Abstract
Over the last decade, there has been increasing interest in the use of probiotic microorganisms. However, certain doubts have arisen around probiotics, because of the beneficial effects of these microorganisms are not clear yet, and in many occasions those beneficial effects have not been proven. Therefore, it would be of interest if these probiotic strains were able to acquire new attributes to allow them improve and increase their beneficial characteristics. Genetic engineering can be used for human applications; for instance, the resistance to antibiotics is removed and the probiotic bacteria are modified in its own DNA. This process can be achieved by: (1) the use of food-grade vectors derived from lactic acid bacteria and/or bifidobacteria cryptic plasmids, (2) the genes integration or deletion in the chromosome of the probiotic strain, by site-specific recombination using the attP/integrase system, or by homologous recombination, using either suicide vectors, (3) using the clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR-associated (Cas) nuclease. Through genetic engineering, the knowledge of probiotic strains as well as its use could be improved, and the doubts about probiotics could be crumped.
Introduction
The World Health Organization defined in 2001 probiotics as ‘live microorganisms which, when administered in adequate amounts, confer a health benefit on the host’. The most common definition of probiotics is ‘live microorganisms that, when administered in adequate amounts, confer a health benefit on the host’. Simply put, probiotics are bacteria that benefit people’s health when in the stomach and intestines. In order to be called probiotics, bacteria must be alive when consumed, and their health benefits must be confirmed through scientific research. Some of their benefits include: (1) promoting intestinal health; (2) improving the immune system to help maintain health; (3) preventing chronic diseases like cancer, high blood pressure, obesity, and diabetes; and (4) improving mental health.
Lactic acid bacteria and bifidobacteria strains are the most common microbes used as probiotics and probiotic potential have been evaluated in research studies in animals and humans with respect to antibiotic-associated diarrhea, travelers’ diarrhea, pediatric diarrhea, irritable bowel syndrome and inflammatory bowel disease, bacterial vaginosis, or as delivery systems for vaccines, immunoglobulins, and other therapies (Borges, Silva, & Teixeira, Citation2014; Clarke, Cryan, Dinan, & Quigley, Citation2012; Iqbal et al., Citation2014). It is commonly admitted that most effects of probiotic are strain-specific and cannot be extended to other probiotics of the same genus or species. Table shows some Lactobacillus and bifidobacteria strains and mixtures of Lactobacillus strains with probiotic properties. In addition, also Bacillus coagulans GBI-30 and Saccharomyces boulardii have probiotic properties (Fitzpatrick, Citation2013).
Table 1. Some Lactobacillus and Bifidobacterium strains considered probiotics.
Lactic acid bacteria and bifidobacteria are Generally Recognized as Safe (GRAS) according to Food and Drug Administration (FDA) and the application of genetic engineering technology to improve characterized strains or develop new strains, is a global area of active research. Advances in gene technology allow us to modify strains by introducing new genes or altering their metabolic functions. Over the last 20 years, numerous ways to introduce heterologous genes into lactic acid bacteria strains have been demonstrated (Bermúdez-Humarán et al., Citation2013; Foligné, Daniel, & Pot, Citation2013; Rondanelli et al., Citation2015). Some described in details techniques could be also useful to introduce into Lactobacillus and bifidobacteria genomes selected genes in order to improve their probiotic activities. However, antibiotic resistance genes have been traditionally used as resistance markers, for this reason, the potential benefit to society and the potential risks associated with the use of antibiotic must be reconsidered. Furthermore, for legal and ethical reasons, transferable genes that confer antibiotic resistance are not acceptable in food and human applications. Further research is needed in order to establish probiotics and evaluate their safety as well as their nutritional aspects
For these reasons exposed above, alternative strategies and/or selection markers should be used in probiotic bacteria to lead improvements in food technology and health (Bermúdez-Humarán et al., Citation2013; LeBlanc, Aubry, & Cortes-Perez, Citation2013; de Vos, Citation1999).
Probiotics today
Consumers can see how probiotics have invaded the market especially in the form of dairy products such as yogurt or kefir, non-dairy fermented products such as kimchi or sauerkraut and other fermented beverages and foods. Moreover, non-dairy and non-fermented probiotics are being developed and introduced recently as new consumer items including breakfast cereals.
Sales of probiotic products have an upward trend from 2010 to 2014, rising globally by 35% from 23.1 billion dollars to 31.3 billion dollars. The main consumers of probiotics in 2014 were Western Europe (8.3 billion dollars) (http://www.statista.com/statistics/252941/probiotic-products-sales-worldwide-by-region/).
Over the last decade, an increasing interest in the study and application of probiotic micro-organisms has been highlighted, and although there are doubts concerning in the use of these micro-organisms to improve both human and animal health, increasing scientific evidence is coming to light which demonstrates the health benefits which probiotic strains provided when consumed in sufficient quantities (Fitzpatrick, Citation2013; Reid, Jass, Sebulsky, & McCormick, Citation2003; Rondanelli et al., Citation2015). This has led to considerable increases in: the number of scientific articles related to probiotics (over de 12,000 articles in the last 10 years in Pubmed); the number of probiotic congresses and societies, as well as investment in research and development by food companies and the pharmaceutical industry. However, the main interest in probiotics has arisen from society in general. Some potential effects of probiotic strains are listed in Table .
Table 2. Potential effects of probiotic strains and some potential effects of probiotics that can acquire or improve with the genetic engineering.
Doubts about probiotics
As has been pointed out, certain doubts have arisen around probiotics because: (1) The beneficial effects of these micro-organisms are not as clear as we would like them to be.
(2) On many occasions, the beneficial effects of certain supposedly probiotic strains have not been proven.
Moreover, in spite of the beneficial effects of probiotics that the scientific community has recognized, up the present, the European Food Safety Authority (EFSA) has not recognized probiotic.
What does appear to be clear is that even among the probiotic micro-organisms that have been shown to be beneficial, these beneficial effects can always be improved upon. Moreover, it would be of interest if these probiotic strains were able to acquire some new properties, which would allow them to improve and increase their probiotic properties, as well as gain recognition from the EFSA.
Some of the characteristics of probiotic micro-organisms which could be improved on in same strains are: adhesion to the intestine; survival under gastrointestinal conditions, and as a greater ability to stimulate the immune system. Some new properties could be, for example, the ability to reduce blood cholesterol levels, to reduce sense of hunger and limit the storage of fat, to prevent Helicobacter pylori infections, to improve the immune system response or to palliate gastrointestinal problems for people with celiac disease. It is also possible that undesirable properties present in these bacteria which could be eliminated, such as the production of biogenic amines.
How can we improve, introduce, or remove these probiotic properties?
Through genetic engineering we can improve, introduce, or remove phenotypes of probiotic bacteria. However, the use of genetic engineering on micro-organisms which can be used on humans are subjected to high controls and are regulated by strict laws; moreover, the use of genetic engineering is not authorized in some countries, or by some social organizations and social groups.
Table 3. Some resistance mechanisms used in food-grade vector.
Even so, genetic engineering can be used for human applications as long as the resistance to antibiotics is removed and the probiotic bacteria are modified with its own DNA or DNA from GRAS organisms. This can be achieved by:
| (1) | The use of food-grade vectors derived from lactic acid bacteria and/or bifidobacteria cryptic plasmids (Landete, Citation2016; Peterbauer, Maischberger, & Haltrich, Citation2011). Food-grade vectors are plasmids in which the resistance to antibiotics has been replaced by resistance mechanisms such as immunity to bacteriocins (for example, the resistance to nisin or lacticin F), resistance to heavy metals (copper or cadmium) or the complementation of auxotrophs (Table ). The presence of bacteriocins or heavy metals would only be needed in the steps prior to the selection of the transformants. | ||||
| (2) | The integration or deletion of genes in the chromosome of the probiotic strain is by site-specific recombination using the attP/integrase system, or by homologous recombination using either suicide vectors, or ones that are temperature sensitive (Gosalbes, Esteban, Galán, & Pérez-Martínez, Citation2000; Lin, Lo, Kuo, & Lin, Citation2013; Martín, Alonso, Suárez, & Alvarez, Citation2000; Song et al., Citation2014). In many cases, the integration on the chromosome is also produced by food-grade vectors (Leenhouts, Bolhuis, Venema, & Kok, Citation1998). | ||||
| (3) | The development of CRISPR-Cas9 system in combination with ssDNA recombineering would allow the edited of genome in probiotic bacteria acquiring some new properties without antibiotic resistance (Jiang, Bikard, Cox, Zhang, & Marraffini, Citation2013; Sander & Joung, Citation2014). | ||||
Figure 1. (A). Example of food-grade vector for lactic acid bacteria based in a replicative plasmid. Plasmid food-grade is built entirely from lactococcal DNA using a replicon for lactic acid bacteria and the resistance to antibiotic is changed by a bacteriocin immunity gene as selection marker. After, transformed Lc. lactis strain must be grown on medium containing bacteriocin for the selection. (B) Example of food-grade vector for lactic acid bacteria based in an integrative plasmid. The food-grade vector contains the Ori+ from pWV01 plasmid, auxotrophic complementation as selectable marker and a DNA fragment of a well-characterized chromosomal region from the Lb. rhamnosus X. The helper strain produce the RepA protein essential for replication of the Ori+ vectors allows the construction and isolation of the integration plasmids from a homologous background. The subsequent purification and transformation of this food-grade vector into Lb. rhamnosus X will produce a single-cross-over integration of the plasmids in Lb. rhamnosus X resulting in amplifications of copies/chromosome after selection of the transformants on auxotrophic complementation.
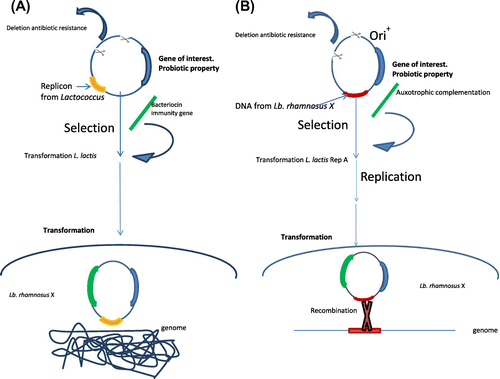
Figure (B) shows a food-grade cloning vector for lactic acid bacteria based in an integrative plasmid. The construction of a food-grade multiple-copy integration system for lactic acid bacteria is one of the best examples of a food-grade vector that integrates into the chromosome allowing the stable integration of multiple plasmid copies in the chromosome. The food-grade vector contains the Ori+ from pWV01 plasmid, auxotrophic complementation as selectable marker, a multiple-cloning site, and a DNA fragment of a well-characterized chromosomal region from the lactic bacteria or bifidobacteria, which be transformed (Lb. rhamnosus X in the Figure ). Resistance to antibiotic is removed from this food-grade vector. The system includes a L. lactis strain, which produce the RepA protein essential for replication of the Ori+ vectors. This helper strain allows the construction and isolation of the replicating form of the integration plasmids from a homologous background. The subsequent purification and transformation of this food-grade vector into a strain which does not produce the protein RepA will produce a single-cross-over integration of the plasmids in Lb. rhamnosus X resulting in amplifications of copies/chromosome after selection of the transformants on auxotrophic complementation.
Figure shows DNA editing could be generated in the chromosome by single-stranded DNA (ssDNA) recombineering (Oh & van Pijkeren, Citation2014; van Pijkeren & Britton, Citation2012; van Pijkeren, Neoh, Sirias, Findley, & Britton, Citation2012). ssDNA recombineering requires inducible expression of a phage-derived ssDNA-binding protein (RecTorBeta). Once the oligonucleotide is in the cell, the ssDNA-binding protein protects the oligonucleotide from degradation by host nucleases and help to form a complex between the oligonucleotide and the lagging strand template DNA. The cotransformation of a recombineering oligonucleotide and a CRISPR-target plasmid, a single-step approach, will yield recombinants when ssDNA recombineering efficiencies are optima (Oh & van Pijkeren, Citation2014; van Pijkeren & Britton, Citation2012; van Pijkeren et al., Citation2012). The prokaryotic CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9, an RNA-guided endonuclease, has been shown to mediate efficient genome editing in a wide variety of organisms. The power of these systems to perform targets, highly efficient alterations of genome sequence and gene expression spured the development of novel molecular therapeutics.
Figure 2. DNA editing by CRISPR-Cas9 in combination with single-stranded DNA (ssDNA) recombineering.ssDNA recombineering requires inducible expression of a phage-derived ssDNA-binding protein When the oligonucleotide is into the cell, the ssDNA-binding protein (Rec T or Beta) protects the oligonucleotide from degradation by host nucleases and help to form a complex between the oligonucleotide and the lagging strand template DNA. The cotransformation of a recombineering oligonucleotide and a CRISPR-target plasmid, a single-step approach, will yield recombinants when ssDNA recombineering efficiencies are optima.
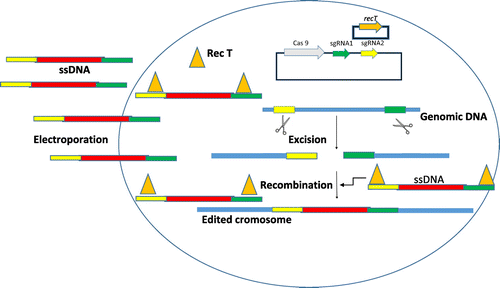
Thus, by means of recombination and later chromosomal integration and food-grade vectors and the development of CRISPR-Cas9 genome editing in combination with ssDNA recombineering, we could express the aggregation factors of L. lactis in the probiotic bacteria with interest, to enhance its adhesion to the intestinal epithelium (Kojic et al., Citation2011). Interleukins or human cytokines in the probiotic strain could be expressed in order to increase immune system stimulation (Li et al., Citation2015); vaccines in probiotic bacteria could be produced (Shi et al., Citation2014); expression of enzymes such as bile salt hydrolase from Lactobacillus fermentum in probiotic bacteria which would also help to reduce cholesterol (Kumar et al., Citation2013); and gastro intestinal problems in patients with celiac disease could be reduced by expressing a prolyl endopeptidase in probiotic bacteria (Alvarez-Sieiro et al., Citation2014; Table ).
Moreover, as many lactic acid bacteria and bifidobacteria genomes have been recently sequenced, we can speculate which same genes that could be used to improve probiotic properties of probiotic strains. Also research on human/animal microbiota has supplied many important information concerning probiotic strains functions.
Up to the present we are not aware of any probiotic bacteria that have been genetically modified and that can be used for probiotic aims in humans. What, then, would be the outcome of the use of genetic engineering on probiotic micro-organisms? We could improve certain characteristics of probiotic bacteria in order to enhance their probiotic role, eliminate undesirable characteristics, and add new probiotic features of interest. We would have a greater number of probiotic strains, with more probiotic effects, which help us to improve and fight many more diseases.
Genetic engineering in probiotic bacteria and biosafety
Food-grade vectors and the integration of genes in the chromosome by site-specific recombination using the attP/integrase system or homologous recombination have been developed to meet the demands of a growing industrial sector to obtain GRAS recombinant products. Although any genetic manipulation of an organism creates a genetic modification of food quality (GMO), i.e. the modification of an organism with its own DNA or with the DNA from GRAS organisms, might not be as ill perceived as a non-food-grade genetic modification (Lu & Kong, Citation2013). Self-cloning, i.e. the re-introduction of DNA from a host that has been modified, or is closely related to the same species strain, was excluded from the European Union on the contained use of genetically modified micro-organisms (CCA-219, 1990) (de Vos, Citation1999). Lactic acid bacteria and bifidobacteria are recognized as GRAS micro-organisms by the US FDA. Thus, organisms that have been modified by self-cloning are not considered as GMOs but are considered safe and suitable for food applications.
The safe use of genetically modified probiotic strains requires the development of food-grade cloning systems composed solely of DNA from the homologous host or GRAS organisms and that condition do not rely on antibiotic markers (Landete, Citation2017). The rationale for the development of cloning vectors derived from lactic acid bacteria and bifidobacteria cryptic plasmids is on the need to achieve new food-grade genetic engineering tools. A strategy to construct vectors is to use the replicons of small cryptic plasmids and incorporate selectable markers. Many cryptic plasmids originally from lactic acid bacteria species, have been isolated and characterized (Shareck, Choi, Lee, & Miguez, Citation2004).
So far, only a few nucleotides have been modified by CRISPR system in lactic bacteria (Oh & van Pijkeren, Citation2014; van Pijkeren & Britton, Citation2012; van Pijkeren et al., Citation2012). However, the development of this technique would allow editing much longer gene sequences, allowing us to introduce new heterologous genes in lactic acid bacteria and bifidobacteria (Stefanovic, Fitzgerald, & McAuliffe, Citation2017). Organisms modified by CRISPR system are not considered as GMOs but are considered safe and suitable for application in food manufacture and human health.
Genetic engineering in probiotic bacteria for therapeutic applications
Modified lactic acid bacteria are used in dairy industry, in other additional types of food fermentations factories, additionally for protein production and for metabolic engineering. Genetically modified lactic acid bacteria have been used for the production of B-vitamins, diacetyl, acetaldehyde, and folate (Sybesma et al., Citation2003). However, the vectors are generally not food-grade. The productions of these metabolites or recombinant proteins do not require food-grade vectors when these metabolites are separated from the modified micro-organisms that produce them. Nonetheless, when they are co-administered to humans or animals these vectors must be of food-grade.
Most applications of food-grade vectors are found in health items. Lactic acid bacteria and bifidobacteria are acid resistant and able to adhere to the mucosal epithelium, therefore, lactic acid bacteria and bifidobacteria strains are good candidates for the development of oral vectors (Guo et al., Citation2015; Hoang et al., Citation2015; Landete, Citation2016). Moreover, these bacteria, that can administrated orally, could harbor genes expressing therapeutic molecule as interleukins (Steidler et al., Citation1998, 2000). Nevertheless, the strains used in these works are not probiotics and/or the vectors are not food-grade. So far, only a potentially probiotic strain Lactobacillus acidophilus NCK56 has been constructed with a deletion of phosphoglycerol transferase after double crossover integration and excision event, thereby eliminating antibiotic resistance (Mohamadzadeh et al., Citation2011). The lipoteichoic acid has a proinflammatory role and the ability of lipoteichoic acid deficient Lb. acidophilus to regulate inflammation and protect against colonic polyposis in a mouse mode was demonstrated by Khazaie et al. (Citation2013).
The majority of food-grade vectors for health are integrated into the chromosome, making them more stable vectors. So, examples of food-grade vectors in clinical application can be found. Martín et al. (Citation2011) describe a chromosomally integrated expression system in Lactobacillus paracasei based on the aggregation-promoting factor gene (apf) of Lactobacillus crispatus (Marcotte et al., Citation2004). Lb. paracasei produced antibodies directed against the rotavirus. Alvarez-Sieiro et al. (Citation2014) engineered a food-grade strain of Lb. casei to deliver Myxococcus xanthus prolyl endopeptidases into the gut environment. Steidler et al. (Citation2003) showed that L. lactis strains expressing and secreting murine interleukin-10 (IL-10) could be used to treat inflammation in mouse colitis models. L. lactis strains secreting human IL-10 was approved by the Dutch authorities for use in a small clinical trial as an experimental therapy for use in humans with inflammatory bowel diseases.
The works done by the Steidler`s group lead the way for the use of genetically modified micro-organisms in treating diseases or improves their symptoms. However, as above mentioned, up to the present we are not aware of any probiotic bacteria that had been genetically modified and that can be used for probiotic ends in humans. Then, the effort must be made in these probiotic bacteria. Although, L. lactis strains and other laboratory strains are easily transformed and their genetic manipulation is easier, efforts should be focused on really probiotic bacteria such as some strains of Lactobacillus and Bifidobacterium (Table ), by use of cryptic plasmids derived from lactic bacteria and bifidobacteria in which antibiotic resistance genes has been removed. Sometimes, the genes of interest can be selected from the probiotic bacteria and in concert with the correct choice of promoter, we would achieve the desired objective.
What problems are related to the use of genetic engineering in probiotic bacteria?
The very same problems arise with those related to food-grade vectors or integration by means of chromosomal recombination. Not all strains can be transformed, and a resistance mechanism must be chosen in order to select the modified strains in the case of food-grade vectors.
Plasmid incompatibility produces a reduction in transformation efficiencies, prevents the co-existence of some plasmids, and the transformants usually lose one or more of the indigenous plasmids (Gravesen, von Wright, Josephsen, & Vogensen, Citation1997). Lactic acid bacteria and bifidobacteria plasmids encode many of the genes with biotechnological properties and industrial strains contain a large number of plasmids, so plasmid loss is usually not acceptable in lactic acid bacteria and bifidobacteria strains with technological properties (Davies & Gasson, Citation1981).
Many industrial strains are nisin resistant, and selection systems based on nisin resistance cannot be used in these cases (Høier et al., Citation1999). When there is not antibiotic resistance, the use of cryptic plasmids is recommended, along with the chromosomal integration of the genes of interest to make them more stable (Gosalbes et al., Citation2000; Lin et al., Citation2013). Plasmids with a thermosensitive replicon could be used to counteract the high transformation efficiencies needed (Maguin, Duwat, Hege, Ehrlich, & Gruss, Citation1992; Song et al., Citation2014). At times, when a high level of expression of the gene of interest is required, it may be preferable to use replicative plasmids instead of chromosomal integration. In such a case, stable theta replication plasmids should be chosen (Emond, Lavallee, Drolet, & Moineau, Citation2001). The choice of the expression mechanism in food-grade vectors may also be of interest. In this way, we can choose promoters that produce a stronger or weaker constitutive expression, or inducible promoters such as the bile inducible promoters.
Finally, while it may seem that there is a rejection of genetically modified organisms, it is assumed that the achievement and development of probiotic strains with relevant and proven benefits, overshadow these rejections in the same way that everyone accepts the use of recombinant insulin.
Conclusions and perspectives
Through genetic engineering, the researchers can improve and increase the characteristics and properties of probiotic strains and then, doubts about probiotics would be eliminated. So far, the few potentially probiotic bacteria that have been genetically modified have not been applied in humans because of gene modifications include resistance to antibiotics. The risk of disseminating the genetic modification through lateral gene transfer is minimized because the gene is integrated in the chromosome of probiotic strains or by the use of food-grade vectors derived from lactic acid bacteria and/or bifidobacteria cryptic plasmids or by the use of CRISPR systems.
CRISPR is an emerging technique. Major efforts should be made in the improvement and application of this method. Sequencing of lactic acid bacteria and bifidobacteria helps a lot in the implementation of this technique for improving the probiotic strains.
We need to combine research into probiotics with genetic engineering, make advances in research into food-grade vectors, and in the creation of compatible vectors. Sequencing the genome of probiotic strains would allow us to detect areas that are susceptible to recombination. And, finally, we must improve the transformation of recalcitrant strains, which have probiotic interest.
In the future, research in biotechnology should clearly show the mechanisms of action of modified lactic acid bacteria and bifidobacteria at cellular and molecular levels in humans. Moreover, research focusing on the effects of modified lactic acid bacteria and bifidobacteria on the proteome and metabolome will be essential.
Future work will investigate further the application of these food-grade vectors in food and health since they would have enormous benefits for the health of individuals, their use in foods, as well as economic benefits.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by INIA, MINECO, Spain [project number RM2012-00004-00-00].
Acknowledgements
J.M. Landete has a postdoctoral contract supported by the research program ‘Ramón y Cajal’ (MINECO, Spain). The authors declare no conflict of interest.
References
- Allison, G.E., & Klaenhammer, T.R. (1996). Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Applied and Environmental Microbiology, 62, 4450–4460.
- Alvarez-Sieiro, P., Martin, M. C., Redruello, B., del Rio, B., Ladero, V., Palanski, B. A., … Alvarez, M. A. (2014). Generation of food-grade recombinant Lactobacillus casei delivering Myxococcus xanthus prolyl endopeptidase. Applied Microbiology and Biotechnology, 98, 6689–6700.10.1007/s00253-014-5730-7
- Benyacoub, J., Bosco, N., Blanchard, C., Demont, A., Philippe, D., Castiel-Higounenc, I., & Guéniche, A. (2014). Immune modulation property of Lactobacillus paracasei NCC2461 (ST11) strain and impact on skin defences. Beneficial Microbes, 5, 129–136.10.3920/BM2013.0014
- Berggren, A., Ahrén, I. L., Larsson, N., & Önning, G. (2011). Randomised, double-blind and placebo-controlled study using new probiotic lactobacilli for strengthening the body immune defence against viral infections. European Journal of Nutrition, 50, 203–210.10.1007/s00394-010-0127-6
- Bermúdez-Humarán, L. G., Aubry, C., Motta, J. P., Deraison, C., Steidler, L., Vergnolle, N., … Langella, P. (2013). Engineering lactococci and lactobacilli for human health. Current Opinion in Microbiology, 16, 278–283.10.1016/j.mib.2013.06.002
- Borges, S., Silva, J., & Teixeira, P. (2014). The role of lactobacilli and probiotics in maintaining vaginal health. Archives of Gynecology and Obstetris, 289, 479–489.10.1007/s00404-013-3064-9
- Canani, R. B., Cirillo, P., Terrin, G., Cesarano, L., Spagnuolo, M. I., De Vincenzo, A., … Guarino, A. (2007). Probiotics for treatment of accute diarrhoea in children: Randomised clinical trial of five different preparations. BMJ, 335, 340. doi:10.1136/bmj.39272.581736.55
- Chen, C.-C., Lin, W.-C., Kong, M.-S., Shi, H. N., Walker, W. A., Lin, C.-Y., … Lin, T.-Y. (2012). Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra-intestinal tissue. British Journal of Nutrition, 107, 1623–1634.10.1017/S0007114511004934
- Clarke, G., Cryan, J. F., Dinan, T. G., & Quigley, E. M. (2012). Review article: Probiotics for the treatment of irritable bowel syndrome–focus on lactic acid bacteria. Alimentary Pharmacology and Therapeutics, 35, 403–413.10.1111/j.1365-2036.2011.04965.x
- Davies, F. L., & Gasson, M. J. (1981). Genetics of lactic acid bacteria. Journal of Dairy Research, 48, 363–376.10.1017/S0022029900021798
- de Vos, W. M. (1999). Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria. International Dairy Journal, 9, 3–10.10.1016/S0958-6946(99)00038-2
- Ducrotté, P., Sawant, P., & Jayanthi, V. (2012). Clinical trial: Lactobacillus plantarum 299v (DSM 9843) improves symptoms of irritable bowel syndrome. World Journal of Gastroenterology, 18, 4012–4018.10.3748/wjg.v18.i30.4012
- El Demerdash, H. A. M., Heller, K. J., & Geis, A. (2003). Application of the shsp gene, encoding a small heat shock protein, as a food-grade selection marker for lactic acid bacteria. Applied and Environmental Microbiology, 69, 4408–4412.10.1128/AEM.69.8.4408-4412.2003
- Emond, E., Lavallee, R., Drolet, G., Moineau, S., & LaPointe, G. (2001). Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Applied and Environmental Microbiology, 67, 1700–1709.10.1128/AEM.67.4.1700-1709.2001
- Fitzpatrick, L. R. (2013). Probiotics for the treatment of Clostridium difficile associated disease. World Journal of Gastrointestinal Pathophysiology, 4, 47–52.10.4291/wjgp.v4.i3.47
- Foligné, B., Daniel, C., & Pot, B. (2013). Probiotics from research to market: The possibilities, risks and challenges. Current Opinion in Microbiology, 16, 284–292.10.1016/j.mib.2013.06.008
- Gao, X. W., Mubasher, M., Fang, C. Y., Reifer, C., & Miller, L. E. (2010). Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and clostridium difficile-associated diarrhea prophylaxis in adult patients. American Journal of Gastroenterology, 105, 1636–1641.10.1038/ajg.2010.11
- Glenting, J., Madsen, S. M., Vrang, A., Fomsgaard, A., & Israelsen, H. (2002). A plasmid selection system in Lactococcus lactis and its use for gene expression in L. lactis and human kidney fibroblasts. Applied and Environmental Microbiology, 68, 5051–5056.10.1128/AEM.68.10.5051-5056.2002
- Gosalbes, M. J., Esteban, C. D., Galán, J. L., & Pérez-Martínez, G. (2000). Integrative food-grade expression system based on the lactose regulon of Lactobacillus casei. Applied and Environmental Microbiology, 66, 4822–4828.10.1128/AEM.66.11.4822-4828.2000
- Gravesen, A., von Wright, A., Josephsen, J., & Vogensen, F. K. (1997). Replication regions of two pairs of incompatible Lactococcal theta-replicating plasmids. Plasmid, 38, 115–127.10.1006/plas.1997.1302
- Groeger, D., O’Mahony, L., Murphy, E. F., Bourke, J. F., Dinan, T. G., Kiely, B., … Quigley, E. M. M. (2013). Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes, 4, 325–329.10.4161/gmic.25487
- Guo, S., Yan, W., McDonough, S. P., Lin, N., He, H., Xianga, M., … Chang, Y.-F. (2015). The recombinant Lactococcus lactis oral vaccine induces protection against C. difficile spore challenge in a mouse model. Vaccine, 33, 1586–1595.10.1016/j.vaccine.2015.02.006
- Hoang, P. M., Cho, S., Kim, K. E., Byun, S. J., Lee, T.-K., & Lee, S. (2015). Development of Lactobacillus paracasei harboring nucleic acid-hydrolyzing 3D8 scFv as a preventive probiotic against murine norovirus infection. Applied Microbiology and Biotechnology, 99, 2793–2803.10.1007/s00253-014-6257-7
- Høier, E., Janzen, T., Henriksen, C. M., Rattray, F., Brockmann, E., & Johansen, E. (1999). The production, application and action of lactic cheese starter cultures. In B. Law (Ed.), The Technology of Cheesemaking (Vol. 99, pp. 99–131). Sheffield: Academic Press.
- Iqbal, M. Z., Qadir, M. I., Hussain, T., Janbaz, K. H., Khan, Y. H., & Ahmad, B. (2014). Review: Probiotics and their beneficial effects against various diseases. Pakistan Journal of Pharmaceutical Science, 27, 405–415.
- Jiang, W., Bikard, D., Cox, D., Zhang, F., & Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology, 31, 233–239.10.1038/nbt.2508
- Khazaie, K., Zadeh, M., Khan, M. W., Bere, P., Gounari, F., Dennis, K., … Mohamadzadeh, M. (2013). Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Procedings of the National Academy of Science, 109, 10462–10467.
- Kojic, M., Jovcic, B., Strahinic, I., Begovic, J., Lozo, J., Veljovic, K., & Topisirovic, L. (2011). Cloning and expression of a novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiology, 11, 265.10.1186/1471-2180-11-265
- Kumar, R., Rajkumar, H., Kumar, M., Varikuti, S. R., Athimamula, R., Shujauddin, M., … Kondapalli, N. (2013). Molecular cloning, characterization and heterologous expression of bile salt hydrolase (Bsh) from Lactobacillus fermentum NCDO394. Molecular Biology Reports, 40, 5057–5066.10.1007/s11033-013-2607-2
- Landete, J. M. (2016). Effector molecules and regulatory proteins: Applications. Trends in Biotechnology, 34(10), 777–780.10.1016/j.tibtech.2016.04.011
- Landete, J. M. (2017). A review of food-grade cloning vectors in lactic acid bacteria: From the laboratory to their application. Critical Review in Biotechnology, 37, 296–308. doi:10.3109/07388551.2016.1144044
- LeBlanc, J. G., Aubry, C., Cortes-Perez, N. G., de Moreno de LeBlanc, A., Vergnolle, N., Langella, P., … Bermúdez-Humarán, L. G. (2013). Mucosal targeting of therapeutic molecules using genetically modified lactic acid bacteria: An update. FEMS Microbiology Letters, 344, 1–9.10.1111/femsle.2013.344.issue-1
- Leenhouts, K., Bolhuis, A., Venema, G., & Kok, J. (1998). Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Applied Microbiology and Biotechnology, 49, 417–423.10.1007/s002530051192
- Li, H.-S., Piao, D.-C., Jiang, T., Bok, J.-D., Cho, C.-S., Lee, Y.-S., … Choi, Y.-J. (2015). Recombinant interleukin 6 with M cell-targeting moiety produced in Lactococcus lactis IL1403 as a potent mucosal adjuvant for peroral immunization. Vaccine, 33, 1959–1967.10.1016/j.vaccine.2015.02.061
- Li, R., Takala, T. M., Qiao, M., Xu, H., & Saris, P. E. J. (2011). Nisin-selectable food-grade secretion vector for Lactococcus lactis. Biotechnology Letters, 33, 797–803.10.1007/s10529-010-0503-6
- Lin, C.-F., Lo, T.-C., Kuo, Y.-C., & Lin, T.-H. (2013). Stable integration and expression of heterologous genes in several lactobacilli using an integration vector constructed from the integrase and attP sequences of phage ΦAT3 isolated from Lactobacillus casei ATCC 393. Applied Microbiology and Biotechnology, 97, 3499–3507.10.1007/s00253-012-4393-5
- Liu, C.-Q., Leelawatcharamas, V., Harvey, M. L., & Dunn, N. W. (1996). Cloning vectors for Lactococci based on a plasmid encoding resistance to cadmium. Current Microbiology, 33, 35–39.10.1007/s002849900070
- Liu, C.-Q., Su, P., Khunajak, N., Deng, Y.-M., Sumual, S., Kim, W. S., … Dunn, N. W. (2005). Development of food-grade cloning and expression vectors for Lactococcus lactis. Journal of Applied Microbiology, 98, 127–135.10.1111/jam.2005.98.issue-1
- Lu, W., Kong, J., & Kong, W. (2013). Construction and application of a food-grade expression system for Lactococcus lactis. Molecular Biotechnology, 54, 170–176.10.1007/s12033-012-9558-z
- Maguin, E., Duwat, P., Hege, T., Ehrlich, S. D., & Gruss, A. (1992). New thermosensitive plasmid for gram-positive bacteria. Journal of Bacteriology, 174, 5633–5638.10.1128/jb.174.17.5633-5638.1992
- Marcotte, H., Ferrari, S., Cesena, C., Hammarström, L., Morelli, L., Pozzi, G., & Oggioni, M. R. (2004). The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. Journal of Applied Microbiology, 97, 749–756.10.1111/jam.2004.97.issue-4
- Martín, M. C., Alonso, J. C., Suárez, J. E., & Alvarez, M. A. (2000). Generation of food-grade recombinant lactic acid bacterium strains by site-specific recombination. Applied and Environmental Microbiology, 66, 2599–2604.10.1128/AEM.66.6.2599-2604.2000
- Martín, M. C., Pant, N., Ladero, V., Günaydın, G., Andersen, K. K., Álvarez, B., … Marcotte, H. (2011). Integrative expression system for delivery of antibody fragments by lactobacilli. Applied and Environmental Microbiology, 77, 2174–2179.10.1128/AEM.02690-10
- Martinez, R. C., Sensey, S. L., Summers, K. L., Nomizo, A., De Martinis, E. C., & Reid, G. (2009). Effect of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on the ability of Candida albicans to infect cells and induce inflammation. Microbiology and Immunology, 53, 487–495.10.1111/mim.2009.53.issue-9
- Mohamadzadeh, M., Pfeiler, E. A., Brown, J. B., Zadeh, M., Gramarossa, M., Managlia, E., … Klaenhammer, T. R. (2011). Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proceding of the National Academy of Science, 108, 4623–4630.10.1073/pnas.1005066107
- Oh, J.-H., & van Pijkeren, J.-P. (2014). CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Research, 42(17), e131. doi:10.1093/nar/gku623
- Peterbauer, C., Maischberger, T., & Haltrich, D. (2011). Food-grade gene expression in lactic acid bacteria. Biotechnology Journal, 6, 1147–1161.10.1002/biot.201100034
- Platteuw, C., van Alen-Boerringter, I., van Schalkwuk, S., & de Vos, W. M. (1996). Food-grade cloning and expression system for Lactococcus lactis. Applied and Environmental Microbiology, 62, 1008–1013.
- Reid, G., Jass, J., Sebulsky, M. T., & McCormick, J. K. (2003). Potential uses of probiotics in clinical practice. Clinical Microbiology Reviews, 16, 658–672.10.1128/CMR.16.4.658-672.2003
- Rondanelli, M., Giacosa, A., Faliva, M. A., Perna, S., Allieri, F., & Castellazzi, A. M. (2015). Review on microbiota and effectiveness of probiotics use in older. World Journal of Clinical Cases, 3, 156–162.10.12998/wjcc.v3.i2.156
- Ross, P., O’Gara, F., & Condon, S. (1999). Thymidilate synthetase gene from Lactococcus lactis as a gene marker: An alternative to antibiotic resistance genes. Applied and Environmental Microbiology, 52, 2164–2169.
- Sander, J. D., & Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology, 32, 347–355.10.1038/nbt.2842
- Sgouras, D. N., Panayotopoulou, E. G., Martinez-Gonzalez, B., Petraki, K., Michopoulos, S., & Mentis, A. (2005). Lactobacillus johnsonii La1 attenuates Helicobacter pylori-associated gastritis and reduces levels of proinflammatory chemokines in C57BL/6 mice. Clinical and Diagnostic Laboratory Immunology, 12, 1378–1386.
- Shareck, J., Choi, Y., Lee, B., & Miguez, C. B. (2004). Cloning vectors based on cryptic plasmids isolated from lactic acid bacteria:Their characteristics and potential applications in biotechnology. Critical Review in Biotechnology, 24, 155–208.10.1080/07388550490904288
- Shi, S.-H., Yang, W.-T., Yang, G.-L., Cong, Y.-L., Huang, H.-B., Wang, Q., … Li, Y. (2014). Immunoprotection against influenza virus H9N2 by the oral administration of recombinant Lactobacillus plantarum NC8 expressing hemagglutinin in BALB/c mice. Virology, 464–465, 166–176.10.1016/j.virol.2014.07.011
- Song, L., Cui, H., Tang, L., Qiao, X., Liu, M., Jiang, Y., … Li, Y. (2014). Construction of upp deletion mutant strains of Lactobacillus casei and Lactococcus lactis based on counterselective system using temperature-sensitive plasmid. Journal of Microbiological Methods, 102, 37–44.10.1016/j.mimet.2014.04.011
- Stefanovic, E., Fitzgerald, G., & McAuliffe, O. (2017). Advances in the genomics and metabolomics of dairy lactobacilli: A review. Food Microbiology, 61, 33–49.10.1016/j.fm.2016.08.009
- Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., … Remaut, E. (2000). Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science, 289, 1352–1355.10.1126/science.289.5483.1352
- Steidler, L., Neirynck, S., Huyghebaert, N., Snoeck, V., Vermeire, A., Goddeeris, B., … Remaut, R. (2003). Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nature Biotechnology, 21, 785–789.10.1038/nbt840
- Steidler, L., Robinson, K., Chamberlain, L., Schofield, K. M., Remaut, E., Le Page, R. W., & Wells, J. M. (1998). Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infection and Immunity, 66, 3183–3189.
- Sybesma, W., Starrenburg, M., Kleerebezem, M., Mierau, I., de Vos, W. M., & Hugenholtz, J. (2003). increased production of folate by metabolic engineering of Lactococcus lactis. Applied and Environmental Microbiology, 69, 3069–3076.10.1128/AEM.69.6.3069-3076.2003
- Szajewska, H., Gyrczuk, E., & Horvath, A. (2013). Lactobacillus reuteri DSM 17938 for the management of infantile colic in breastfed infants: A randomized, double-blind, placebo-controlled trial. The Journal of Pediatrics, 162, 257–262.10.1016/j.jpeds.2012.08.004
- Valeur, N., Engel, P., Carbajal, N., Connolly, E., & Ladefoged, K. (2004). Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Applied and Environmental Microbiology, 70, 1176–1181.10.1128/AEM.70.2.1176-1181.2004
- van Pijkeren, J.-P., & Britton, R. A. (2012). High efficiency recombineering in lactic acid bacteria. Nucleic Acids Research, 40(10), e76. doi:10.1093/nar/gks147
- van Pijkeren, J.-P., Neoh, K. M., Sirias, D., Findley, A. S., & Britton, R. A. (2012). Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered, 3(4), 209–217.10.4161/bioe.21049
- Walker, S. A., & Klaenhammer, T. R. (1998). Molecular characterization of phage inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage 31. Journal of Bacteriology, 180, 921–931.
