ABSTRACT
Background: Rotavirus C (RVC), a known etiological agent of diarrheal outbreaks, mainly inflicts swine population globally with sporadic incidence in human, cattle, ferret, mink and dog.
Objective: To demonstrate the presence of RVC in Indian swine population and characterization of its selected structural (VP6) and non-structural (NSP4 and NSP5) genes.
Methods: A total of 108 diarrheic samples from different regions of India were used. Isolated RNA was loaded onto polyacrylamide gel to screen for the presence of RVs through the identification of specific electrophoretic genomic migration pattern. To characterize the RVC strains, VP6 gene and NSP4 and NSP5 genes were amplified, sequenced and analyzed.
Results: Based on VP6 gene specific diagnostic RT-PCR, the presence of RVC was confirmed in 12.0% (13/108) piglet fecal specimens. The nucleotide sequence analysis of VP6 gene, encoding inner capsid protein, from selected porcine RVC (PoRVC) strains revealed more than 93% homologies to human RVC strains (HuRVC) of Eurasian origin. These strains were distant from hitherto reported PoRVCs and clustered with HuRVCs, owning I2 genotype. However, the two non-structural genes, i.e. NSP4 and NSP5, of these strains were found to be of swine type, signifying a re-assortment event that has occurred in the Indian swine population.
Conclusion: The findings indicate the presence of human-like RVC in Indian pigs and division of RVC clade with I2 genotype into further sub-clades. To the best of our knowledge, this appears to be the first report of RVC in Indian swine population. Incidence of human-like RVC VP6 gene in swine supports its subsequent zoonotic prospective.
1. Introduction
Rotaviruses (RVs) are important pathogens of the family Reoviridae that cause acute gastroenteritis in neonates of all warm-blooded animals, birds and man (Estes and Kapikian Citation2007). Genome of RVs consists of 11 double-stranded RNA segments which encode for 6 structural (VP1-4, VP6-8) and 5/6 non-structural proteins (NSP1-NSP5/6). Based on the inner capsid protein, i.e. VP6 (group-specific protein), eight groups of RVs (A–H) are reported from different host species till to date. Recently, group I (RVI) is detected in dogs and cats (Mihalov-Kovács et al. Citation2015; Phan et al. Citation2017), and proposed as a new group for consideration by the Rotavirus Classification Working Group (RCWG). Among these, RVA remains the most important due to its wide host range and higher prevalence. In the animal kingdom, RVA, rotavirus B (RVB) and RVC infect domesticated animals, while strains from Rotavirus D, F and G are solely reported from birds (Otto et al. Citation2012; Kattoor et al. Citation2013). Among domesticated animals, the swine population witnesses infection by RVs such as RVA, RVB, RVC, RVE and RVH (Estes and Kapikian Citation2007; Wakuda et al. Citation2011). Hitherto reports mention co-infection of different RV genotypes in pigs (Nyaga et al. Citation2015). Hence, a chance for re-assortment in swine cannot be overlooked until the extensive screening of pig population for the presence of re-assorted or new RV types across the globe (Steyer et al. Citation2007; Médici et al. Citation2011; Sharma et al. Citation2013; Navarro et al. Citation2017).
After its discovery in swine population (Saif et al. Citation1980), RVC has been detected in human, ferret, mink, cattle and dog from many countries (Torres-Medina Citation1987; Tsunemitsu et al. Citation1991; Saif and Jiang Citation1994; Otto et al. Citation1999; Mawatari et al. Citation2004; Wise et al. Citation2009; Marton et al. Citation2015). It is now a well-established etiological agent of diarrheal outbreaks for neonatal and adult gastroenteritis (Araújo et al. Citation2011; Kumazaki and Usuku Citation2014). It is reported in both symptomatic as well as asymptomatic pigs of all age groups (Morin et al. Citation1990; Kim et al. Citation1999; Collins et al. Citation2008; Marthaler et al. Citation2013). Published reports based on the sequence analysis of VP7 and VP4 genes from RVC reveal the circulation of a genetically heterogeneous group of isolates worldwide (Martella et al. Citation2007; Jeong et al. Citation2015; Moutelíková et al. Citation2015). Based on the VP6 sequence analysis, a total of 11 I genotypes are confirmed in RVC (Suzuki et al. Citation2014; Moutelíková et al. Citation2015), whereas based on VP4 gene, P1–P8 types, and on VP7 gene, G1–G11 types are identified (Jeong et al. Citation2015; Moutelíková et al. Citation2015) and approved by the RCWG. Recently, two new genotypes (G12 and G13) are also reported from Japan (Niira et al. Citation2016). An increasing number of sequence-based detection of RVC isolates from different parts of the world reveal evolution of several new genotypes gradually, which further needs approval from the RCWG.
Hitherto epidemiological studies targeting RVC depict a wide prevalence ranging from 1% to 46% in humans and animals around the world (Tsunemitsu et al. Citation1996; Sanchez-Fauquier et al. Citation2003; Bányai et al. Citation2009; Marthaler et al. Citation2013). The virus is detected with a very high prevalence (up to 78%) in neonatal piglets from the USA (Marthaler et al. Citation2013). Relevance of RVC is being augmented because piggery constitutes the main livelihood framework of rural and tribal population in many developing countries along with the possibility of zoonotic potential as indicated in earlier studies (Iturriza-Gómara et al. Citation2004; Gabbay et al. Citation2008). Detection of porcine-type RV in bovine sample (Chang et al. Citation1999) and bovine-type isolate in porcine sample (Jeong et al. Citation2009) also support the possibility of species jumping or re-assortment events. In India, only human RVCs are identified till to date (Brown et al. Citation1988) and this group of RV could not mark its presence in other species before. Here, we report the presence of RVC in Indian swine population for the first time. The findings reveal that identified RVC isolates carry human-like VP6 gene, while NSP4 and NSP5 genes of swine origin.
2. Materials and methods
2.1. Collection of samples and RNA-PAGE analysis
A total of 108 diarrheic samples were collected from northeastern, northern and southern regions of India, where a substantial number of small-scale piggeries are present. Diarrheic fecal samples were made into 10% (w/v) suspension with phosphate-buffered saline (pH 7.2). Total RNA from suspended fecal samples was isolated using Qiazol reagent (Qiagen GmbH, Hilden, Germany) as per the manufacturer's protocol and quantified on Nanodrop spectrometer. Isolated RNA (1 μg) was loaded on to polyacrylamide gel to screen the presence of RVs through the identification of specific electrophoretic genomic migration pattern.
2.2. Random priming diagnostic RT-PCR
Extracted RNA (500 ng) was used to prepare a pool of first-strand cDNA by random-priming reverse transcription (RT) using recombinant MMLV-RT (Promega Corporation, Madison, WI, USA) and random hexamer (Qiagen GmbH, Hilden, Germany) at 37 °C. To detect the presence of RVC in diarrheic piglet fecal specimens, diagnostic RT-PCR based on VP6 gene was performed using RVC-VP6-DF; 5′-ARTCHGTTCTATGYGATTC-3′ (custom synthesized) and BMJ44; 5′-AGCCACATAGTTCACATTTC-3′ (Sanchez-Fauquier et al. Citation2003). The diagnostic primer was expected to amplify a 335 bp amplicon. PCR conditions were standardized to amplify the product at initial single cycle of 95 °C for 5 minutes followed by 35 cycles of 94 °C for 20 seconds, 48 °C for 20 seconds, 72 °C for 20 seconds and final extension cycle of 72 °C for 5 minutes using SapphireAmp Fast PCR master mix (Takara Bio Inc, Shiga, Japan).
2.3. Amplification of genes, cloning and sequencing
To characterize the RVC strains, structural (VP6 gene) and non-structural genes (NSP4 and NSP5 genes) were amplified. Porcine RVC VP6 gene specific primer pair (amplicon size: 1353 bp) reported by Cooke et al. (Citation1992) and custom-synthesized pair, viz. RVC VP6 PF 5′-TCATACTGGGGCATTGGAAC-3′ and RVC VP6 PR 5′-GCCAAGTGTTTGATTATTAGG-3′ (amplicon size: 584 bp) were used for the amplification of VP6 gene in samples that were confirmed positive in diagnostic RT-PCR. To amplify the coding region of NSP4 gene, custom-synthesized primer pair, viz. RVC-NSP4-CDS-FP 5′-CTCTACGAAGCAATGGAGTTCATCA A-3′ and RVC-NSP4-CDS-RP 5′-AGCGCAGAAGATTCATAGACA-3′ (amplicon size: 477 bp) was used. Likewise, full-length NSP5 was amplified using another custom-synthesized primer pair, i.e. RVC-NSP5-Full-FP 5′-TGCGACAATGTCCATTTCG-3′ and RVC-NSP5-Full-RP 5′-GGCAAGTACTCGACAAATTTAC-3′ (amplicon size: 650 bp). Amplified products were cloned into pDRIVE (Qiagen GmbH, Hilden, Germany) cloning vector and transformed. Recombinant plasmids were isolated using GeneJET plasmid Miniprep kit (Thermo Fisher Scientific, Waltham, MA ,Vilnius,, Lithuania) and analyzed through restriction analysis. Positive recombinant clones were sequenced by Bigdye terminator Sanger sequencing method in ABI 3730xl sequencer (Eurofins Genomic India Ltd, Bangalore, India).
2.4. Pairwise distance and phylogenetic analysis
To determine the identity of strains confirmed as RVC during the study and their divergence with other groups, viz. RVA, RVB and RVH, cut-off-based identity analysis was done as described previously (Matthijnssens et al. Citation2012) at nucleotide level using MegAlign tool. The distance among the nucleotide sequences of VP6, NSP4 and NSP5 genes was calculated. For genetic relatedness study, representative full-length genes (VP6: n = 68, NSP4: n = 67, NSP5: n = 60) of human, cattle, pig and dog RVC strains were retrieved from the NCBI database. Phylogenetic reconstruction was performed using maximum likelihood method (1500 bootstrap replicates) in MEGA 6 software (Tamura et al. Citation2013). The suitable dendrogram reconstruction model was identified using Find best DNA/protein model tool available in MEGA 6 which was confirmed with FindModel online tool (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html).
2.5. Sequence submissions in GenBank
The full-length nucleotide sequences of VP6 genes were submitted to GenBank under accession numbers KT932962-63 and partial genes as KX374486, KX374489 and KX374492. The RVC NSP4 genes were submitted to GenBank with accession numbers KY783645, KY783646, KY783647, KY783648 and KY783649. Accession numbers obtained for three NSP5 sequences were KY783653, KY783654 and KY783655.
3. Results
3.1. Detection of RVC
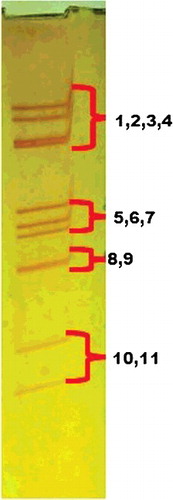
One fecal specimen among 108 samples exhibited electrophoretic pattern of RVC (). However, a total of 13 fecal specimens (12.0%) were found positive for the RVC in VP6 gene based diagnostic RT-PCR.
3.2. Sequence and phylogenetic analysis of structural gene (VP6)
We could amplify full-length VP6 gene (1353 bp) from two RVC-positive fecal specimens and these were named as RVC/Pig-wt/IND/ASM-132/2013/GXP[X]-I2 and RVC/Pig-wt/IND/ASM-140/2013/GXP[X]-I2 using the reported primer pair (Cooke et al. Citation1992). Custom-synthesized primer pair could amplify partial length of VP6 gene (584 bp) from three more positive fecal specimens and were designated as RVC/Pig-wt/IND/UP-404/2016/GXP[X]-I2, RVC/Pig-wt/IND/TRI-542/2015/GXP[X]-I2 and RVC/Pig-wt/IND/KL-224/2015/GXP[X]-I2. These strains are described as ASM-132, ASM-140, UP-404, TRI-542 and KL-224, respectively, in the manuscript.
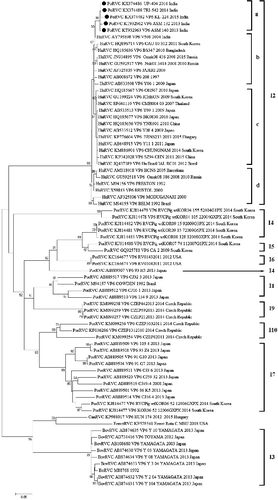
PoRVC strains identified in India exhibited maximum identity of their VP6 gene with cognate gene of different RVC isolates (average value = 81.93%) and very low identity with other RV groups in cut-off-based pairwise identity analysis (). PoRVC strains shared high homologies, i.e. >93.9% with cognate gene of human RVC strain/isolates () from Eurasian continental plate (Russia, India, Bangladesh, Bristol, Barcelona). Homology with PoRVC strains/isolates ranged within 78.3%–81.1%, 79.6%--82.2%, 78.7%--82.2%, 77.7%--82% and 76.6%--80.7% for ASM-132, ASM-140, UP-404, TRI-542 and KL-224 strain, respectively, as determined through the sequence distance in MegAlign software. Upon phylogenetic reconstruction based on VP6 gene of newly identified PoRVC strains along with other representative strains/isolates (n = 68) from the NCBI database, the formation of two clusters could be observed; one major cluster containing strains of human and porcine origin, while another minor cluster containing RVC strains of cattle (I3), dog and ferret origin (). The major cluster was further divided into different clades representing human or human-like PoRVC strains (I2a–d), and porcine RVC strains (I1, I4–10). Indian PoRVC strains constituted independent sub-clade (I2a) within I2 clade of HuRVCs from Russia, India, Bangladesh and South Korea (). Within I2 clade, different putative sub-clades (I2a–d) were also observed representing strains/isolates from Eurasian continental plate, South Asia, East Asian Islands, Brazil and Hungary (). In contrast to Indian PoRVC strains, other reported PoRVC strains/isolates formed different sub-clades with varied I genotype suggesting marked differences in the nucleotide sequences of inner capsid gene. RVC strains from dog and ferret formed independent sub-clade within the minor cluster ().
Table 1. Percentage identity of VP6 gene from PoRVC strains (ASM-132 and ASM-140) identified in the study with cognate gene from different types of rotaviruses.
Figure 2. Phylogenetic analysis of group C rotavirus based on VP6 genes of human, swine, cattle, dog and ferret origin at nucleotide level. Phylogenetic reconstruction was performed using maximum likelihood method (1000 bootstrap replicates) in MEGA 6 software. The Tamura-3+G algorithm was identified using Find best DNA/protein model tool available in MEGA 6 which was confirmed with FindModel online tool (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html). Numbers on branches indicate percentages of bootstrap support from 1000 replicates. VP6 gene based I typing of RVC is denoted along with clusters. Sub-clades within human type RVC are depicted in square brackets. Host species depicted are human (Hu); cattle (Bo); pig (Por); dog (Can) and ferret. Strains/isolates are represented according to their host species, accession number, gene, strain, year of isolation and country of origin. Isolates of the current study are denoted by solid dots.
3.3. Sequence and phylogenetic analysis of non-structural genes (NSP4 and NSP5)
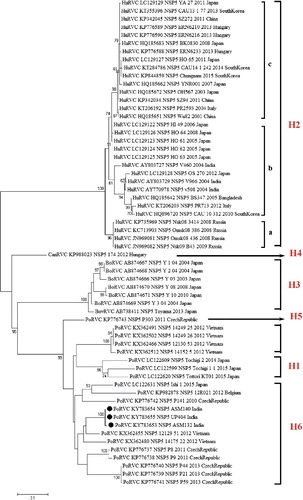
The coding regions of NSP4 genes (amplicon size 477 bp) were amplified from all the five PoRVC strains of this study, whereas full-length NSP5 genes (amplicon size: 650 bp) could be amplified only in three PoRVC strains (ASM-132, ASM-140 and UP-404) using custom-synthesized primer pair. The NSP4 and NSP5 genes exhibited high homologies, i.e. >74% and >81%, respectively, with cognate genes of PoRVCs, whereas the maximum percent identities with cognate genes of HuRVCs at nucleotide level were found only 63.9% (NSP4) and 73.4% (NSP5), respectively (data not shown). Upon phylogenetic reconstruction of NSP4 genes, two major clusters were formed. The first cluster was exclusive for human strains/isolates, whereas the second one incorporated all animal RVCs isolated till to date. The animal RVC cluster was subdivided into two clades encompassing bovine and porcine strains. Porcine clade further showed marked differentiations into sub-clades representing diverse strains from different countries/regions. Indian PoRVC strains exhibited significant divergence from the reference strain (Cowden strain) and clustered into two sub-clades. Strains, viz. KL-224 and TRI-542, exhibited some closeness to those from Japan, while other three, i.e. ASM-132, ASM-140 and UP-404, constituted a separate sub-clade (). Phylogenetic reconstruction of RVC isolates based on NSP5 gene revealed colossal genetic divergence in the same gene of different species. Like the phylogram based on NSP4 gene, formation of two major clusters representing RVC strains of human and animal origin, respectively, was also observed. Owing to their diversity, RVC strains of bovine and porcine origin constituted separate clades. The Indian PoRVC strains clustered along with other PoRVC strains. The only known RVC strain from dog formed independent clade and was found to be placed between the two major clusters having characteristics of human and animal origin ().
Figure 3. Phylogenetic analysis of group C rotavirus based on NSP4 genes of human, swine, cattle, dog and ferret origin at nucleotide level. Phylogenetic reconstruction was performed using maximum likelihood method (1000 bootstrap replicates) in MEGA 6 software. The Tamura-3+G algorithm was identified using Find best DNA/protein model tool available in MEGA 6 which was confirmed with FindModel online tool (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html). Numbers on branches indicate percentages of bootstrap support from 1000 replicates. NSP4 gene based E typing of RVC is denoted along with clusters. Host species depicted are human (Hu); cattle (Bo); pig (Por); dog (Can) and ferret. Strains/isolates are represented according to their host species, accession number, gene, strain, year of isolation and country of origin. Isolates of current study are denoted by solid dots. Established E genotypes are given in black, whereas probable new genotypes based on percent divergence at nucleotide level are marked in red color alongside the isolates.

Figure 4. Phylogenetic analysis of group C rotavirus based on NSP5 genes of human, swine, cattle, dog and ferret origin at nucleotide level. RVC strains of human, bovine and pig origin constituted separate clusters based on nucleotide sequences of NSP5 gene owing to their diverse genotypic configuration. Probable new H genotypes are marked in red color alongside the isolates.
4. Discussion
A number of studies on RVC from all over the world by several researchers prove that this group is as important as RVA and causes severe acute gastroenteritis in human and swine (Rahman et al. Citation2005; Meleg et al. Citation2008; Moon et al. Citation2011; Marthaler et al. Citation2013; Suzuki et al. Citation2014). In a routine electrophoretic screening survey for RVA in swine population, we found an atypical migration pattern of genomic RNA segments from a piglet fecal specimen (), resembling established electrophoretic pattern (4-3-2-2) of RVC (Desselberger Citation2014). Although HuRVC had been detected long back in India (Brown et al. Citation1988), PoRVC is never identified from any part of the country. Non-availability of PoRVC sequences from Indian subcontinent does not support the fact that RVC is not prevalent in Indian pig population, rather implicates the absence of sensitive, specific and reliable diagnostic techniques, which can be opted for an early detection and eventually control of RVC in swine industry. This study establishes the presence of RVC in pigs of India. VP6 gene based diagnostic RT-PCR used in this study reveals the presence of RVC (up to 12%) in diarrheic pig samples. This detection is not region specific as almost an equal number of RVC-positive fecal specimens are detected from different regions of India. Moreover, nucleotide sequences of VP6 gene from Indian PoRVC strains are highly homologous (>97%) to each other irrespective of the size (full length and partial) and collection place. It is expected that the RVC prevalence might further increase, if a national surveillance program is launched for RVC in Indian swine population. A low rate of RVC detection is documented by many researchers (Sanchez-Fauquier et al. Citation2003; Khamrin et al. Citation2008). Nevertheless, low detection rate of the virus might be attributed to the absence of intact virus particle or degraded viral RNA in repeated freeze–thawed fecal samples. The latter can hinder amplification of specific gene products as said in different reports (Esona et al. Citation2008; Moon et al. Citation2011; Matthijnssens and Theuns Citation2015). In this study also, we could amplify only two full-length VP6 genes from 13 RVC-positive fecal specimens with reported primer pair, while with the custom-synthesized primer pair, we could amplify VP6 gene of partial length from three other specimens. It indicates sequence variation in VP6 gene of Indian PoRVCs. Furthermore, difficulty in cultivation of RVC in cell culture makes molecular characterization of fully formed virus a tedious process (Kusanagi et al. Citation1992).
Based on the VP6 gene sequence, till to date, 11 I genotypes have been identified (Moutelíková et al. Citation2015; Zhirakovskaia et al. Citation2016). Of these, HuRVCs exclusively possess I2 genotype, whereas bovine RVCs have I3 genotype and PoRVCs exhibit a number of diverse genotypes with huge sequence diversity (). Our observations are in agreement with recent phylogenetic study involving RVC strains of different origin (Zhirakovskaia et al. Citation2016). Of note, PoRVC strains identified in this study group within HuRVC clade possess VP6 of I2 genotype. It is the first incidence of PoRVC with I2 genotype, which otherwise is exclusive for HuRVCs. Our results indicate the occurrence of one of the two putative evolutionary events, viz. ‘re-assortment’, i.e. transmission of one/a few genes or ‘species-jumping’, i.e. transmission of whole HuRVC to swine population, in the country. Thus, we tried to amplify other structural and non-structural genes to gain better understanding of RVC in Indian pig population. Unfortunately, despite several efforts we could not amplify VP7 and VP4 genes in any of the identified PoRVC strains. It might be attributed to the degradation of genomic RNA within fecal specimens or due to huge genomic diversity in Indian PoRVC strains. Albeit, we could amplify NSP4 genes in all five strains, while NSP5 genes could be amplified in only three strains. Amplified genes were used to make phylogram along with those retrieved from the database. In contrast to the result obtained in phylogenetic analysis based on VP6 gene, the NSP4 and NSP5 genes of Indian PoRVC strains cluster with that of pig origin ( and ). A marked differentiation is visible as RVC strains of human, bovine and pig origin constitute separate clusters based on nucleotide sequences of non-structural genes. From the phylodendrograms, it is vivid that non-structural genes in Indian PoRVC strains are also diverse among them. They can be considered as highly evolving as they constitute separate sub-clades within clade harboring sequences of pig origin. Sequence-based genotyping as done for other genes (VP6, VP4 and VP7) of RVC are not available for NSP5 gene. Our results suggested diverse genotype for NSP5 genes with human and animal origin and need for classification.
RVCs also exhibit genotypic diversity in other genes, as a total of 11 G genotypes (VP7 gene) and 9 P genotypes (VP4 gene) are identified including 7 porcine-specific G genotypes (Marthaler et al. Citation2013). This broad diversity of G, P and I types makes researchers to think upon development of a polyvalent vaccine against RVC in swine. It is noteworthy that no human-like PoRVC is detected as of now (Jeong et al. Citation2015) and also the zoonotic potential of PoRVC is not strongly supported by published literature as documented for PoRVA (Alfieri et al. Citation1996). Detection of human-like VP6 gene in PoRVCs is in agreement with the report of Gabbay et al. (Citation2008). It is possible that animals might serve as reservoir host for re-assortment of RVs from different host origin (Mascarenhas et al. Citation2007; Gabbay et al. Citation2008; Navarro et al. Citation2017). RVC infection in pigs becomes more important as the main livelihood framework of rural and tribal population that dominates the northeastern states of India is piggery. The human–pig ratio in Northeastern India is much higher as compared to national ratio (Njuki et al. Citation2010), which might help in transmission also. From this study, it is obvious that the PoRVCs that are not clustered into I2 type have marked their presence in I2 clade along with human strains which can substantiate the speedy variability in gene sequence of PoRVCs. Moreover, only VP6 gene of Indian PoRVCs is of human type whereas other genes sequenced (NSP4 and NSP5) show their origin from swine. Finding from the genes sequenced shows a re-assortment event occurred in the Indian swine population. Furthermore, this study emphasizes the need for molecular epidemiological studies focusing on the emergence of new RV groups/genotypes in swine population of India among others. The current study supports the ‘interspecies’ transmission of RVC as evidenced earlier. A thorough screening of fecal samples for RVC is needed to substantiate the probability of species jumping.
Acknowledgments
The authors are thankful to Indian Council of Medical Research, India, for junior research fellowship to Kattoor JJ, and Indian Council of Agricultural Research, India, for National Fellowship to Malik YS. The authors are also grateful to the authority of ICAR-Indian Veterinary Research Institute, Bareilly, India, for providing infrastructure facilities.
Disclosure statement
The authors declare that there is no conflict of interest that could possibly arise.
Additional information
Funding
References
- Alfieri AA, Leite JP, Nakagomi O, Kaga P, Woods PA, Glass RI, Gentsch JR. 1996. Characterization of human rotavirus genotype P [8]G5 from Brazil by probe-hybridization and sequence. Arch Virol. 141:2353–2364.
- Araújo IT, Heinemann MB, Fialho AM, Leite JPG. 2011. Detection and molecular characterization of human group C rotavirus in Brazil. Intervirology. 54:261–267.
- Bányai K, Bogdán Á, Domonkos G, Kisfali P, Molnár P, Tóth A, Melegh B, Martella V, Gentsch JR, Szűcs G. 2009. Genetic diversity and zoonotic potential of human rotavirus strains, 2003–2006, Hungary. J Med Virol. 81:362–370.
- Brown D, Mathan M, Mathew M, Martin R, Beards G, Mathan V. 1988. Rotavirus epidemiology in Vellore, South India: group, subgroup, serotype, and electropherotype. J Clin Microbiol. 26:2410–2414.
- Chang KO, Nielsen PR, Ward LA, Saif LJ. 1999. Dual infection of gnotobiotic calves with bovine strains of group A and porcine like group C rotaviruses influences pathogenesis of the group C rotavirus. J Virol. 73:9284–9293.
- Collins P, Martella V, O'Shea H. 2008. Detection and characterization of group C rotaviruses in asymptomatic piglets in Ireland. J Clin Microbiol. 46:2973–2979.
- Cooke SJ, Clarke IN, Freitas RB, Gabbay YB, Lambden PR. 1992. The correct sequence of the porcine group C/Cowden rotavirus major inner capsid protein shows close homology with human isolates from Brazil and the U.K. Virology. 190:531–537.
- Desselberger U. 2014. Rotaviruses. Virus Res. 190:75–96.
- Esona MD, Humphrey CD, Dennehy PH, Jiang B. 2008. Prevalence of group C rotavirus among children in Rhode Island, United States. J Clin Virol. 42:221–224.
- Estes MK, Kapikian AZ. 2007. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 5th ed. Philadelphia (PA): Wolters Kluwer Health/Lippincott Williams & Wilkins; p. 1917–1974.
- Gabbay YB, Borges AA, Oliveira DS, Linhares AC, Mascarenhas JD, Barardi CR, Simões CM, Wang Y, Glass RI, Jiang B. 2008. Evidence for zoonotic transmission of group C rotaviruses among children in Belém, Brazil. J Med Virol. 80:1666–1674.
- Iturriza-Gómara M, Kang G, Gray J. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 31:259–265.
- Jeong YJ, Matthijnssens J, Kim DS, Kim JY, Alfajaro MM, Park JG, Hosmillo M, Son KY, Soliman M, Baek YB. 2015. Genetic diversity of the VP7, VP4 and VP6 genes of Korean porcine group C rotaviruses. Vet Microbiol. 176:61–69.
- Jeong YJ, Park SI, Hosmillo M, Shin DJ, Chun YH, Kim HJ, Kwon HJ, Kang SY, Woo SK, Park SJ. 2009. Detection and molecular characterization of porcine group C rotaviruses in South Korea. Vet Microbiol. 138:217–224.
- Kattoor JJ, Malik YS, Sharma K, Kumar N, Batra M, Jindal N, Yadav AS. 2013. Molecular evidence of group D rotavirus in commercial broiler chicks in India. Avian Biol Res. 6:313–316.
- Khamrin P, Peerakome S, Malasao R, Mizuguchi M, Okitsu S, Ushijima H, Maneekarn N. 2008. Genetic characterization of group C rotavirus isolated from a child hospitalized with acute gastroenteritis in Chiang Mai, Thailand. Virus Gen. 37:314–321.
- Kim Y, Chang K-O, Straw B, Saif LJ. 1999. Characterization of group C rotaviruses associated with diarrhea outbreaks in feeder pigs. J Clin Microbiol. 37:1484–1488.
- Kumazaki M, Usuku S. 2014. Epidemiological and genetic analysis of human group C rotaviruses isolated from outbreaks of acute gastroenteritis in Yokohama, Japan, between 2006 and 2012. Arch Virol. 159:761–771.
- Kusanagi K, Kuwahara H, Katoh T, Nunoya T, Ishikawa Y, Samejima T, Tajima M. 1992. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J Vet Med Sci. 54:313–318.
- Martella V, Ciarlet M, Banyai K, Lorusso E, Arista S, Lavazza A, Pezzotti G, Decaro N, Cavalli A, Lucente M. 2007. Identification of group A porcine rotavirus strains bearing a novel VP4 (P) genotype in Italian swine herds. J Clin Microbiol. 45:577–580.
- Marthaler D, Rossow K, Culhane M, Collins J, Goyal S, Ciarlet M, Matthijnssens J. 2013. Identification, phylogenetic analysis and classification of porcine group C rotavirus VP7 sequences from the United States and Canada. Virology. 446:189–198.
- Marton S, Mihalov-Kovács E, Dóró R, Csata T, Fehér E, Oldal M, Jakab F, Matthijnssens J, Martella V, Bányai K. 2015. Canine rotavirus C strain detected in Hungary shows marked genotype diversity. J Gen Virol. 96:3059–3071.
- Mascarenhas JD, Linhares AC, Gabbay YB, Lima CS, Sylvia de Fátima SG, Soares LS, Oliveira DS, Lima JC, Macêdo O, Leite JPG. 2007. Molecular characterization of VP4 and NSP4 genes from rotavirus strains infecting neonates and young children in Belem, Brazil. Virus Res. 126:149–158.
- Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol. 157:1177–1182.
- Matthijnssens J, Theuns S. 2015. Minutes of the 7th Rotavirus Classification Working Group (RCWG) meeting. 12th International dsRNA Virus Symposium; Oct. 9; Goa, India. https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/minutes-of-the-7th-rcwg-meeting.
- Mawatari T, Taneichi A, Kawagoe T, Hosokawa M, Togashi K, Tsunemitsu H. 2004. Detection of a bovine group C rotavirus from adult cows with diarrhea and reduced milk production. J Vet Med Sci. 66:887–890.
- Médici KC, Barry AF, Alfieri AF, Alfieri AA. 2011. Porcine rotavirus groups A, B, and C identified by polymerase chain reaction in a fecal sample collection with inconclusive results by polyacrylamide gel electrophoresis. J Swine Health Prod. 19:146–150.
- Meleg E, Bányai K, Martella V, Jiang B, Kocsis B, Kisfali P, Melegh B, Szűcs G. 2008. Detection and quantification of group C rotaviruses in communal sewage. Appl Environ Microbiol. 74:3394–3399.
- Mihalov-Kovács E, Gellért Á, Marton S, Farkas SL, Fehér E, Oldal M, Jakab F, Martella V, Bányai K. 2015. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg Infect Diseases. 21:660–663.
- Moon S, Humphrey C, Kim J, Baek L, Song JW, Song KJ, Jiang B. 2011. First detection of group C rotavirus in children with acute gastroenteritis in South Korea. Clin Microbiol Infect. 17:244–247.
- Morin M, Magar R, Robinson Y. 1990. Porcine group C rotavirus as a cause of neonatal diarrhea in a Quebec swine herd. Can J Vet Res. 54:385.
- Moutelíková R, Prodělalová J, Dufková L. 2015. Diversity of VP7, VP4, VP6, NSP2, NSP4, and NSP5 genes of porcine rotavirus C: phylogenetic analysis and description of potential new VP7, VP4, VP6, and NSP4 genotypes. Arch Virol. 160:1715–1727.
- Navarro R, Aung MS, Brenes KZ, Ketzis J, Gallagher CA, Beierschmitt A, Malik YS, Kobayashi N, Ghosh S. 2017. Whole genome analysis provides evidence for porcine-to-simian interspecies transmission of rotavirus-A. Infect Genet Evol. 49:21–31.
- Niira K, Ito M, Masuda T, Saitou T, Abe T, Komoto S, Sato M, Yamasato H, Kishimoto M, Naoi Y. 2016. Whole genome sequences of Japanese porcine species C rotaviruses reveal a high diversity of genotypes of individual genes and will contribute to a comprehensive, generally accepted classification system. Infect Genet Evol. 44:106–113.
- Njuki J, Pali P, Mburu S, Poole J. 2010. Pig production, management and marketing in the North East Indian State of Nagaland. [accessed 2016 July 25]. https://cgspace.cgiar.org/bitstream/handle/10568/29056/elks_nagaland.pdf?sequence=1.
- Nyaga MM, Jere KC, Esona MD, Seheri ML, Stucker KM, Halpin RA, Akopov A, Stockwell TB, Peenze I, Diop A. 2015. Whole genome detection of rotavirus mixed infections in human, porcine and bovine samples co-infected with various rotavirus strains collected from sub-Saharan Africa. Infect Genet Evol. 31:321–334.
- Otto PH, Ahmed MU, Hotzel H, Machnowska P, Reetz J, Roth B, Trojnar E, Johne R. 2012. Detection of avian rotaviruses of groups A, D, F and G in diseased chickens and turkeys from Europe and Bangladesh. Vet Microbiol. 156:8–15.
- Otto P, Schulze P, Herbst W. 1999. Demonstration of group C rotaviruses in fecal samples of diarrheic dogs in Germany. Arch Virol. 144:2467–2473.
- Phan TG, Leutenegger CM, Chan R, Delwart E. 2017. Rotavirus I in feces of a cat with diarrhea. Virus Genes. 53:487–490.
- Rahman M, Banik S, Faruque AS, Taniguchi K, Sack DA, Van Ranst M, Azim T. 2005. Detection and characterization of human group C rotaviruses in Bangladesh. J Clin Microbiol. 43:4460–4465.
- Saif LJ, Bohl EH, Theil KW, Cross RF, House JA. 1980. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol. 12:105–111.
- Saif LJ, Jiang B. 1994. Nongroup A rotaviruses of humans and animals. Curr Top Microbiol Immunol. 185:339–371.
- Sanchez-Fauquier A, Roman E, Colomina J, Wilhelmi I, Glass R, Jiang B. 2003. First detection of group C rotavirus in children with acute diarrhea in Spain. Arch Virol. 148:399–404.
- Sharma R, Bora DP, Chakraborty P, Das S, Barman NN. 2013. Circulation of group Aotaviruse amet of human, cow and pig: study from Assam, a north eastern state of India. Indian J Virol. 24:250–255.
- Steyer A, Poljšak-Prijatelj M, Barlič-Maganja D, Jamnikar U, Mijovski JZ, Marin J. 2007. Molecular characterization of a new porcine rotavirus P genotype found in an asymptomatic pig in Slovenia. Virology. 359:275–282.
- Suzuki T, Hasebe A, Miyazaki A, Tsunemitsu H. 2014. Phylogenetic characterization of VP6 gene (inner capsid) of porcine rotavirus C collected in Japan. Infect Genet Evol. 26:223–227.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Torres-Medina A. 1987. Isolation of an atypical rotavirus causing diarrhea in neonatal ferrets. Lab Anim Sci. 37:167–171.
- Tsunemitsu H, Jiang B, Saif LJ. 1996. Sequence comparison of the VP7 gene encoding the outer capsid glycoprotein among animal and human group C rotaviruses. Arch Virol. 141:705–713.
- Tsunemitsu H, Saif LJ, Jiang B, Shimizu M, Hiro M, Yamaguchi H, Ishiyama T, Hirai T. 1991. Isolation, characterization, and serial propagation of a bovine group C rotavirus in a monkey kidney cell line (MA104). J Clin Microbiol. 29:2609–2613.
- Wakuda M, Ide T, Sasaki J, Komoto S, Ishii J, Sanekata T, Taniguchi K. 2011. Porcine rotavirus closely related to novel group of human rotaviruses. Emerg Infect Dis. 17:1491–1493.
- Wise A, Smedley R, Kiupel M, Maes R. 2009. Detection of group C rotavirus in juvenile ferrets (Mustela putorius furo) with diarrhea by reverse transcription polymerase chain reaction: sequencing and analysis of the complete coding region of the VP6 gene. Vet Pathol Online. 46:985–991.
- Zhirakovskaia E, Tikunov A, Klemesheva V, Loginovskikh N, Netesov S, Tikunova N. 2016. First genetic characterization of rotavirus C in Russia. Infect Gen