 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Acute respiratory distress syndrome (ARDS) is a respiratory failure syndrome characterized by hypoxemia and changes in the respiratory system. ARDS is the most common cause of death in COVID-19 deaths was ARDS. In this study, we explored the role of miR-223 in exosomes in ARDS.
Methods
Exosomes were purified from the supernatants of macrophages. qPCR was used to detect relative mRNA levels. A luciferase reporter assay was performed to verify the miRNA target genes. Western blotting was used to detect the activation of inflammatory pathways. Flow cytometry was performed to assess apoptosis. An LPS-induced ARDS mouse model was used to assess the function of miR-223 in ARDS.
Results
Exosomes secreted by macrophages promoted apoptosis in A549 cells. Macrophages and exosomes contain high levels of miR-223. Exogenous miR-223 can decrease the expression of insulin-like growth factor 1 receptor (IGF-1R) in A549 and promote the apoptosis of A549.Transfection of anti-miR223 antisense nucleotides effectively reduced the level of miR-223 in macrophages and exosomes and eliminated the pro-apoptotic effect of A549. In vivo, LPS stimulation increased inflammatory cell infiltration in the lungs of mice, whereas knockdown of miR-223 in mice resulted in significantly reduced eosinophil infiltration.
Conclusions
Macrophages can secrete exosomes containing miR-223 and promote apoptosis by targeting the IGF-1R/Akt/mTOR signaling pathway in A549 cells and mouse models, suggesting that miR-223 is a potential target for treating COVID-19 induced ARDS.
Introduction
Acute lung injury was first defined in a series of cases in 1967,Citation1 and the understanding and treatment of ARDS have evolved since then. Although progress has been made in the understanding of the pathophysiological cascade leading to ARDS,Citation2 there is currently no specific drug therapy for this condition. COVID-19 induced severe ARDS, and the most common immediate cause of death in COVID-19 deaths was ARDS, accounting for 50% of all deaths.Citation3 Interventions to improve the prognosis of ARDS remain clinical management strategies, such as lung-protective mechanical ventilation and prone position. Although these interventions have improved outcomes in patients with ARDS to some extent, the burden of lung injury remains severe with high morbidity and mortality. Recently, with the outbreak of Covid -19, the treatment and mechanism of ARDS has gained renewed attention.Citation4,Citation5 Ashvattha Therapeutics Co., Ltd. developed an N-acetyl cysteine conjugated to a hydroxyl dendrimer drug (OP-101) targeting macrophages, which has been shown to be effective in sever COVID-19 induced ARDS (clinical trials: NCT04458298). This suggests that macrophages play an important role in COVID-19 induced ARDS and could be a target for ARDS drug therapy.
The insulin-like growth factor-1 (IGF-1) signaling pathway plays a critical role in lung development.Citation6 Studies have found significant changes in the IGF-1 signaling pathway in various lung injury models.Citation7 It has also been suggested that IGF-1 signaling may play an important role in injury repair, a process that is often impaired in premature infants at risk of respiratory distress syndrome (RDS).Citation8 A mechanistic study showed that after IGF-1 binds to IGF1 receptor (IGF-1R), PI3K/AKT and mTOR signaling pathways are activated to regulate cell proliferation.Citation9
MicroRNAs (miRNAs) are endogenous single-stranded small (19-21nt) noncoding RNAs that post-transcriptionally regulate gene expression in both plants and animals.Citation10,Citation11 It is well known that miRNAs play key roles in various cellular processes, such as differentiation, proliferation, maturation, and apoptosis, and therefore, in various diseases, such as cancer, diabetes, and auto-immune diseases.Citation12–14
An increasing number of studies have suggested that cells selectively secrete miRNAs via exosomes in response to various stimuli.Citation15 Exosomes containing cell-secreted miRNAs can be taken up by recipient cells, where miRNAs can be released to silence their target genes, thus affecting the recipient cell function.Citation16 Therefore, exosome-encapsulated miRNAs may serve as a novel class of signaling molecules that remotely mediate intercellular communication.
MiR-223 was first characterized in the hematopoietic system, where it is mainly expressed in myeloid, granulocytic, and monocytic compartments but not in B and T lymphocytes.Citation17,Citation18 miR-223 is significantly upregulated in bladder cancer and recurrent ovarian cancer.Citation19,Citation20 The function of miR-223 remains unclear and controversial. Although miR-223 was reported to promote granulocytic differentiation and maturation,Citation21 some studies have reported that miR-223 negatively regulates granulocyte differentiation in miR-223 −/Y transgenic mice.Citation22 However, the expression and function of miR-223 in ARDS progression remain unclear.
In this study, we examined miR-223 expression in macrophages and its potential role in ARDS. Our study showed that macrophages have high expression levels of miR-223 both intracellularly and in exosomes. We showed that exosomes effectively delivered miR-223 into A549 cells, in which macrophage-derived miR-223 targeted IGF-1R and promoted A549 apoptosis.
Materials and methods
Plasmid construction
The 3′ UTR fragment of IGF1-R (encoded by gene IGF1R, from 1 to 2000 bp in the 3′ UTR of IGF1R) and mutant within the perfect binding site with the miR-223 seed sequence were amplified by PCR using the following primers: forward,5′-GATCGGATCCTCCTTGGATCCTGAATCTGTGC-3′ and reverse,5′-GATCGTCGACAACGCCAGTGCGAAGGAG-3′ and cloned into the pGL3-promoter. The IGF1R 3′-UTR was cloned downstream of the firefly luciferase gene, allowing the expression of a firefly transcript with the 3′-UTR of IGF1R. The pGL3-Renilla plasmid was co-transfected and Renilla luciferase activity was used as a control.
To overexpress IGF-1R, we constructed the pLVX-IGF-1R vector. The following primers were used: forward,5′-GATCGAATTCATGAAGTCTGGCTCCGGAGGAGGG-3′; reverse,5′-GATCGGATCCTCAGCAGTCGAAGACTGGGGCAGCGGC-3′
We designed siRNA to silence IGF-1R expression, siRNA sequence: siIGF-1R:5′-GUUGAG GAUCAGCGAGAAU-3′; scrambled sequence sense:5′-UCGUACGUACCUGUCACGU-3′.
Cell culture, lentivirus packaging and infection
Human alveolar basal epithelial cells (A549) mimic lung injury. Therefore, we used A549 cells to investigate ARDS in this study. A549 cells were cultured in DMEM supplemented with 10% fetal calf serum (Gibco). A total of 12 µg of the plasmids was co-transfected with 30 µl LipofectamineTM2000 (Invitrogen) into 80-90% confluent A549 cells in a 10-cm diameter plastic culture dish. After 48 h of incubation, the supernatant was collected and filtered using a filter with 0.22 µm pore size filter.
THP-1 cells were cultured in DMEM (GIBCO, Grand Island, NY) containing 10% FBS (GIBCO, Grand Island, NY) in 5% CO2 at 37 °C°C. The cells were treated with 500 nM PMA for 4 h to stimulate their differentiation into macrophages. Next, the cells were washed 3 times with phosphate-buffered saline (PBS) and incubated for 24 h to exclude PMA interference.
THP-1 differentiated macrophages were cultured in DMEM supplemented with 10% fetal calf serum (Gibco). For conditioned medium and exosome isolation, macrophages were cultured in DMEM for 24 h, and supernatants were collected and filtered with a 0.22 µm pore size filter.
A549 were infected with viruses in the presence of 8 mg/ml polybrene (Sigma-Aldrich, St. Louis, MO, USA) for 12 h, and the medium was refreshed. Forty-eight hours after infection, the efficiency of infection was measured by western blotting.
Exosome purification
Exosomes were purified from the cell culture supernatant of the macrophages by ultracentrifugation. Supernatant media collected from 24-h macrophage cultures were filtered through 0.22-μm pore filters, followed by ultracentrifugation at 10,000 × g for 40 min and then 100,000 × g for 14 h. Total protein concentration in each exosome sample was quantified using the Bradford assay (Bio-Rad) and expressed as micrograms of containing proteins.
Luciferase reporter assay
A luciferase reporter assay was performed on the A549 cells. Forty-eight hours after transfection, the cells were washed twice with PBS and lysed in 100 µL Passive Lysis Buffer (Promega), and luciferase activity was measured from 20 µL of lysate using the Dual-Luciferase Reporter Assay Kit (Promega) on an illuminometer (Promega). All data were obtained by averaging the results obtained from three independent replicates.
RNA isolation and qPCR
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen), and reverse transcription was performed using the RT Kit according to the manufacturer’s protocol (Invitrogen). qPCR was performed to detect mature miR-223 with specific primers, using 2 × SYBR Green Master Mix (Toyobo). The primers used in this experiment were as follows:
miR-223: forward:5′-GTGCAGGGTCCGAGGT-3′; reverse:5′-CGGGCTGTCAGTTTGTCA-3′; U6: forward:5′-CTCGCTTCGGCAGCACA-3′, reverse:5′-AACGCTTCACGAATTTGCGT-3′; β-actin: forward:5′-GAGCTACGAGCTGCCTGACG-3′, reverse:5′-CCTAGAAGCATTTGCGGTGG-3′; IGF-1R: forward:5′-GGACAGGTCAGAGGGTTTC-3′, reverse:5′-CTCGTAACTCTTCTCTGTGCC-3′. All experiments were carried 3 independent times.
Western blot
Cells were lysed in RIPA buffer with proteinase cocktail (Thermo Fisher Scientific), and 50 µg of total protein was loaded onto 10% SDS–PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). After blocking with 5% nonfat milk in TBST, the membranes were incubated with primary antibodies against CD63 (Abcam), IGF-1R, AKT, phosphorylated AKT, mTOR, phosphorylated mTOR, BCL2 (Cell Signaling Technology), and GAPDH (Chemicon). After 1 h of incubation with the HRP-conjugated secondary antibody at room temperature, the membrane was developed using an enhanced chemiluminescence detection kit (Thermo).
Apoptosis assay
Cell Apoptosis assays were performed using the CaspACE FITC-VAD-FMK In Situ Marker (Promega), following the manufacturer’s instructions. Briefly, A549 (1.0 x105 in 100 μl) were seeded into a 96-well plate and allowed to attach for 16 h at 37 °C in 5% CO2. The cells were then treated accordingly. At the end of the assay, the substrate was added to the wells and cultured for 4 h at 37 °C, and the absorbance was recorded at 520 nm using a 96-well plate reader.
Apoptosis was assessed by flow cytometry. The A549 cells were digested into single cells and counted. 100,000 cells were resuspended with 195 μl Annexin V-FITC (Beyoutime) binding solution, then 5 μl Annexin V-FITC and 10 μl propidium iodide were added. The cells were incubated at room temperature in the dark for 15 min and analyzed using flow cytometry.
Antagomir of miR-223
Antagomirs targeting miR-223 and a disordered control antagomir designed to not target any known miRNA sequences were purchased from Sangon Biotech (Shanghai, PR. China) Co., Ltd. The sequence is ‘UGUCAGUUUGUCAAAUACCCCA’
Mice model
Mice were intranasally administered 50 μg antagomir in 50 μL sterile saline 24 h before the initial LPS attack. This dose was repeated daily throughout the model. The 60 mice were divided equally into three groups: nasal control antagomir only, nasal control antagomir and LPS-induced acute lung injury group, and nasal targeting miR-223 antagomir and LPS-induced acute lung. This study was approved by the Animal Ethics Committee of he Fourth Affiliated Hospital Zhejiang University School of Medicine. And the experiment design and procedure accomplied with 3 R principle.
Statistical analysis
Statistical analysis was performed using SPSS Statistical Package version 16 and expressed as means S.E.) using the Student’s t-test. Statistical significance was set at p < 0.05.
Results
Exosomes from macrophage promote apoptosis of A549
To assess the function of macrophages in the progression of ARDS, conditioned medium (CM) of macrophages was co-cultured with endothelial A549 cells for 3 days. An apoptosis assay was performed using A549 cells to examine the caspase activity. The results showed that the CM from macrophages promoted caspase activity ( A) in A549 cells, indicating the promotion of apoptosis. BCL2 is an antiapoptotic protein. Consistently, western blotting against BCL2 was downregulated in A549 cells after co-culture with the CM of macrophages compared to that in the control ( B).
Figure 1. Exosomes from macrophage promotes apoptosis of A549. A. Apoptosis assay by CaspACE FITC-VAD-FMK In Situ Marker showed CM from macrophage could promote apoptosis of A549, compared with A549 control. CM: conditioned medium from macrophage. *** P < 0.001. B. Western blot shows BCL2 downregulation in A549 cells after co-culture with CM of macrophage compared with control. CM: conditioned medium from macrophage. C. Western blot for exosomes marker CD63, actin from the lysate of cells from which exosomes were isolated as the loading control. Exo: exosomes. GW4689 is an exosome generation inhibitor. The exosomes generated by macrophage was effectively inhibited by GW4689. D. Apoptosis assay by CaspACE FITC-VAD-FMK In Situ Marker showed that exosomes isolated from macrophage could promote apoptosis of A549, compared with exosomes from macrophage treated with PBS and A549 control. *** P < 0.001. E. Western blot for BCL2 in A549, using actin as the loading control. Exo: exosomes. Each experiment was repeated at least 3 times. GW4689 is an exosome generation inhibitor. In GW4689 treated macrophage group, the BCL2 was upregulated which indicated that apoptosis was inhibited.
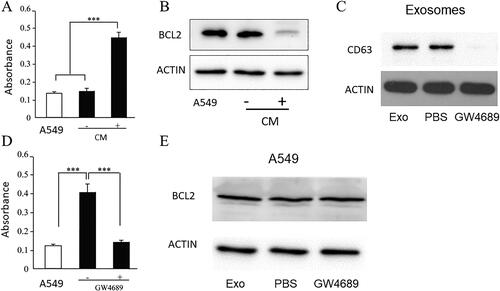
To assess whether exosomes mediate A549 apoptosis, exosomes were isolated from the CM of macrophages by ultracentrifugation. The presence of exosomes was confirmed by western blotting for exosomal markers ( C). Incubation of A549 cells with the isolated exosomes resulted in an apoptotic phenotype ( D). We then treated macrophages with GW4689, which inhibits neutral sphingomyelinase, to block exosome formation and isolate exosomes. Western blotting ( C) showed significant downregulation of exosome markers in isolated exosomes after GW4689 treatment. Exosomes isolated from the CM of macrophages were cocultured with A549 cells. In the absence of exosomes, CM did not induce apoptosis ( D) of A549. Western blotting against BCL2 showed that in GW4689 treated macrophage, exosomes did not downregulate BCL2, which confirmed exosome-induced apoptosis of A549 ( E).
MiR-223 is highly presented in exosomes and is effectively transferred into A549
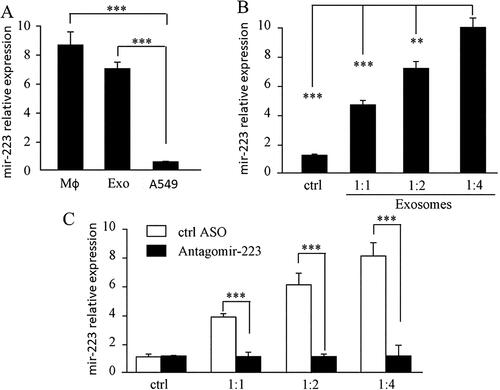
We compared the miR-223 levels in macrophages, A549 cells, and exosomes using QPCR. The results clearly showed that the miR-223 levels in macrophages and exosomes were much higher than those in A549 ( A). To determine the potential biological functions of miR-223 in exosomes, we studied the delivery of miR-223 into A549 cells using exosomes isolated from macrophages. qPCR results showed that after incubation with different amount of exosomes (ratio of the volume of media vs. exosomes), the expression of miR-223 was highly upregulated in A549 cells compared to that in the parental A549 via a dose-dependent manner ( B). To confirm that the upregulation of miR-223 was not due to de novo miRNA biosynthesis in A549 cells, we treated macrophages with anti-miR-223 or ASO control and then isolated the exosomes and incubated them with A549 cells, which showed that the expression of miR-223 in A549 cells was not changed compared to that in parental A549 cells, but the exosomes of non-antagomir-223 treated macrophage increased miR-223 in A549 cells ( C).
Figure 2. MiR-223 Is highly presented in exosomes and is effectively transferred into A549.
A. QPCR showed miR-223 level in macrophage, isolated exosomes and A549. *** P < 0.001. B. QPCR showed the expression of miR-223 in A549. Ctrl bar is the miR-223 level in A549 without treatment, the graph showed the expression of miR-223 in A549 increased via a dose-dependent manner. C. QPCR showed the expression of miR-223 in A54. Ctrl bar is the miR-223 level in A549 which treated with different dosages of exosomes from macrophages. Antagomir-223 bar is miR-223 level in A549 which treated with exosomes from macrophages of being transfected with Antagomir-223. Mφ: macrophage, Exo: exosomes. *** P < 0.001 each experiment was repeated at least 3 times.
MiR-223 promotes epithelial cell apoptosis through targeting IGF-1R
To determine whether exogenous miR-223 delivered by isolated exosomes resulted in the apoptosis of A549 cells, miR-223 overexpression in A549 cells via transfection with miR-223 clearly showed that miR-223 strongly promoted cell apoptosis in a dose-dependent ( A) and time-dependent manner ( B). However, exosomes secreted by macrophages treated with antagomir-223 did not induce apoptosis in A549 ().
Figure 3. MiR-223 promotes endothelial cell apoptosis through targeting IGF-1R.
A. Isolated exosomes from CM of macrophage treated with antisense oligonucleotide against miR-223 couldn’t promote apoptosis of A549, CM without ASO as control. Overexpression of miR-223 in A549 promotes apoptosis of A549, empty vector as control. *** P < 0.001 EV: empty vector. ASO: antisense oligonucleotide. B. Isolated exosomes from CM of macrophage treated with antisense oligonucleotide against miR-223 couldn’t promote apoptosis of A549, CM without ASO as control. Overexpression of miR-223 in A549 promotes apoptosis of A549 as early as 12 h after incubation, empty vector as control. *** P < 0.001 EV: empty vector. ASO: antisense oligonucleotide. C. Bioinformatics analysis showed that miR-223 could bind to IGF-1R in 3’UTR. D. Western blot for IGF-1R in A549, using actin as the loading control. Exo: exosomes. EV: empty vector. E. Luciferase activity in A549 cells with co-transfection of miR-223 and empty vector or WT or mutant UTR 3’UTR of IGF-1R. *** P < 0.001. EV: empty vector. F. Apoptosis assay by CaspACE FITC-VAD-FMK In Situ Marker. A549 Cells were transfected with miR-223. Using the transfected Cells, the caspase activity of overexpression IGF1R and knock down of IGF1R were tested. The absorbance value stands for the activity of caspase and higher absorbance stands for higher activity of caspase. *** P < 0.001. SCR: Control group.
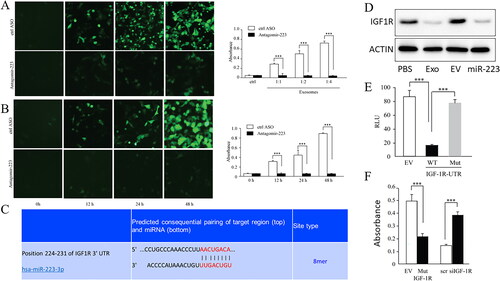
We then attempted to further study the target of miR-223, and previous data showed that IGF-1R is a potential target of miR-223 18. Through bioinformatics prediction, we found that miR-233 could conservatively bind to the 3′-UTR region of IGF-1R ( C). Similarly, Western blotting showed a significant decrease in IGF-1R expression in A549 cells overexpressing miR-223 or cultured with miR-223-containing exosomes ( D). Furthermore, the Luciferase reporter assay showed that the WT IGF-1R 3′ UTR was downregulated by miR-223, but not by the mutated 3′ UTR of IGF-1R at the miR-223 binding site ( E). These results suggested that miR-223 targets IGF-1R in A549 cells.
To determine whether miR-223 inhibited A549 proliferation by targeting IGF-1R, we overexpressed IGF-1R with mutated 3′-UTR by transfection. Caspase activity assays showed that 3’UTR mutated IGF-1R overexpression rescued the apoptosis induced by miR-223. At the same time, knockdown of IGF-1R in A549 could induce apoptosis ( F).
MiR-223 inhibits IGF-1R/akt/mTOR signaling pathway
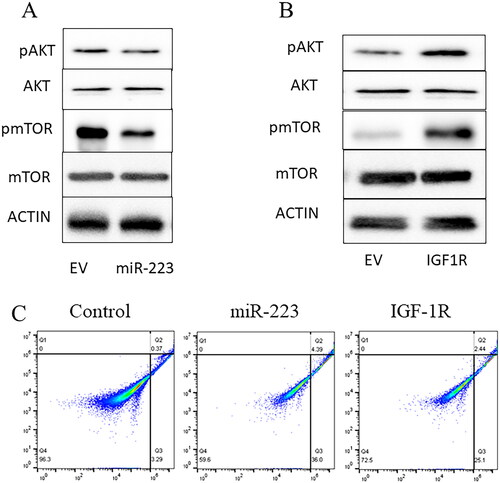
We further examined whether the IGF-1R downstream signaling pathway was affected by miR-223. Western blotting results showed that phosphorylated AKT was reduced in the miR-223 overexpression group, while total AKT was unaffected ( A). To further investigate the pathway alteration of miR-223, we found that phosphorylation of mTOR was greatly reduced in the miR-223 overexpressing group compared with the control, but total mTOR was unaffected ( A). Moreover, overexpression of IGF-1R by transfection reversed the inhibition of the AKT/mTOR signaling pathway ( B). Flow cytometric analysis also showed that IGF-1R overexpression reversed miR-223-induced apoptosis, reducing the rate of apoptosis from 40.4% to 27.5% ().
Figure 4. MiR-223 inhibits IGFR-1R/AKT/mTOR signaling pathway.
A. Western blot for AKT/mTOR signaling pathway in A549 overexpressing miR-223, using actin as the loading control. B. Western blot for AKT/mTOR signaling pathway in A549 overexpressing IGF-1R, using actin as the loading control. EV: empty vector. Each experiment was repeated at least 3 times. C. A549 apoptosis rate was detected by flow cytometry. Non-exosomes treated A549 cells used as control group. The miR-223 picture stands for the apoptosis rate of miR-223 overexpressed A549 cells and IGF-1R picture stands for the apoptosis rate of IFG-1R and miR-223 overexpressed A549 cells. The apoptosis rate was calculated by Q2 + Q3.
Inhibition of miR-233 expression down-regulates the inflammation
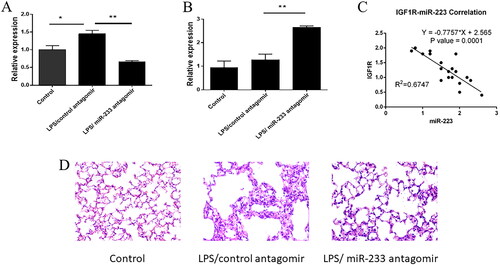
We used an LPS acute lung injury model to further explore the role of miR-233 in vivo. The expression of miR-233 was significantly elevated in LPS-treated mice and was restored to baseline levels after intranasal administration of antagomir, which targets miR-233 ( A). We then examined the expression of IGF1R in the lungs of mice and found that there was no significant difference in the expression of IGF1R in the lungs of mice treated with LPS induction compared to the control group, but the expression of IGF1R in the lungs of mice treated with LPS induction after knocking down miR-233 by nasal drip treatment was significantly higher ( B). Consistent with the cellular data, miR-233 and IGF1R levels were negatively correlated in response to LPS stimulation ( C). The effect of miR-223 on lung eosinophil inflammation was assessed by counting eosinophil infiltration in hematoxylin and eosin (H&E)-stained lung sections under a light microscope. Eosinophils were significantly increased in the lungs of LPS-treated mice compared to those in the control group, and knockdown of miR-233 significantly reduced this phenomenon ( D). As expected, LPS stimulation increased inflammatory cell infiltration in the lungs of mice, while miR-233 knockdown in mice resulted in significantly reduced eosinophil infiltration.
Figure 5. MiR-223 inhibits IGF-1R expression in acute lung injury.
A. Relative expression of miR-223 in the lungs of wild-type mice in a model of LPS-induced acute lung injury, treated with control anticoagulants or anticoagulants targeting miR-223 (n = 20). B. Expression of IGF1R in the lungs of wild-type mice treated with control anticoagulant or anticoagulant against miR-223 in a model of LPS-induced acute lung injury (n = 20). C. Spearman’s correlation between miR-223 and IGF1R in mouse lungs in a model of LPS-induced acute lung injury. D. Determination of eosinophils in the alveolar epithelial cell layer by light microscopic analysis of H&E-stained mouse lung sections.
Discussion
This study investigated the role of macrophage-derived exosomes containing miR-223 in regulating apoptosis of human alveolar basal epithelial (A549) cells. We demonstrated that miR-223-containing exosomes induce apoptosis in A549 cells by targeting the IGF-1R/AKT/mTOR signaling pathway. These findings suggest a potential role for miR-223 as a therapeutic target for ARDS.
Several studies have highlighted the role of exosomes in ARDS pathogenesis. It has been well known that macrophage can release large numbers of exosomes particularly under inflammatory conditions.Citation23 Ashvattha’s developing of COVID-19 induced ARDS drug inspired us to hypothesize that exosomes from macrophage may play an important role in ARDS. Exosomes can mediate intercellular communication by transferring various molecules, including miRNAs. Our study identified miR-223 as a key player in this process, suggesting that targeting miR-223-containing exosomes might be a promising therapeutic strategy.
We identified IGF-1R/AKT/mTOR signaling pathway as the primary target of miR-223-induced apoptosis in A549 cells. This pathway plays a crucial role in cell survival and proliferation. Abnormal IGF-1R signaling can lead to cell growth regulation and pro-apoptotic responses. To support the role of miR-223–targeting IGF-1R in promoting A549 apoptosis, direct silencing of IGF-1R expression using IGF-1R siRNA resulted in increased apoptosis in A549 cells (). Depletion of miR-223 in macrophages by antisense oligonucleotides largely reversed the phenotype of A549 apoptosis induced by exosomes, indicating that miR-223 in exosomes of macrophages plays a critical role in modulating A549 apoptosis (). By targeting IGF-1R and suppressing this signaling pathway, miR-223 promotes apoptosis, potentially limiting the progression of lung injury in ARDS. This result is consistent with recent studies showing that cell-secreted exosomes serve as physiological carriers of miRNAs to exchange genetic material and signaling molecules between cells.Citation24 Shi and colleagues have demonstrated that miR-223 deficiency exacerbated hypoxia- and load-induced right ventricular failure, while upregulation of miR-223 attenuated these effects through downregulation of IGF-1R.Citation25 Gao and colleagures suggest that IGF-1R is the functional target for miR-223 promotion of cell apoptosis, and its downstream PI3K/Akt signaling pathway was suppressed by miR-223 through targeting of IGF-1R.Citation26
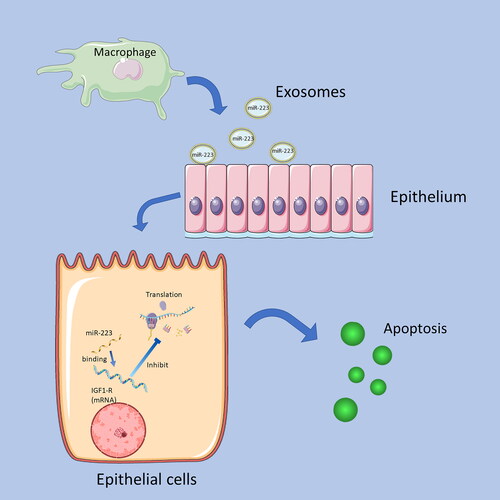
Figure 6. Summary of the mechanism of MiR-223 inhibits IGF1-R.
Exosomes of macrophages contain miR-223, and when miR-223 is transported to epithelial cells by Exosomes, the miR-223 binding to the mRNA of IGF1-R receptors and inhibits its translation. Decreased IGF1-R expression promotes epithelial cell apoptosis.
Our findings suggest that miR-223 could be a valuable biomarker for diagnosing and monitoring ARDS induced by COVID-19 by targeting macrophages. Additionally, targeting miR-223-containing exosomes could be a novel therapeutic strategy for this critical condition.
Despite the promising results, several challenges need to be addressed before miR-223-based therapies can be translated into clinical practice. Optimizing delivery methods for miR-223 is crucial to ensure efficient and targeted delivery to the lungs. Additionally, minimizing potential off-target effects and identifying the optimal dose and timing of miR-223 administration are essential for safe and effective therapy. Further research is needed to explore the long-term effects of miR-223-based therapies and assess their efficacy in animal models of ARDS. Furthermore, clinical trials are crucial to evaluate the safety and effectiveness of these therapies in humans.
Overall, our study provides compelling evidence for the potential of miR-223-containing exosomes as a therapeutic target for ARDS. Further research and development are needed to overcome existing challenges and translate this promising approach into effective clinical interventions.
Acknowledgment
None
Declaration of interest
No potential conflict of interest was reported by the author(s).
Data availability statement
The data are deposited in Figure share with DOI:10.6084/m9.figshare.23546049 and 10.6084/m9.figshare.23546199.
Additional information
Funding
References
- Ashbaugh DG, Bigelow DB, Petty TL, Levine BE. Acute respiratory distress in adults. Lancet. 1967;2(7511):319–323. doi:10.1016/s0140-6736(67)90168-7.
- Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. Aug 10 2017;377(6):562–572. doi:10.1056/NEJMra1608077.
- von Stillfried S, Bülow RD, Röhrig R, Boor P, First report from the German COVID-19 autopsy registry. Lancet Reg Health Eur. 2022;15:100330.
- Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. Jul 2020;213(2):54–56 e1. doi:10.5694/mja2.50674.
- Whyte CS, Morrow GB, Mitchell JL, Chowdary P, Mutch NJ. Fibrinolytic abnormalities in acute respiratory distress syndrome (ARDS) and versatility of thrombolytic drugs to treat COVID. JThrombHaemost. 2020;18(7):1548–1555. doi:10.1111/jth.14872.
- Narasaraju TA, Chen H, Weng T, et al. Expression profile of IGF system during lung injury and recovery in rats exposed to hyperoxia: a possible role of IGF-1 in alveolar epithelial cell proliferation and differentiation. J Cell Biochem. 2006;97(5):984–998. doi:10.1002/jcb.20653.
- Qian L, Liu H, Yu W, et al. Effects of positive end-expiratory pressure, inhaled nitric oxide and surfactant on expression of proinflammatory cytokines and growth factors in preterm piglet lungs. Pediatr Res. 2008;64(1):17–23. doi:10.1203/PDR.0b013e31817330a6.
- Chetty A, Andersson S, Lassus P, Nielsen HC. Insulin-like growth factor-1 (IGF-1) and IGF-1 receptor (IGF-1R) expression in human lung in RDS and BPD. Pediatr Pulmonol. 2004;37(2):128–136. doi:10.1002/ppul.10415.
- LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat. 2008;11(1–2):32–50. doi:10.1016/j.drup.2007.11.003.
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi:10.1038/ncb0309-228.
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi:10.1038/nrc3932.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 23 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5.
- Davis-Dusenbery BN, Hata A. MicroRNA in cancer: the involvement of aberrant MicroRNA biogenesis regulatory pathways. Genes Cancer. Nov 2010;1(11):1100–1114. doi:10.1177/1947601910396213.
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. Mar 2010;16(3):506–515. doi:10.1261/rna.1952110.
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi:10.1038/nri855.
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi:10.1093/jb/mvj128.
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi:10.1126/science.1091903.
- Jia CY, Li HH, Zhu XC, et al. MiR-223 suppresses cell proliferation by targeting IGF-1R. PLOS One. 2011;6(11):e27008. doi:10.1371/journal.pone.0027008.
- Laios A, O’Toole S, Flavin R, et al. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7(1):35. doi:10.1186/1476-4598-7-35.
- Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25(5):387–392. doi:10.1016/j.urolonc.2007.01.019.
- Felli N, Pedini F, Romania P, et al. MicroRNA 223-dependent expression of LMO2 regulates normal erythropoiesis. Haematologica. 2009;94(4):479–486. doi:10.3324/haematol.2008.002345.
- Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–1129. doi:10.1038/nature06607.
- Tang N, Sun B, Gupta A, Rempel H, Pulliam L. Monocyte exosomes induce adhesion molecules and cytokines via activation of NF-kappaB in endothelial cells. Faseb J. 2016;30(9):3097–3106. doi:10.1096/fj.201600368RR.
- Saha B, Momen-Heravi F, Kodys K, Szabo G. MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages. J Biol Chem. 2016;291(1):149–159. doi:10.1074/jbc.M115.694133.
- Shi L, Kojonazarov B, Elgheznawy A, et al. miR-223-IGF-IR signalling in hypoxia- and load-induced right-ventricular failure: a novel therapeutic approach. Cardiovasc Res. 2016;111(3):184–193. doi:10.1093/cvr/cvw065.
- Gao H, Deng H, Xu H, et al. MicroRNA-223 promotes mast cell apoptosis by targeting the insulin-like growth factor 1 receptor. Exp Ther Med. 2016;11(6):2171–2176. doi:10.3892/etm.2016.3227.
