citation2015; Mardanov et al. Citation2016; Sato et al. Citation2019). Desulfosporosinus members are also able to use Fe(III) as an electron acceptor with some organic electron donors (Sánchez-Andrea et al. Citation2015) conferring them with the metabolic flexibility to thrive in these settings. Notwithstanding, the possibility that this microorganism could survive in the form of spores should not be ruled out (Suzuki et al. Citation2003), probably these spores being transported from another place in the tunnel where the redox potential is lower, and in which sulfate reduction metabolisms could occur successfully.
The remarkable yellow/orange color in sediments of PM3 () is an indicative signal of an important Fe(III) oxide deposition (González-Toril et al. Citation2011). Although the iron oxidizer Ferritrophicum (Betaproteobacteria) is normally reported as a scarce member in some AMD sites (Grettenberger et al. Citation2017), OTUs corresponding to this genus were dominant in this study (20.5 ± 13.1%). Type strain, Ferritrophicum radicicola has been reported as a chemolithoautotrophic aerobe, isolated from the rhizosphere of wetland plants in an AMD system (Weiss et al. Citation2007). Similar abundance of sequences affiliated to the Ferritrophicales order has been informed in AMD originated from mercury sulfides (Jew et al. Citation2014). An interesting OTU found as rare taxon in this site was Methylobacter (Gammaproteobacteria), a heterotroph reported in cold mining water and involved in methane oxidation (Bomberg et al. Citation2015). OTUs related to SRB were not detected in these samples (in accordance with the negative results of microcosms experiments with glycerol as electron donor). In contrast, the acid-tolerant Burkholderia (Betaproteobacteria), which is featured as an oligotrophic bacterium, can grow chemolithoautotrophically on thiosulfate (Bhowal and Chakraborty Citation2011) contributing to sulfate that can be used by SRB (like Desulfosporosinus in water samples) as an electron acceptor.
Sulfate reduction activity and microbial communities in the microcosms
Microcosms with glycerol (GlycPM1) as electron donor showed sulfidogenic activity with moderately acidic sediments from mine tunnels of Hualgayoc district (Cajamarca, Peru) at the lowest pH tested in the enrichment assay (pH 3.4), supporting an acidophilic nature of the native sulfate reducers. Low pH sulfate reduction coupled to glycerol as electron donor has been reported previously (Alazard et al. Citation2010; Kimura et al. Citation2006; Sánchez-Andrea et al. Citation2013, Citation2015). Although sulfate reduction employing ionic (lactate, acetate, succinate, etc.) or nonionic substrates (glycerol, methanol, H2) as electron donors has been extensively studied at circumneutral pH, there is little information at acidic conditions. Sulfate reduction can be energetically more favorable at pH 3 (ΔG0= −198 kJ mol−1) than in neutral conditions (ΔG0= −152 kJ mol−1) with H2 as electron donor (Meier et al. Citation2012). On the other hand, a good deal of that energy gain might have to go to proton export and homeostasis.
An increment of pH and a blackish color (typical of iron sulfides) were noticed together with sulfide production in all microcosm assays. These features are good indicators of the occurrence of sulfate reduction (Sánchez-Andrea et al. Citation2013). Further, iron sulfides formation (EquationEquation 1(1)
(1) ) can prevent inhibition by hydrogen sulfide (toxic at low pH in concentrations ranging from 1.2 to 3.6 mM (Kaksonen et al. Citation2004)) (Meier et al. Citation2012; Xia et al. Citation2019). In addition to sulfide, SRB produce bicarbonate ions when performing incomplete (EquationEquation 2
(2)
(2) ) or complete (EquationEquation 3
(3)
(3) ) oxidation of electron donors such as lactate, which further raise the pH of the media (Ayangbenro et al. Citation2018; Kaksonen et al. Citation2004). Additionally, sulfate reduction is a proton consuming reaction when H2 is involved (EquationEquation 4
(4)
(4) ) (Muyzer and Stams Citation2008; Rabus et al. Citation2013).
(1)
(1)
(2)
(2)
(3)
(3)
(4)
(4)
Among all assays, hydrogen-containing microcosms (HydMix) showed the highest increment of pH, in line with previous studies reporting increase of up to 2 pH units in enrichments including hydrogen cultures at pH 4 (Sánchez-Andrea et al. Citation2013). Accordingly, HydMix microcosms also showed the maximum concentration of sulfide (1.3 mM in the enrichment, and 4.9 mM in the transfer). Chemolithotrophic growth with hydrogen is theoretically ideal at a low pH because this substrate is not toxic, it is a thermodynamically favorable metabolism, and there are few competitors such as hydrogenotrophic methanogens (Kaksonen and Puhakka Citation2007; Koschorreck Citation2008), which are easily outcompeted in the studied conditions (low pH, high sulfide concentrations).
Glycerol (GlycPM1/PM2) and acetate (AceMix) microcosms of the transfers share similar microbial community composition at the genus level despite having different inoculum sources and sulfate depletion rates. Sulfate reduction rate in GlycPM2 (0.2 mM day−1 at pH 5) was similar to other surveys (Sánchez-Andrea et al. Citation2013) and higher than AceMix (0.03 mM day−1 at pH 5.2). Sulfate reduction in these microcosms was detected at day 12 in GlycPM2 and at day 4 in AceMix, and the detection of sequences related to Desulfosporosinus-like bacteria (14.4% in GlycPM2 and 8.2% in AceMix) confirm that the presence of aSRB. Desulfosporosinus spp. have been isolated from different acidic sediments (Alazard et al. Citation2010; Sánchez-Andrea et al. Citation2013). They are able to grow at pH lower than 4.0, and have been detected at even lower pH conditions (2.9–4.0) in the water column of several acidic pit lakes (Sánchez-España et al. Citation2020; van der Graaf et al. Citation2020). However, this genus was not detected in the sediments of the sampling sites in this study. Similarly, Sato et al. (Citation2019) did not find Desulfosporosinus sequences in the inoculum used in bioreactors assays, but they found them in water samples in quite low relative abundance (0.0025 − 0.0069%). We ignore why sulfate reduction took relevance during the final stage of the assay in GlycPM2, but it has been reported that the onset of sulfate reduction can be delayed by the presence of Fe(III) in microcosms assays with organic electron donors (Xia et al. Citation2019). On the other hand, detection of Desulfosporosinus in AceMix was unexpected considering that the different isolated species cannot employ acetate as an electron donor. The presence of Desulfosporosinus in the AceMix microcosms could point toward a phylotype able to perform complete oxidation (EquationEquation 5(5)
(5) ), a very convenient feature in AMD treatment.
(5)
(5)
The establishment of additional microbial proton-releasing metabolisms (like fermentation) in GlycPM1/PM2 and AceMix microcosms, was demonstrated by the detection of Microbacter sp. (isolated from acidophilic sulfidogenic enrichments (Sánchez-Andrea et al. Citation2014b)) and Caproiciproducens spp. (Kim et al. Citation2015), possibly contributing to the additional volatile fatty acids (VFA) production and also explaining the relatively stable pH in these microcosms despite sulfate reduction in the transfers. Although more analyses are needed to demonstrate the shift of microbial metabolisms in our study, many other surveys had reported these variations in bioreactors, mainly controlled by pH and temperature settings (Santos and Johnson Citation2017), by the substrate concentration (Dar et al. Citation2008) or even by the incubation time (Xia et al. Citation2019) allowing a specific group of microorganisms to predominate.
Around 8.1 from 9.2 mM of sulfate was metabolized in the HydMix transfer in 16 days, a really fast rate of sulfate consumption (0.5 mM day−1 at pH 5.1) in comparison with other assays (Meier et al. Citation2012). The higher activity of HydMix microcosms is supported by the occurrence of Desulfosporosinus and Desulfovibrio-like bacteria. OTUs of Desulfosporosinus (60.3%) was an important member of these, as confirmed by the clear patterns of pH increment and sulfide production commented above. In addition to Desulfosporosinus, sequences affiliated to Desulfovibrio was also found to be abundant (17.8%). Desulfovibrio spp., which belongs to Deltraproteobacteria group, is an intensively studied sulfate reducer able to grow chemolithotrophically with H2 and acetate (Heidelberg et al. Citation2004; Rabus et al. Citation2013), although it has been recently discovered that Desulfovibrio desulfuricans also perform CO2 fixation via the reductive glycine pathway (Sánchez-Andrea et al. Citation2020). The development of both sulfate reducers together has been reported before in H2-fed microcosms, confirming that H2 is an essential electron donor to both SRB (Fichtel et al. Citation2012). Desulfovibrio spp. was also dominant in sulfidogenic enrichments with high metal content (Zampieri, personal communication). On the other hand, the fastest sulfate consumption in HydMix microcosms could be explained by the outcompetition of other microbial metabolisms. It is possible that SRB was not performed in parallel by members of both taxa but sequentially. It is more likely that Desulfosporosinus, which have been broadly isolated at low pH (Karnachuk et al. Citation2015; Sánchez-Andrea et al. Citation2015), started performing sulfate reduction and at certain pH rise, Desulfovibrio, a generally faster grower (Yu et al. Citation2020) could have started the consumption of the remaining sulfate.
Additional sequences affiliated to microorganisms with bioremediation capabilities and with fermentative metabolisms were found in HydMix microcosms. Microbacter, Cellulomonas and Clostridium OTUs were found together in a similar way to the other assays (González et al. Citation2019; Sánchez-Andrea et al. Citation2013), which could be involved in additional acetate production. Sequences identified as an uncultured Desulfitobacterium spp., were also found in low abundance. Church et al. (Citation2007) reported this bacterium together to Desulfosporosinus in an experiment with anaerobic sediments at a low pH. When checking closely the sequence of these OTUs, it was observed that they do not really belong to Desulfitobacterium genus, but represent a new cluster, Desulfobacillus gen. nov, isolated before (Sánchez-Andrea et al. Citation2013).
Despite the low pH conditions, the community profiles in LacMix microcosms were similar to HydMix microcosms. Sequences related to Desulfosporosinus-like bacteria represented 23.8% of the total sequences count. Although we could not measure sulfate depletion and acetate production in these microcosms, we assume that sulfate reduction coupled to the oxidation of lactate could have started on day 9, when hydrogen sulfide was detected. This delayed in sulfate reduction was perhaps overcome by the alkalinization generated by the Fe(III) reduction metabolism of Cellulomonas-like bacteria (Sani et al. Citation2002), whose OTUs were detected. Naturally, the late development of Desulfosporosinus in these microcosms might be related to the lower pH (3.5) onset of the culture and its relation with organic acids behavior. Sánchez-Andrea et al. (Citation2013) showed that at low pH, little amounts of lactate are mandatory to stimulate growth on SRB (1 mM of lactate at pH 4), otherwise an inhibitory effect was observed. Worth noting that in additional assays at the same initial pH we found that hydrogen sulfide was produced in less time when a new enrichment started (data not shown), indicating the ability of this bacterial genus to adapt to changing acidic conditions. Additionally, the presence of non-sulfate reducers (mainly fermentative bacteria) was detected in LacMix microcosms. For example, sequences identified as Acidocella sp., an acidophilic heterotrophic bacterium capable to oxidize acetate to H2 and CO2 was found. This genus of bacteria is normally found together with sulfate reducers (A. aromatica and D. acididurans (Kimura et al. Citation2006)) in a syntrophic relationship since Acidocella can oxidize the acetate produced by Desulfosporosinus to hydrogen, and Desulfosporosinus can use the latest as electron donor (Kimura et al. Citation2006).
Acetate was overproduced in all microcosms of the transfer (up 8.1 mM in GlycPM2, 10.1 mM in AceMix, and 5.7 mM in HydMix microcosms). In GlycPM2 microcosms, the early acetate production (day 4) uncoupled to sulfate reduction is well explained by the fermentation of glycerol as observed in other studies in which heterotrophic sulfate metabolism has been evaluated (Dar et al. Citation2008; González et al. Citation2019). OTUs belonging to the non-sulfate reducer Clostridium genus were found as the most abundant in these microcosms and probably performing the incomplete oxidation of glycerol onset of the culture. Clostridium is a very heterogeneous group concerning their metabolic capabilities (Schnürer et al. Citation1996). Generally, Clostridium spp. perform fermentation of a plethora of utilizable substrates like carbohydrates and amino acids, producing acetate as the main product (Madigan et al. Citation2015). Once sulfate consumption started in these microcosms more acetate was produced, indicating that sulfate reduction with this electron donor was incomplete (EquationEquation 6(6)
(6) ), as other studies show (Kimura et al. Citation2006; Sánchez-Andrea et al. Citation2013). Worth noting, sulfate reduction starting at higher acetate concentrations is not commonly found, taking into account that a concentration of 54 mg L−1 (0.9 mM) of this organic acid is quite toxic to SRB (decreases of 50% of the bacterial growth rate) in the pH range 5.8–7 (Reis et al. Citation1990).
(6)
(6)
In AceMix microcosms, the unexpected production of acetate at day 8 might be related to the detection of Clostridium-affiliated sequences in these cultures. Clostridium might be related to the production of more acetate, possibly via chemolithotrophic acetogenesis as defined for C. ultunense (Karakashev et al. Citation2006; Schnürer et al. Citation1996), but also by heterotrophic acetogenesis, likely to the presence of organic matter in sediment samples. Dar et al. (Citation2008) reported this acetogenic metabolism joined to heterotrophic sulfate reduction in a syntrophic way, although in reactors that had been fed with anaerobic sediments in sulfate-limiting conditions (electron donor/sulfate = 1.94). In addition, some Clostridium members such as C. sulfidigenes can produce hydrogen sulfide when L-cysteine is used as a carbon and energy source (Sallam and Steinbüchel Citation2009); since L-cysteine was used in this study as reducing agent in this step, this might explain the slight increase of this reduced form of sulfur once sulfate reduction stopped.
In HydMix microcosms, the accumulation of acetate is a common feature in hydrogen-fed microcosms and not only attributed to the presence of acetogenic bacteria (like Microbacter OTUs found in these microcosms), which have been shown coexisting with SRB when H2/CO2 are the sole substrates (Weijma et al. Citation2002), but also by SRB themselves, which produce acetate when growing chemolithotrophically (Sánchez-Andrea et al. Citation2020).
Conclusion
The geochemical features of the mine tunnels from Hualgayoc have allowed the establishment of unique prokaryotic communities at each sampling site. Special characteristics such as the low depth of water runoff confer a very close connection between water and sediment samples. Here, carbon and nitrogen fixers appear as key representatives driving primary production supporting thus the development of heterotrophic populations. Metabolisms related to the oxidation-reduction of iron were found in all sampling sites indicating the active cycling of this element in all the AMD sites. On the other hand, sulfur oxidation was found to be an important bacterial metabolism in all samples, possibly contributing to additional steps in the sulfur cycle. Conversely, typical sulfate reducers were not detected in sediments by culture-independent methods used in this study. Well-known AMD microorganisms were found potentially driving these biogeochemical cycles, but some other uncommon members were also found with not yet defined roles in the microbial community thriving in these AMD systems.
A critical step of the sulfur cycle, the dissimilatory sulfate reduction, has been demonstrated by microcosms assays at low pH, with AMD sediments from Hualgayoc mine district (Cajamarca, Peru) as inoculum and with different organic substrates and hydrogen as electron donors. Indigenous aSRB from AMD sites showed high metabolic activity due to their ability to use ionic and nonionic electron donors, hydrogen being the most preferable one. However, with some electron donors, sulfate reduction metabolism seemed to be promoted by the previous colonization of other microbial metabolisms, such as Fe reduction, which could alkalinize the medium. Microbial community analysis revealed that electron donors induced a convergence of composition. Even if their apparent microbial community differed significantly, their rare microbial populations overcome in the right settings. Although typical SRBs inhabited all the analyzed microcosms (Desulfosporosinus and Desulfovibrio), the non-detection of these bacteria in sediments suggests a low cell density of these prokaryotes in the environmental samples.
In the future, microorganisms found in this study such as aSRB and other sulfur/iron cycling bacteria can be used as inocula for biotechnological applications, especially in bioremediation of AMD detrimental effects in the surrounding ecosystems of many active or abandoned mining areas around the world.
Supplementary_information.docx
Download MS Word (1.1 MB)Acknowledgments
The authors want to thank Nohemi Campos Quevedo for their help with the basis of microcosms experiments and DNA extraction process, Prokopis Konstanti for their support with the DNA library preparation and Conall Holohan for the logistic part before sequencing. Buenaventura S.A. mining company is also acknowledged for allowing sample collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alazard D, Joseph M, Battaglia-Brunet F, Cayol JL, Ollivier B. 2010. Desulfosporosinus acidiphilus sp. nov.: a moderately acidophilic sulfate-reducing bacterium isolated from acid mining drainage sediments: new taxa: Firmicutes (Class Clostridia, Order Clostridiales, Family Peptococcaceae). Extremophiles 14(3):305–312.
- Auld RR, Mykytczuk NCS, Leduc LG, Merritt TJS. 2017. Seasonal variation in an acid mine drainage microbial community. Can J Microbiol 63(2):137–152.
- Ayangbenro AS, Olanrewaju OS, Babalola OO. 2018. Sulfate-reducing bacteria as an effective tool for sustainable acid mine bioremediation. Front Microbiol 9:1986.
- Baker BJ, Banfield JF. 2003. Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44(2):139–152.
- Bartsch S, Gensch A, Stephan S, Doetsch A, Gescher J. 2017. Metallibacterium scheffleri: genomic data reveal a versatile metabolism. FEMS Microbiol Ecol 93(3):fix011.
- Battaglia-Brunet F, Achbouni HE, Quemeneur M, Hallberg KB, Kelly DP, Joulian C. 2011. Proposal that the arsenite-oxidizing organisms Thiomonas cuprina and “‘Thiomonas arsenivorans' be reclassified as strains of Thiomonas delicata, and emended description of Thiomonas delicata”. Int J Syst Evol Microbiol 61(Pt 12):2816–2821.
- Bhowal S, Chakraborty R. 2011. Five novel acid-tolerant oligotrophic thiosulfate-metabolizing chemolithotrophic acid mine drainage strains affiliated with the genus Burkholderia of Betaproteobacteria and identification of two novel soxB gene homologues. Res Microbiol 162(4):436–445.
- Bomberg M, Arnold M, Kinnunen P. 2015. Characterization of the bacterial and sulphate reducing community in the alkaline and constantly cold water of the closed Kotalahti mine. Minerals 5(3):452–472.
- Bomberg M, Mäkinen J, Salo M, Kinnunen P. 2019. High diversity in iron cycling microbial communities in acidic, iron-rich water of the Pyhäsalmi mine, Finland. Geofluids 2019:1–17.
- Brantner JS, Haake ZJ, Burwick JE, Menge CM, Hotchkiss ST, Senko JM. 2014. Depth-dependent geochemical and microbiological gradients in Fe(III) deposits resulting from coal mine-derived acid mine drainage. Front Microbiol 5:215.
- Bryan CG, Marchal M, Battaglia-Brunet F, Kugler V, Lemaitre-Guillier C, Lièvremont D, Bertin PN, Arsène-Ploetze F. 2009. Carbon and arsenic metabolism in Thiomonas strains: differences revealed diverse adaptation processes. BMC Microbiol 9:127.
- Canchaya S. 1990. In Stratabound Ore Deposits in the Andes. New York (NY): Springer-Verlag Berlin Heidelberg. p569–582.
- Chen L, Huang L, Méndez-García C, Kuang J, Hua Z, Liu J, Shu W. 2016. Microbial communities, processes and functions in acid mine drainage ecosystems. Curr Opin Biotechnol 38:150–158.
- Church CD, Wilkin RT, Alpers CN, Rye RO, Blaine RB. 2007. Microbial sulfate reduction and metal attenuation in pH 4 acid mine water. Geochem Trans 8:10.
- Cline JD. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14(3):454–458.
- Dar SA, Kleerebezem R, Stams AJM, Kuenen JG, Muyzer G. 2008. Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio. Appl Microbiol Biotechnol 78(6):1045–1055.
- Diez-Ercilla M, Sánchez-España J, Yusta I, Wendt-Potthoff K, Koschorreck M. 2014. Formation of biogenic sulphides in the water column of an acidic pit lake: biogeochemical controls and effects on trace metal dynamics. Biogeochemistry 121(3):519–536.
- Dold B. 2014. Evolution of acid mine drainage formation in sulphidic mine tailings. Minerals 4(3):621–641.
- Dopson M, Holmes DS. 2014. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl Microbiol Biotechnol 98(19):8133–8144.
- [DIGESA] Dirección General de Salud ambiental. 2012. Accessed September 10, 2021. Available at http://www.digesa.minsa.gob.pe/DEPA/vigilancia_recursos_hidricos.asp
- Eisen S, Poehlein A, Johnson DB, Daniel R, Schlömann M, Mühling M. 2016. Genome sequence of the acidophilic iron oxidizer Ferrimicrobium acidiphilum strain T23T. Genome Announc 3(2):383–398.
- Falagán C, Johnson DB. 2014. Acidibacter ferrireducens gen. nov., sp. nov.: an acidophilic ferric iron-reducing gammaproteobacterium. Extremophiles 18(6):1067–1073.
- Falagán C, Sánchez-España J, Johnson DB. 2014. New insights into the biogeochemistry of extremely acidic environments revealed by a combined cultivation-based and culture-independent study of two stratified pit lakes. FEMS Microbiol Ecol 87(1):231–243.
- Fang Y, Xu M, Chen X, Sun G, Guo J, Wu W, Liu X. 2015. Modified pretreatment method for total microbial DNA extraction from contaminated river sediment. Front Environ Sci Eng. 9(3):444–452.
- Ferreira ML, Ramirez SA, Vullo DL. 2018. Chemical characterization and ligand behaviour of Pseudomonas veronii 2E siderophores. World J Microbiol Biotechnol 34(9):1–12.
- Fichtel K, Mathes F, Könneke M, Cypionka H, Engelen B, Teske A, Loy A. 2012. Isolation of sulfate-reducing bacteria from sediments above the deep-subseafloor aquifer. Front Microbiol 3:65.
- García-Moyano A, Austnes A, Lanzén A, González-Toril E, Aguilera Á, Øvreås L. 2015. Novel and unexpected microbial diversity in acid mine drainage in Svalbard (78° N), revealed by culture-independent approaches. Microorganisms 3(4):667–694.
- Gavrilov SN, Korzhenkov AA, Kublanov IV, Bargiela R, Zamana LV, Popova AA, Toshchakov SV, Golyshin PN, Golyshina OV. 2019. Microbial communities of polymetallic deposits’ acidic ecosystems of continental climatic zone with high temperature contrasts. Front Microbiol 10:1573.
- Giovannoni SJ, Schabtach E, Castenholz RW. 1987. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch Microbiol 147(3):276–284.
- González D, Liu Y, Villa Gomez D, Southam G, Hedrich S, Galleguillos P, Colipai C, Nancucheo I. 2019. Performance of a sulfidogenic bioreactor inoculated with indigenous acidic communities for treating an extremely acidic mine water. Min Eng 131:370–375.
- González-Toril E, Aguilera Á, Souza-Egipsy V, López Pamo E, Sánchez España J, Amils R. 2011. Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain). Appl Environ Microbiol 77(8):2685–2694.
- Grettenberger CL, Pearce AR, Bibby KJ, Jones DS, Burgos WD, Macalady JL. 2017. Efficient low-pH iron removal by a microbial iron oxide mound ecosystem at scalp level run. Appl Environ Microbiol 83(7):e00015–17.
- Hallbeck L, Pedersen K. 2014. The family Gallionellaceae. In: Rosenberg, E, DeLong, EF, Lory, S, Stackebrandt, E, Thompson, F, editors. The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. 4th ed. London: Springer-Verlag Berlin Heidelberg, p853–858.
- Hausmann B, Pelikan C, Herbold CW, Köstlbacher S, Albertsen M, Eichorst SA, Glavina del Rio T, Huemer M, Nielsen PH, Rattei T, et al. 2018. Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J 12(7):1729–1742.
- Hausmann B, Pelikan C, Rattei T, Loy A, Pester M, Mark Bailey EJ. 2019. Long-term transcriptional activity at zero growth of a cosmopolitan rare biosphere member. mBio 10(1):e02189–18.
- Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, et al. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol 22(5):554–559.
- Iakovleva E, Mäkilä E, Salonen J, Sitarz M, Wang S, Sillanpää M. 2015. Acid mine drainage (AMD) treatment: Neutralization and toxic elements removal with unmodified and modified limestone. Ecol Eng 81:30–40.
- Islam FS, Boothman C, Gault AG, Polya DA, Lloyd JR. 2005. Potential role of the Fe(III)-reducing bacteria Geobacter and Geothrix in controlling arsenic solubility in Bengal delta sediments. Mineral Mag 69(5):865–875.
- Jew AD, Behrens SF, Rytuba JJ, Kappler A, Spormann AM, Brown GE. Jr. 2014. Microbially enhanced dissolution of HgS in an acid mine drainage system in the California coast range. Geobiology 12(1):20–33.
- Johnson DB, Hallberg KB. 2003. The microbiology of acidic mine waters. Res Microbiol 154(7):466–473.
- Kaksonen AH, Franzmann PD, Puhakka JA. 2004. Effects of hydraulic retention time and sulfide toxicity on ethanol and acetate oxidation in sulfate-reducing metal-precipitating fluidized-bed reactor. Biotechnol Bioeng 86(3):332–343.
- Kaksonen AH, Puhakka JA. 2007. Sulfate reduction based bioprocesses for the treatment of acid mine drainage and the recovery of metals. Eng Life Sci 7(6):541–564.
- Karakashev D, Batstone DJ, Trably E, Angelidaki I. 2006. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl Environ Microbiol 72(7):5138–5141.
- Karimian N, Johnston SG, Burton ED. 2018. Iron and sulfur cycling in acid sulfate soil wetlands under dynamic redox conditions: a review. Chemosphere 197:803–816.
- Karnachuk OV, Kurganskaya IA, Avakyan MR, Frank YA, Ikkert OP, Filenko RA, Danilova EV, Pimenov NV. 2015. An acidophilic Desulfosporosinus isolated from the oxidized mining wastes in the Transbaikal area. Microbiology 84(5):677–686.
- Kelly DP, Uchino Y, Huber H, Amils R, Wood AP. 2007. Reassessment of the phylogenetic relationships of Thiomonas cuprina. Int J Sys Evol Microbiol. 57(11):2720–2724.
- Kim BC, Jeon BS, Kim S, Kim H, Um Y, Sang BI. 2015. Caproiciproducens galactitolivorans gen. nov., sp. nov., a bacterium capable of producing caproic acid from galactitol, isolated from a wastewater treatment plant. Int J Syst Evol Microbiol 65(12):4902–4908.
- Kimura S, Hallberg KB, Johnson DB. 2006. Sulfidogenesis in low pH (3.8–4.2) media by a mixed population of acidophilic bacteria. Biodegradation 17(2):159–167.
- Koschorreck M. 2008. Microbial sulphate reduction at a low pH. FEMS Microbiol Ecol 64(3):329–342.
- Lee KC, Dunfield P F, Stott MB. 2014. The Phylum Armatimonadetes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: Other Major Lineages of Bacteria and The Archaea. 4th ed. London: Springer-Verlag Berlin Heidelberg; p. 447–458.
- Lu S, Gischkat S, Reiche M, Akob DM, Hallberg KB, Küsel K. 2010. Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl Environ Microbiol 76(24):8174–8183.
- Lucheta AR, Otero XL, Macías F, Lambais MR. 2013. Bacterial and archaeal communities in the acid pit lake sediments of a chalcopyrite mine. Extremophiles 17(6):941–951.
- Madigan MT, Martinko JM, Bender KS, Buckley DH, Stahl D. 2015. Brock biology of microorganisms. 14th ed. USA: Pearson Education, Inc.; p493–497.
- Mahmood Q, Zheng P, Hu B, Jilani G, Azim MR, Wu D, Liu D. 2009. Isolation and characterization of Pseudomonas stutzeri QZ1 from an anoxic sulfide-oxidizing bioreactor. Anaerobe 15(4):108–115.
- Mardanov AV, Panova IA, Beletsky AV, Avakyan MR, Kadnikov VV, Antsiferov DV, Banks D, Frank YA, Pimenov NV, Ravin NV, et al. 2016. Genomic insights into a new acidophilic, copper-resistant Desulfosporosinus isolate from the oxidized tailings area of an abandoned gold mine. FEMS Microbiol Ecol 92(8):fiw111.
- Meier J, Piva A, Fortin D. 2012. Enrichment of sulfate-reducing bacteria and resulting mineral formation in media mimicking pore water metal ion concentrations and pH conditions of acidic pit t lakes. FEMS Microbiol Ecol 79(1):69–84.
- Méndez-García C, Peláez AI, Mesa V, Sánchez J, Golyshina OV, Ferrer M. 2015. Microbial diversity and metabolic networks in acid mine drainage habitats. Front Microbiol 6:475.
- Mesa V, Gallego JLR, González-Gil R, Lauga B, Sánchez J, Méndez-García C, Peláez AI. 2017. Bacterial, archaeal, and eukaryotic diversity across distinct microhabitats in an acid mine drainage. Front Microbiol 8:1756.
- Muyzer G, Stams AJM. 2008. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6(6):441–454.
- Neal AL, Techkarnjanaruk S, Dohnalkova A, McCready D, Peyton BM, Geesey GG. 2001. Iron sulfides and sulfur species produced at hematite surfaces in the presence of sulfate-reducing bacteria. Geochim Cosmochim Acta 65(2):223–235.
- Nordstrom DK, Alpers CN. 1999. Geochemistry of acid mine waters. In: Plumlee, GS, Logsdon, MJ, editors. The Environmental Geochemistry of Mineral Deposits, Part A: Processes, Techniques, and Health Issues. Littleton: Society of Economic Geologists; p133–160.
- Portielje R, Lijklema L. 1995. The effect of reaeration and benthic algae on the oxygen balance of an artificial ditch. Ecol Model 79(1–3):35–48.
- Prieto-Barajas CM, Valencia-Cantero E, Santoyo G. 2018. Microbial mat ecosystems: structure types, functional diversity, and biotechnological application. Electron J Biotechnol 31:48–56.
- Puente-Sánchez F, Arce-Rodríguez A, Oggerin M, García-Villadangos M, Moreno-Paz M, Blanco Y, Rodríguez N, Bird L, Lincoln SA, Tornos F, et al. 2018. Viable cyanobacteria in the deep continental subsurface. Proc Natl Acad Sci USA. 115(42):10702–10707.
- Rabus R, Hansen TA, Widdel F. 2013. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In: Rosenberg, E, DeLong, EF, Lory, S, Stackebrandt, E, Thompson, F, editors. The Prokaryotes: Prokaryotic Physiology and Biochemistry. 4th ed. London: Springer-Verlag Berlin Heidelberg; p309–404.
- Ramiro-Garcia J, Hermes GDA, Giatsis C, Sipkema D, Zoetendal EG, Schaap PJ, Smidt H. 2016. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Res 5:1791.
- Reis MAM, Lemos PC, Almeida JS, Carrondo MJT. 1990. Influence of produced acetic acid on growth of sulfate reducing bacteria. Biotechnol Lett 12(2):145–148.
- Rowe OF, Sánchez-España J, Hallberg KB, Johnson DB. 2007. Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ Microbiol 9(7):1761–1771.
- Sajjad W, Zheng G, Zhang G, Ma X, Xu W, Ali B, Rafiq M. 2018. Diversity of prokaryotic communities indigenous to acid mine drainage and related rocks from Baiyin open-pit copper mine stope, china. Geomicrobiol J 35(7):580–600.
- Sallam A, Steinbüchel A. 2009. Clostridium sulfidigenes sp. nov., a mesophilic, proteolytic, thiosulfate- and sulfur-reducing bacterium isolated from pond sediment. Int J Syst Evol Microbiol 59(Pt 7):1661–1665.
- Sánchez España J, López Pamo E, Santofimia E, Aduvire O, Reyes J, Barettino D. 2005. Acid mine drainage in the Iberian pyrite belt (Odiel river watershed, Huelva, SW Spain): geochemistry, mineralogy and environmental implications. J Appl Geochem 20(7):1320–1356.
- Sánchez-Andrea I, Guedes IA, Hornung B, Boeren S, Lawson CE, Sousa DZ, Bar-Even A, Claassens NJ, Stams AJM. 2020. The reductive glycine pathway allows autotrophic growth of Desulfovibrio desulfuricans. Nat Commun 11(1):1–12.
- Sánchez-Andrea I, Knittel K, Amann R, Amils R, Luis Sanz J. 2012. Quantification of Tinto River sediment microbial communities: importance of sulfate-reducing bacteria and their role in attenuating acid mine drainage. Appl Environ Microbiol 78(13):4638–4645.
- Sánchez-Andrea I, Rodríguez N, Amils R, Sanz JL. 2011. Microbial diversity in anaerobic sediments at Rio Tinto, a naturally acidic environment with a high heavy metal content. Appl Environ Microbiol 77(17):6085–6093.
- Sánchez-Andrea I, Sanz JL, Bijmans MF, Stams AJ. 2014a. Sulfate reduction at low pH to remediate acid mine drainage. J Hazard Mater 269:98–109.
- Sánchez-Andrea I, Sanz JL, Stams AM. 2014b. Microbacter margulisiae gen. nov., sp. nov., a propionigenic bacterium isolated from sediments of an acid rock drainage pond. Int J Syst Evol Microbiol 64(Pt 12):3936–3942.
- Sánchez-Andrea I, Stams AJM, Hedrich S, Ňancucheo I, Johnson DB. 2015. Desulfosporosinus acididurans sp. nov.: an acidophilic sulfate-reducing bacterium isolated from acidic sediments. Extremophiles 19(1):39–47.
- Sánchez-Andrea I, Stams AJM, Amils R, Sanz JL. 2013. Enrichment and isolation of acidophilic sulfate-reducing bacteria from Tinto River sediments. Environ Microbiol Rep 5(5):672–678.
- Sánchez-España J, Yusta I, Ilin A, van der Graaf C, Sánchez-Andrea I. 2020. Microbial geochemistry of the acidic saline pit lake of Brunita mine (La Unión, SE Spain). Mine Water Environ 39(3):535–555.
- Sani RK, Peyton BM, Smith WA, Apel WA, Petersen JN. 2002. Dissimilatory reduction of Cr(VI), Fe(III), and U(VI) by Cellulomonas isolates. Appl Microbiol Biotechnol 60(1–2):192–199.
- Santos AL, Johnson DB. 2017. The effects of temperature and pH on the kinetics of an acidophilic sulfidogenic bioreactor and indigenous microbial communities. Hydrometallurgy 168:116–120.
- Sato Y, Hamai T, Hori T, Aoyagi T, Inaba T, Kobayashi M, Habe H, Sakata T. 2019. Desulfosporosinus spp. were the most predominant sulfate-reducing bacteria in pilot- and laboratory-scale passive bioreactors for acid mine drainage treatment. Appl Microbiol Biotechnol 103(18):7783–7793.
- Schippers A, Breuker A, Blazejak A, Bosecker K, Kock D, Wright TL. 2010. The biogeochemistry and microbiology of sulfidic mine waste and bioleaching dumps and heaps, and novel Fe(II)-oxidizing bacteria. Hydrometallurgy 104(3–4):342–350.
- Schnürer A, Schink B, Svensson BH. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int J Syst Bacteriol 46(4):1145–1152.
- Stams AJ, Van Dijk JB, Dijkema C, Plugge CM. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol 59(4):1114–1119.
- Suzuki Y, Kelly SD, Kemner KM, Banfield JF. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl Environ Microbiol 69(3):1337–1346.
- Teitzel GM, Parsek MR. 2003. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 69(4):2313–2320.
- Tuffnell S. 2017. Acid drainage: the global environmental crisis you’ve never heard of. [UK]: The Conversation. Accessed January 30, 2020. Available at https://www.history.ox.ac.uk/article/acid-drainage-global-environmental-crisis-youve-never-heard.
- van der Graaf CM, Sánchez-España J, Yusta I, Ilin A, Shetty SA, Bale NJ, Villanueva L, Stams AJM, Sánchez-Andrea I. 2020. Biosulfidogenesis mediates natural attenuation in acidic mine pit lakes. Microorganisms 8(9):1275.
- Ward N, Staley JT, Fuerst JA, Giovannoni S, Schlesner H, Stackebrandt E. 2006. The order Planctomycetales, including the genera Planctomyces, Pirellula, Gemmata and Isosphaera and the candidatus genera Brocadia, Kuenenia and Scalindua. Prokaryotes 7:757–793.
- Weijma J, Gubbels F, Hulshoff Pol LW, Stams AJ, Lens P, Lettinga G. 2002. Competition for H2 between sulfate reducers, methanogens and homoacetogens in a gas-lift reactor. Water Sci Technol. 45(10):75–80.
- Weiss JV, Rentz JA, Plaia T, Neubauer SC, Merrill-Floyd M, Lilburn T, Bradburne C, Megonigal JP, Emerson D. 2007. Characterization of Neutrophilic Fe(II)-Oxidizing Bacteria Isolated from the Rhizosphere of Wetland Plants and Description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol J 24(7–8):559–570.
- Widdel F, Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows, A, Trüper, HG, Dworkin, M, Harder, W, Schleifer, KH, editors. The Prokaryotes. 2nd ed. New York: Springer Science Bussiness Media; p3352–3378.
- Xia D, Yi X, Lu Y, Huang W, Xie Y, Ye H, Dang Z, Tao X, Li L, Lu G. 2019. Dissimilatory iron and sulfate reduction by native microbial communities using lactate and citrate as carbon sources and electron donors. Ecotoxicol Environ Saf 174:524–531.
- Yu H, Jiang Z, Lu Y, Yao X, Han C, Ouyang Y, Wang H, Guo C, Ling F, Dang Z. 2020. Transcriptome analysis of the acid stress response of Desulfovibrio vulgaris ATCC 7757. Curr Microbiol 77(10):2702–2711.
- Ziegler S, Waidner B, Itoh T, Schumann P, Spring S, Gescher J. 2013. Metallibacterium scheffleri gen. nov., sp. nov., an alkalinizing gammaproteobacterium isolated from an acidic biofilm. Int J Syst Evol Microbiol 63(Pt_4):1499–1504.
