Abstract
This study exploits the virulent bacteriophages phi 6 (dsRNA) and MS2 (ssRNA) as surrogates for airborne RNA viruses. Two different filter types, polytetrafluoroethylene (PTFE) and polycarbonate (PC), were tested for their efficiency in collecting aerosolized RNA phages. Two commercial kits were tested for total RNA isolation. Also, heat shock treatments were performed in three different media to obtain the most favorable conditions for reverse transcription assays of dsRNA. Our findings suggest that PC filters are more suitable to recover infectious airborne RNA viruses as determined by plaque assays. Both types of filters were equally efficient in recovering RNA from aerosolized phage phi 6 as established by qRT-PCR. Viral samples should be treated with QIAamp Viral RNA Mini Kit and a 5 min heat shock treatment at 110°C in TE buffer before RT-PCR to maximize detection of phage phi 6. Overall, the infectivity of the recovered phages was severely affected by the aerosolization/air sampling process and the presence of RNA viruses in air samples should be determined by qRT-PCR.
INTRODUCTION
RNA viruses such as influenza viruses, severe acute respiratory syndrome coronavirus (SARS-CoV), and respiratory syncytial virus (RSV) are leading causes of respiratory illnesses and infectious disease-related deaths amongst young children and the elderly worldwide (WHO 2009). Airborne transmission of these viruses often occurs but this phenomenon is still misunderstood. A problem encountered when studying bioaerosol transmission is the hazards to the laboratory staff while manipulating pathogenic strains. Therefore, the use of proper surrogates could reduce the safety risks and facilitate in-depth investigations.
Phages pose no significant risk to humans, they are easy to propagate in laboratory and their similarities with eukaryote viruses make them interesting models for aerovirology research. For these reasons, the coliphage MS2 (CitationGolmohammadi et al. 1993), a small non-enveloped bacterial virus (Leviviridae family) with a capsid diameter of 25 nm and a genome made of a single-stranded RNA (ssRNA) molecule (3569 nucleotides), has been used as an airborne viral model in many aerosol studies (CitationBarker and Jones 2005; CitationBurton et al. 2007; CitationEninger et al. 2009; CitationGrinshpun et al. 2007; CitationHogan et al. 2005; CitationHogan et al. 2006; CitationHogan et al. 2004; CitationHolton and Webb 1994; CitationKettleson et al. 2009; CitationLi et al. 2009; CitationPerrott et al. 2009; CitationTseng and Li, 2005; Citation2006; CitationWalker and Ko, 2007; CitationWang and Brion 2007). The Pseudomonas syringae virulent phage phi 6 is another well characterized bacterial virus that has some interesting features as a surrogate for RNA viruses, although it has been used only sporadically in aerosol studies (CitationEllis and Schlegel 1974). This virion is spherical and approximately 85 nm in diameter (Cystoviridae family). It possesses a lipid-containing envelope exterior to nucleocapsid (CitationEllis and Schlegel 1974) as well as a double-stranded RNA (dsRNA) genome separated into three segments of 6354 bp (segment L, (CitationMindich et al. 1988)), 4063 bp (segment M, (CitationGottlieb et al. 1988)), and 2948 bp (segment S, (CitationMcGraw et al. 1986)). Overall, it has similar structural organization with other dsRNA viruses such as rotaviruses and reoviruses (CitationBamford et al. 2002; CitationPoranen and Tuma 2004), as well as with some ssRNA viruses (HIV and hepatitis C) (CitationButcher et al. 2001).
Detection of airborne viral particles is greatly influenced by three main factors: the concentration of airborne viruses, the efficiency of the air-sampling system to recover airborne particles, and the sensitivity of the diagnostic assays used to detect the target in the sample (CitationHermann et al. 2006). Therefore, protocol optimization based on these factors is crucial for better environmental sampling analysis, especially when encountering low viral concentrations.
A wide variety of air samplers using various physical proprieties are available for recovering airborne viruses and they include cyclone samplers, liquid impingers, slit samplers, electrostatic precipitators, and filters (CitationVerreault et al. 2008). No perfect system is available as they have each their advantages and pitfalls. For example, filters are attractive due to their high efficiency in trapping particles with an aerodynamic size of less than 500 nm (CitationVerreault et al. 2008). Additionally, filters can be used to collect air samples for at least 40 h (CitationMyatt et al. 2003), thus enhancing the probability of recovering low viral concentrations. However, it was previously reported that filters are less efficient for recovering infectious viral particles (CitationTseng and Li 2005). Filter sampling coupled with PCR-based technology is considered a powerful method for detecting low concentrations of airborne viruses and is becoming highly recommended to evaluate bioaerosol contamination levels in different environments (CitationAintablian et al. 1998; CitationMcCluskey et al. 1996; CitationMyatt et al. 2003; CitationTsai et al. 2006). Consequently, the development of sensitive techniques for airborne viral detection should be a priority.
The aim of this study was to develop and optimize the detection of airborne RNA viruses using virulent phages MS2 and phi 6 as surrogates. First, the viral particle collection efficiencies of PTFE and PC filters were compared using qRT-PCR and plaque assays. Then, the sampling stress on the infectivity of two test phages was determined. The efficacy of Qiagen's QIAamp Viral RNA Mini Kit and Invitrogen's TRIzol® LS Reagent for total RNA extraction from viral sample was also examined. Finally to increase the detection limit, the effects of a heat shock treatment on dsRNA.
MATERIALS AND METHODS
Bacteriophages
Phage phi 6 (HER 102) and its bacterial host Pseudomonas syringae (HER 1102) were provided by the Felix d’Hérelle Reference Center for Bacterial Viruses (www.phage.ulaval.ca). Bacteriophage MS2 (ATCC 15597-B1) and its bacterial host Escherichia coli (ATCC 15597) were obtained from the American Type Culture Collection (ATCC). Phage MS2 was propagated at 37°C under agitation in flasks containing ATCC medium 271 and its E. coli host. The phage was added when OD600nm reached 0.1 and the incubation was carried on for 2 to 3 h until most cells had lysed. Cells and debris were removed from the phage lysate by centrifugation at 3500 RPM for 10 min at room temperature. The phage-containing supernatant was filtered (0.45 μm) and then kept at 4°C until use. For phage phi 6, 10-fold dilutions of the phage stock prepared in Tryptic Soy Broth (TSB) (Becton, Dickinson and Company, Sparks, MD) were mixed with an overnight (O/N) culture of P. syringae in TSB and soft agar (TSB supplemented with 0.7% agar), then the mixtures were poured on the surface of TSB plates (1.5% agar) and incubated overnight at 25°C. After incubation, the soft agar containing the phages was added to tubes containing 5 ml of TSB. Tubes were placed on a shaker adjusted for slow rotations for 20 min. Finally, centrifugation and filtration were performed as described for phage MS2. Phage amplifications produced approximately 1010 PFU per ml as determined by plaque assay (CitationAdams, 1959).
Test Filters
Two types of 37 mm filters were used, polytetrafluoroethylene filters with 0.3 μm porosity (PTFE; SKC Inc., Eighty Four, PA) and polycarbonate filters with 0.4 μm porosity (PC; SKC Inc.). The filters were mounted on cellulose support pads placed in 37 mm clear styrene 3-piece cassettes (SureSeal, SKC Inc.).
Aerosol Generation and Viral Particle Sampling
Aerosolization was performed in a customized SCL-GenaMINI chamber (SCL Medtech Inc., Montreal, QC, Canada). The experimental design for aerosol production and sampling was successfully used previously with DNA phages and is described elsewhere (CitationVerreault et al. 2010). A nebulizer (Single-Jet Atomizer, model 9302, TSI Inc., Shoreview, MN) generated the aerosols at a rate of 3 L/min. The nebulized solution was composed of 69 ml of phage buffer (Tris-HCl 10 mM, pH 7.4, NaCl 100 mM, MgSO410 mM) and 1 ml of amplified phage lysate. The final concentration in the nebulizer for each phage was approximately 108 PFU per ml. The liquid flow rate of the nebulized solution was of 0.15 ml/min. The generated aerosol passed through a desiccator (model 3062, TSI Inc., Shoreview, MN) to remove the excess of water before penetrating the chamber and to allow the formation of droplet nuclei. The desiccant blue beads (EMD Chemicals Inc., Gibbstown, NJ, Cat. # EM-DX0017-3) were used in the desiccator in order to eliminate unwanted background created by the original silica desiccant. While entering the chamber, the aerosol was diluted with HEPA-filtered dry air. The quantity of air added was calculated based on the quantity of air pumped out of the chamber. A total of 6 filters (3 PC and 3 PTFE) as well as an Aerodynamic Particle Sizer (APS; model 3321, TSI Inc.) were connected to the chamber. Each filter system was connected to a GILAIR-5 constant flow air sampling pump (Sensidyne Inc., Clearwater, FL) functioning at a rate of 2 L/min. Pumps were calibrated with a DryCal DC-1 Flow Calibrator (Bios International Corporation, Butler, NJ) before each experiment. The APS aspirated air at rate of 5 L/min and the total amount of aerosol pumped out of the chamber was 17 L/min. Consequently, the total volume of aerosol entering the chamber was 20 L/min (3 L/min generated by the nebulizer and 17 L/min by the HEPA filtered air). Simultaneously, the vacuum connected to the chamber aspirated automatically the excess air that could not be pumped out of the chamber so that the pressure inside the chamber would remain nil. The aerosolization and collection times were of 20 min and prior to aerosol collection, a period of 5 to 10 min was used to stabilize the aerosol generation, which was monitored with the APS.
Extraction of Viral Particles from Filters
Immediately after the viral particle sampling, 5 ml of TSB was deposited on each filter by removing the top part of the cassette and then closing it tight enough to avoid spills. While previous studies used sterile water with or without a detergent to elute the particles from the filters (CitationPalmgren et al. 1986; CitationTseng and Li, 2005), it was necessary to use TSB for phage phi 6 in order to recover evenly dispersed particles (data not shown). Then, the filters were placed on a shaker at maximum velocity for 20 min at room temperature (RT) to elute the phage particles from the filters.
Extraction Efficiency
After collecting airborne particles on filters, the extraction process plays an important role on the recovery efficiency of infectious particles and of RNA. To study the efficiency of the extraction step, 10 μl of a known quantity of viruses was deposited on both filter types and air-dried (for approx. 20 min) in fully mounted open-face cassettes. Then, the previously described extraction protocol (see previous section) was applied. Results of the air-dried particle recovery were compared to samples in which the air-drying step was omitted. In this case, the phages were not deposited on the membrane and air-dried but spiked into the 5 ml of TSB, and the cassettes were shaken. The phages were measured by plaque assays to count infectious units and by RT-qPCR to estimate RNA recovery. Furthermore, results were compared to liquid samples to determine the consequences of agitation on viral recovery. Experiments were performed in triplicate. The plaque assays were performed in triplicate and qRT-PCR in duplicate.
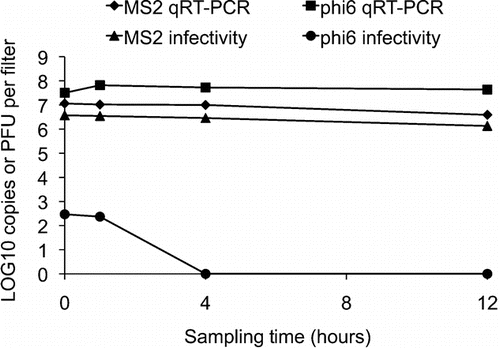
Impact of Sampling Stress on Phage Recovery
For this part of the study, PC filters were chosen as models. Sampling stress was evaluated by passing HEPA filtered air at 2 L/min through closed-face cassettes containing PC filters after a 20 min previous chamber sampling (see Aerosol generation and viral particle sampling section). Samples were taken at 0, 1, 4, and 12 h for analysis by plaque assays and qRT-PCR. Sampling stress on viral RNA was evaluated by comparing the quantity of viral genome copies (qRT-PCR) per filter at a given time divided by the quantity of viral genome copies per filter at time 0 (directly after the 20 min sampling period). Infectivity rate at each sampling time was also calculated by dividing the quantity of infectious phage (PFU) filter by the quantity of viral genome copies per filter at the same given time. Experiment was performed once and measurements were done in triplicates for plaque assays and duplicates for qRT-PCR.
RNA Extraction with the Qiagen's QIAamp Viral RNA Mini Kit
When using the QIAamp mini-columns, we noticed that the use of 5.6 μg of carrier RNA suggested by manufacturer's protocol (Qiagen Canada Inc., Mississauga, ON, Canada) decreased the recovery of viral RNA from the two phages used in this study, 5-fold for MS2 and more than 10-fold for phi 6 as compared to no carrier RNA (data not shown). Therefore, RNA was extracted from samples using the Qiagen's QIAamp Viral RNA Mini Kit but without adding carrier RNA to the AVL buffer. Briefly, 140 μl of each sample to be tested was used for the extraction and a double elution using 2 × 40 μl was done for maximum RNA recovery as recommended by the manufacturer.
RNA Extraction with the Invitrogen's TRIzol® LS Reagent
RNA extraction was performed according to the instructions provided with Invitrogen's TRIzol® LS Reagent (Invitrogen Inc., Burlington, ON, Canada). For better comparison with Qiagen's QIAamp Viral RNA Mini Kit, 140 μl of samples were used for RNA extraction and isolation. At the end of the procedure, RNA was vacuum-dried for 10 min before resuspending the RNA in 80 μl of IDTE buffer (Integrated DNA Technologies Inc., Coralville, IA).
cDNA Synthesis
Prior to cDNA synthesis, heat shock treatments were performed on phi 6 genome to denature the dsRNA segments. Samples were either used without any heat shock treatment or heated at 95°C or 110°C for 5 min before being placed on ice for 5 min. Different solutions used to resuspend phage RNA were also tested including RNase free Sigma water (Sigma Aldrich Inc., St-Louis, MO), AVE buffer (Qiagen's QIAamp Viral RNA Mini Kit's elution buffer) and IDTE buffer (10 mM Tris, pH 8, 0.1 mM EDTA). cDNA synthesis was achieved by using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Five μl of extracted RNA was used as a template for cDNA synthesis with 4 μl of 5x iScript Reaction Mix, 1 μl of iScript Reverse Transcriptase, and 10 μl of Nuclease-free water for a total volume of 20 μl. The reaction was performed according to the manufacturer's protocol.
Primers and Probes
The primers and probes used for this study were designed using the Beacon Designer 4.02 software (Premier Biosoft International, Palo Alto, CA). For phage MS2, the primers used were forward primer (5′-GTCCATACCTTAGATGCG TTAGC-3′, positions 1261–1284 in the genome of phage MS2), reverse primer (5′-CCGTTAGCGAAGTTGCTTGG-3′, positions 1401–1420), and dual labeled probe (5′-/56-FAM/
ACGTCGCCAGTTCCGCCATTGTCG/3BH, positions 1367-1391). The forward primer was within the mat gene coding for the maturation protein while the other two primers were targeting the cp gene coding for the coat structural protein. The amplified region corresponded to a cDNA fragment of 160 bp. For phage phi 6, the primers used were the forward primer (5′-TGGCGGCGGTCAAGAGC-3′, positions 430–446), reverse primer (5′-GGATGATTCTCCAGAAGCTGCTG-3′, positions 506–530), and dual labeled probe (5′-/5HEX/CGGTC GTCGCAGGTCTGACACTCGC/3BH, positions 450–475). All three primers targeted the phi-6S_1 gene (coding for the P8 protein) located on the S segment. The amplified region corresponded to a cDNA fragment of 232 bp.
qPCR
All PCR reactions were performed with the DNA Engine Opticon 2 Real-Time PCR Detection System (Bio-Rad Laboratories). Samples were analyzed using the Opticon Monitor Software version 2.02.24. The PCR reaction mix contained 1X final concentration of iQ Supermix (Bio-Rad Laboratories), 1 μM of forward and reverse primers, 150 nM of MS2 probe or 300 nM of phi 6 probe, and 2 μl of the desired cDNA template. The qPCR samples were adjusted to a final volume of 25 μl with sterile Sigma water. The amplification cycle was: 94°C for 3 min followed by 35 cycles of 94°C for 15 s and 60°C for 1 min with a plate read after the elongation step. All qPCR assays were performed in triplicates.
Construction of Plasmid Standards for qRT-PCR
PCR was performed on the cDNA of each viral sample with the same specific primers used in the qRT-PCR assays and with the same conditions as described above. The desired amplicon was purified using QIAquick PCR purification kit (Qiagen) and cloned into pCR4-TOPO with TOPO TA Cloning kit (Invitrogen). Transformation was done into One Shot (Invitrogen) chemically competent E. coli and plasmid DNA was purified using QIAprep Spin Miniprep Kit (Qiagen). Serial dilutions from the resulting clones were used as standard curves, each containing a known amount of input copy number.
Calculation of Viral Recovery
In order to compare the recovery efficiency of the filters, relative recovery (RR) parameters were used. RR was defined as the concentration of phages found on filters (PFU or genome copies per L of air) divided by the concentration of phages aerosolized (which is the product of the nebulizer liquid flow rate, 0.15 ml/min, and the PFU or genome copy concentrations in the nebulizer solution, divided by the total amount of aerosol generated after 20 min, i.e., 400 L). We defined RRp as the relative recovery determined by plaque assay and RRq as the relative recovery determined by qRT-PCR.
Statistical Analyses
Data were expressed using mean and 95% confidence intervals (CI) and analyzed using the two-way ANOVA. For viral recovery data, two experimental factors were defined, one associated to the comparison between the two viruses (MS2 vs. phi 6) and one associated to the comparison between the two filter types (PC vs. PTFE). For sampling stress, one factor was associated to the comparison between the two viruses and one associated to the comparison between the different sampling times (0–12 h). For RNA extraction data, the same statistical approach was used as one factor was linked to the viruses and one to the comparison of the two techniques (TRIzol® versus QIAamp). For heat shock measurements, one factor was associated to the media comparisons (water, AVE, and TE buffer) and the other to the temperatures (no heat shock, 95 and 110°C). All values were log transformed to stabilize variances and the reported P-values were based on theses transformations. Posteriori comparisons were performed using the Tukey's technique. The normality assumptions were verified with the Shapiro-Wilk tests. The Brown and Forsythe's variation of Levene's test was used to verify the homogeneity of variances. The results were considered significant with P-values ⩽0.05. All analyses were conducted using the statistical package SAS, version 9.1.3 (SAS Institute Inc, Cary, NC).
RESULTS
Characteristics of Airborne Viral-Laden Particles
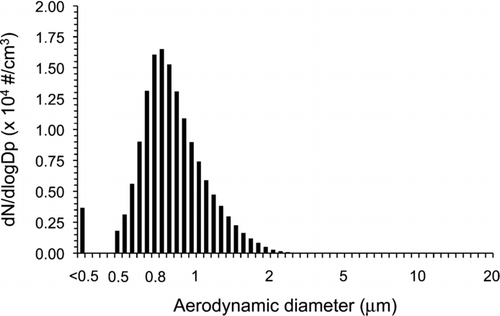
During duplicate experiments where the 20 min aerosol sampling was performed for filter comparison purposes, the mass median aerodynamic diameter (MMAD) of the dried viral-laden particles obtained after nebulization was 1.02 and 1.01 μm and the total concentrations were 6.37 × 103 and 6.29 × 103 particles/cm3 (). Each time, temperature and relative humidity (RH) inside the chamber were of 30 ± 1°C and 20 ±
5%, respectively. The quantity of phages in the nebulization medium was measured by plaque assays and qRT-PCR. Phage MS2 titer was 1.1 × 108 PFU per ml (95% CI: 8.8 × 107– 1.4 × 108), while phage phi 6 was 1.5 × 108 PFU per ml (95% CI: 1.5 × 108– 1.6 × 108). The corresponding infective aerosol generated inside the chamber, considering no aerosol loss before penetrating the chamber, was of 8.3 × 105 PFU per L of air for MS2 and of 1.1 × 106 for phi 6. Interestingly, the genome copies were much higher in the nebulization medium as determined by qRT-PCR than the plaque counts. For phage MS2, 1.4 × 109 genome copies (ssRNA) per ml (95% CI: 9.7 × 108– 1.9 × 109) were detected in the nebulization medium while 1.2 × 1010 copies (dsRNA) per ml (95% CI: 7.6 × 109– 1.6 × 1010) were detected for phage phi 6. The corresponding genome copies aerosolized inside the chamber were of 1.1 × 107 copies per L of air for MS2 and of 9.0 × 107 for phi 6. These data suggest that a significant number of phages are either not infectious in the lysate or that several copies of the phage genomes are not packaged during phage multiplication and are liberated in the lysate after cell lysis.
Determination of the Viral Recovery by Filters using qRT-PCR
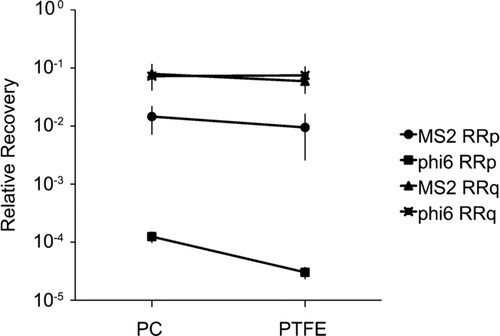
No statistical difference was observed for viral recovery when comparing RRq for the two filters and the two phages (P > 0.05, n = 6) (). Nonetheless for the recovery of phage MS2, PC filters were slightly more efficient (1.3-fold) than PTFE filters. Indeed, 8.3 × 105 (95% CI: 2.6 × 105– 1.4 × 106) viral genome copies per liter of air were quantified on PC filters while 6.2 × 105 (95% CI: 3.1 × 105– 9.2 × 105) genome copies per liter of air were found on PTFE filters. The recovery of phage phi 6 was virtually the same with both filters, 6.5 × 106 (95% CI: 3.5 × 106– 9.5 × 106) genome copies per liter of air were quantified on PC filters while 6.7 × 106 (95% CI: 3.2 × 106– 1.0 × 107) genome copies per liter of air were detected on PTFE filters.
Determination of the Viral Recovery by Filters using Plaque Assays
For phage MS2, PC filters recovered 1.5-fold more infectious bacterial viruses than PTFE filters (P < 0.05, n = 6) (). A total of 1.2 × 104 (95% CI: 1.6 × 103– 2.2 × 104) PFU per liter of air were recovered from PC filters while the average phage count was 7.8 × 103 (95% CI: 1.6 × 103– 1.7 × 104) PFU per liter of air on PTFE filters. By comparing the plaque assays results with qRT-PCR data, PC and PTFE filters recovered relatively the same amount of infectious MS2 particles (P > 0.05), 18% and 16%, respectively.
For phage phi 6, PC filters recovered 4-fold more infectious phages than PTFE filters (P < 0.05, n = 6) (). However, the phage counts were much lower with 1.4 × 102 (95% CI: 8.4 × 101– 2.0 × 102) PFU per liter of air with PC filters and 3.4 × 101 (95% CI: 1.8 × 101– 5.0 × 101) PFU/L air with PTFE filters. By comparing with the results obtained with the qRT-PCR, PC filters recovered significantly more (P < 0.05) infectious phi 6 particles. Indeed, PC filters recovered 0.17% of infectious particles whereas PTFE filters recovered only 0.04%.
TABLE 1 Ratios of RNA and infectious bacterial viruses recovery after 20 min of agitation in closed-face cassettes containing the phage mix in 5 ml of TSB
Extraction Efficiency After Agitation
In order to evaluate the effect of the sampling and extraction processes on phage recovery (without the input of the aerosolization process), filters were used to test the air-drying desiccation and agitation effects. Filters were spiked with known amounts of viruses, air-dried or not and agitated or not (liquid sample). The extraction efficiencies of the two tested filters in recovering total RNA and infectious phages after agitation are summarized in . For phage MS2, no statistical difference was observed between PC and PTFE filters for recovering infectious particles after 20 min of vigorous agitation of the closed-face cassettes containing 5 ml of TSB (P > 0.05, n = 6). We recovered over 85% of infectious bacterial viruses from both type of filters. A total of 2.1 × 105 (95% CI: 1.8 × 105– 2.3 × 105) PFU per ml of phage mix were recovered prior to agitation compared to 1.9 × 105 (95% CI: 1.7 × 105– 2.1 × 105) PFU per ml on PC filters and 1.8 × 105 (95% CI: 1.7 × 105– 2.0 × 105) PFU per ml on PTFE filters after agitation. For RNA recovery from MS2, again no statistical difference was observed between PC and PTFE filters after agitation of the phage mix in closed-face cassettes. In fact, we recovered over 100% of viral genomes from both type of filters. A total of 1.0 × 107 (95% CI: 8.2 × 106– 1.2 × 107) copies per ml of phage mix were recovered prior to agitation compared to 1.4 × 107 (95% CI: 1.1 × 107– 1.6 × 107) copies per ml on PC filters and 1.0 × 107 (95% CI: 9.2 × 106– 1.1 × 107) copies per ml on PTFE filters after agitation.
For phage phi 6, we recovered over 100% of infectious bacterial viruses from PC filters while we recovered approximately 30% from PTFE filters after 20 min of vigorous agitation of the phage mix in closed-face cassettes containing 5 ml of TSB (P < 0.05, n = 6). A total of 5.6 × 105 (95% CI: 5.3 × 105– 6.0 × 105) PFU per ml of phage mix were recovered prior to agitation compared to 6.2 × 105 (95% CI: 6.0 × 105– 6.4 × 105) PFU per ml on PC filters and 1.5 × 105 (95% CI: 1.3 × 105– 1.7 × 105) PFU per ml on PTFE filters after agitation. For RNA recovery from phage phi 6, we recovered a much greater amount of RNA after agitation as compared to prior agitation. A total of 2.4 × 107 (95% CI: 2.1 × 107 – 2.7 × 107) copies per ml of phage mix were recovered prior to agitation compared to 7.4 × 107 (95% CI: 6.9 × 107 – 8.0 × 107) copies per ml on PC filters and 5.6 × 107 (95% CI: 4.2 × 107 – 7.0 × 107) copies per ml on PTFE filters after agitation.
TABLE 2 Ratios of RNA and infectious bacterial viruses recovery after desiccation of phage mix on filters and 20 min of agitation in closed-face cassettes containing 5 ml of TSB
Extraction Efficiency After Desiccation and Agitation
The extraction efficiencies of the two tested filters in recovering total RNA and infectious phages after desiccation and agitation are summarized in . For phage MS2, no statistical difference was observed between PC and PTFE filters for recovering infectious phage particles after desiccation and 20 min of agitation of the phages in cassettes containing 5 ml of TSB (P > 0.05, n = 6). We recovered approximately 30% of infectious phages with both filters. A total of 2.1 × 105 (95% CI: 1.8 × 105– 2.3 × 105) PFU per ml of phage mix were recovered prior desiccation and agitation compared to 5.4 × 104 (95% CI: 5.0 × 104– 5.8 × 104) PFU per ml on PC filters and 5.4 × 104 (95% CI: 5.1 × 104– 5.7 × 104) PFU per ml on PTFE filters after desiccation and agitation. Similarly, for total RNA recovery from MS2 phage, no statistical difference was observed between PC and PTFE filters after desiccation. Both filters recovered approximately 70% of total RNA. A total of 1.0 × 107 (95% CI: 8.2 × 106– 1.2 × 107) phage genome copies per ml of phage mix were recovered prior to desiccation and agitation compared to 7.3 × 106 (95% CI: 5.9 × 106– 8.7 × 106) copies per ml on PC filters and 7.3 × 106 (95% CI: 5.5 × 106– 9.0 × 106) copies per ml on PTFE filters.
For phage phi 6, PC filters recovered 4% of infectious bacterial viruses while PTFE filters recovered 0.09% after desiccation and agitation in closed-face cassettes. A total of 5.6 × 105 (95% CI: 5.3 × 105– 6.0 × 105) PFU per ml were recovered prior to desiccation and agitation while we recovered 2.0 × 104 (95% CI: 1.0 × 104– 3.0 × 104) PFU per ml on PC filters and 5.0 × 102 (95% CI: 3.0 × 102– 7.0 × 102) PFU per ml on PTFE filters after desiccation and agitation. As for the recovery of phi 6 RNA, we picked up 1.5-fold more phi 6 RNA with PTFE filters than with PC filters after desiccation and agitation in cassettes. Again, we recovered a much greater amount of RNA after desiccation and agitation as before desiccation and agitation (). A total of 2.4 × 107 (95% CI: 2.1 × 107– 2.7 × 107) copies per ml of phage mi× were recovered prior desiccation and agitation compared to 4.5 × 107 (95% CI: 3.2 × 107– 4.8 × 107) copies per ml on PC filters and 6.6 × 107 (95% CI: 4.4× 107– 8.9 × 107) copies per ml on PTFE filters.
Sampling Stress
shows the effect of the sampling stress on the two viruses. For phage MS2, there was a decrease of more than 60% in infectivity and also for RNA detection after 12 h of sampling pure air at 2 L/min through the filter. A total of 1.2 × 107 (95% CI: 8.1 × 106– 1.5 × 107) genome copies per filter were quantified at time 0 compared to 3.9 × 106 (95% CI: 2.9 × 106– 5.0 × 106) copies per filter at time 12 h. A total of 3.7 × 106 (95% CI: 3.0 × 106– 4.5 × 106) PFU per filter were quantified at time 0 as compared to 1.4 × 106 (95% CI: 1.1 × 106– 1.6 × 106) PFU per filter at time 12 h.
For phage phi 6, there was no decrease in RNA detection after 12 h of sampling pure air at 2 L/min through the filter (P > 0.05, n = 2). A total of 3.2 × 107 (95% CI: 2.9 × 107– 3.5 × 107) genome copies per filter were quantified at time 0 compared to 6.9 × 107 (95% CI: 6.3 × 107– 7.6 × 107) copies per filter at time 1 h and 4 × 107 (95% CI: 3 × 107– 6 × 107) copies per filter at time 24 h. However, we observed a significant loss of infectivity with this phage (). When comparing the total PFU obtained per filter to the total copies of phage genomes per filter, only 0.0009% of phi 6 particles were infectious at time 0 and no infectious phi 6 particles were detected after 4 h of sampling stress. This suggests that the aerosolization and the 20 min sampling already significantly damaged the virus.
Comparison of RNA Extraction Methods
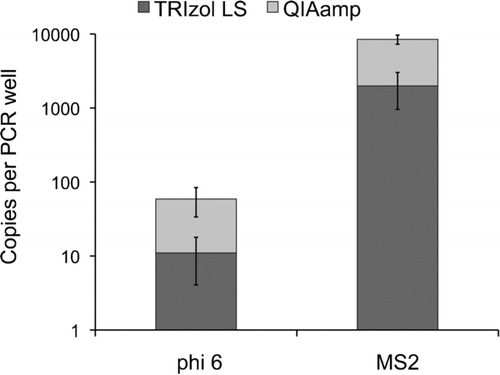
QIAamp mini-columns recovered 3.8-fold more MS2 RNA and 3.5-fold more phi 6 RNA than the TRIzol® LS reagent (P < 0.05, n = 6) (). For phage MS2, 6.5 × 103 (95% CI: 4.1 × 103– 8.9 × 103) copies per PCR well were quantified by qRT-PCR using QIAamp mini-columns while 1.7 × 103 (95% CI: 7.1 × 102– 4.1 × 103) copies per PCR well was detected using the TRIzol® LS reagent. For phage phi 6, 3.8 × 101 (95% CI: 1.1 × 101– 6.5 × 101) copies per PCR well were quantified using QIAamp mini-columns as compared to 1.1 × 101 (95% CI: 2.9 × 100– 2.5 × 101) copies per PCR well when using TRIzol® LS reagent.
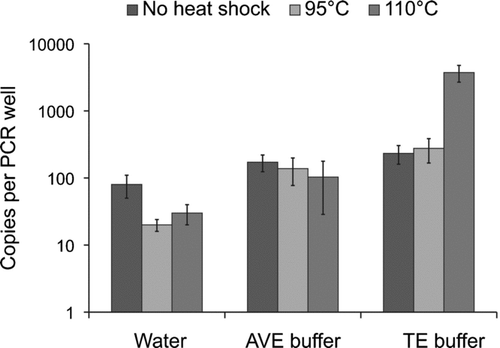
Heat Shock Treatments
For phage phi 6, a 5 min heat shock treatment of 110°C in TE buffer was the optimal condition to denature dsRNA prior to reverse transcriptase (). At this condition, detection was increased 14-fold and 20-fold compared to the 95°C heat shock and the treatment without heat shock in TE buffer (P < 0.05, n = 3). When comparing the optimal condition to AVE buffer and water without heat shock treatment, the detection was increased 36-fold and 46-fold, respectively (P < 0.05).
DISCUSSION
Comparison of PC and PTFE Filters for Airborne Viral Recovery
In a previous study (CitationVerreault et al. 2010), two DNA-containing phages (phiX174/ssDNA and P008/dsDNA) underwent the same aerosolization and sampling process to compare the efficiency of PC and PTFE filters. The qPCR and plaque assay results demonstrated superior sampling/elution efficiency with PC filters as compared to PTFE filters for both types of DNA phages. In this study, our qRT-PCR results did not show any statistical differences between PC and PTFE filters except for a slight tendency for PC filters to increase the recovery of MS2 particles. However, we recovered much more RNA from phage phi 6 after agitation of the samples during the elution process.
When using plaque assays to monitor airborne viral recovery, PC filters recovered more infectious particles than PTFE filters (1.5-fold and 4-fold for MS2 and phi 6, respectively). However, we observed a significant loss of infectivity that was caused by the elution method. This is likely due to the structure of the filters. In a previous study (CitationVerreault et al. 2010), scanning electron micrographs showed that PTFE filters have complex microscopic structures where phages could be trapped or broken during the elution step. In contrast, PC filters have a smooth surface that could be less damageable to viruses. Furthermore, other filter properties are known to affect the physical recovery of viral aerosols such as the filter's surface electrostatic charge (CitationLi et al. 2009) or whether the material is hydrophobic (PTFE) or hydrophilic (PC) (CitationReitz 2005). However, our qRT-PCR results show no significant differences between the two tested filters for the airborne viral recovery of the two tested bacteriophages suggesting that the overall particle collection efficiency is not significantly altered by the composition of the filter when using our experimental settings (RH, temperature, MMAD of particles and sampling time).
Comparison between qPCR and Plaque Assays
When comparing relative recoveries using qRT-PCR and plaque assay, more than 80% of MS2 virions and more than 99.8% for phage phi 6 lost infectivity after aerosolization and sampling. Similar observations were made when analyzing airborne DNA phages (CitationVerreault et al. 2010). One possible explanation could be the presence of phage aggregates that could lead to under estimation of phage titers. The low recovery of infectious phi 6 particles after aerosolization is comparable with results obtained by others (CitationTseng and Li, 2005). It is likely that the fragile lipid containing envelope of phage phi 6, necessary for infecting the bacterial host (CitationOjala et al. 1990), is sensitive to the aerosolization. In fact, the sampling stress assays confirmed that phage phi 6 was significantly affected by this process.
qRT-PCR Optimization
While the two RNA extraction methods performed in this study use guanidine isothiocyanate to denature proteins and liberate viral RNA, Qiagen's QIAamp Viral RNA Mini Kit uses silicon binding column-based method to recover RNA whereas Invitrogen's TRIzol® LS Reagent uses organic solvents. A previous study comparing a TRIzol® -chloroform based method to the QIAamp viral RNA columns with the use of carrier RNA showed that the TRIzol® -chloroform extraction recovered 2.5 times more influenza viral RNA (CitationFabian et al. 2009). Our results showed that the use of Qiagen's QIAamp Viral RNA Mini Kit without carrier RNA led to greater quantities of RNA than Invitrogen's TRIzol® LS Reagent (3.8- and 3.5-fold more for MS2 and phi 6, respectively). For this reason, the use of carrier RNA should carefully be examined before application with any diagnostic system. Of note, the total RNA isolated from phi 6 could be increased by 2- to 3-fold when agitating filled-cassette for 20 min prior to using QIAamp viral RNA columns.
Others have also previously used heat shock treatments at 95°C to denature viral dsRNA (CitationMittal et al. 2005; CitationRimstad et al. 1990) while others opted for 100°C and above (CitationDavis and Boylet, 1990; CitationEtten et al. 1974). Our results showed that it was better to heat RNA samples to 110°C instead of 95°C to get a more sensitive and accurate quantification of phage phi 6 dsRNA by qRT-PCR. Moreover, the melting process of dsRNA is known to be dependent on the ionic strength of the medium (CitationSteger et al. 1980). In our case, TE buffer was the best solution among the three tested.
CONCLUSIONS
We have used two non-pathogenic viruses, phages phi 6 and MS2, as surrogates for studying airborne RNA pathogenic viruses. Our results support the use of PCR-based technology to increase the sensitivity of viral detection in air samples. The choice of filter type should depend on the type of virus to sample and the method used to quantify viral particles. A protocol using a commercial kit and a heat shock treatment was also optimized to extract phage RNA from aerosolized samples. The method described in this article for detecting airborne RNA viruses could be applicable to other RNA viruses in different types of environment and could help answer questions related to their airborne transmission and risk assessment either in industrial settings as well as indoors and outdoors.
Acknowledgments
We express appreciations to Serge Simard for statistical analysis and to Geneviève Rousseau for technical advice. L.G. received a Canadian Center for Health and Safety in Agriculture (CCHSA) summer training award. C.D. received funding from the Natural Sciences and Engineering Research Council (NSERC) of Canada. C.D. is also the recipient of an FRSQ Junior 2 scholarship as well as CCHSA Time-release support. S.M. and C.D are members of the FRSQ Respiratory Health network. This work was funded by a collaborative health research project grant from NSERC of Canada to S.M and C.D.
REFERENCES
- Adams , M. H. 1959 . Bacteriophages , New York : Interscience Publishers .
- Aintablian , N. , Walpita , P. and Sawyer , M. 1998 . Detection of Bordetella Pertussis and Respiratory Syncytial Virus in Air Samples from Hospital Rooms . Infection Control and Hospital Epidemiology. , 19 : 918 – 923 .
- Bamford , D. H. , Burnett , R. M. and Stuart , D. I. 2002 . Evolution of Viral Structure . Theoretical Population Biology. , 61 : 461 – 470 .
- Barker , J. and Jones , M. V. 2005 . The Potential Spread of Infection Caused by Aerosol Contamination of Surfaces after Flushing a Domestic Toilet . J. Applied Microbiology. , 99 : 339 – 347 .
- Burton , N. C. , Grinshpun , S. A. and Reponen , T. 2007 . Physical Collection Efficiency of Filter Materials for Bacteria and Viruses . Ann. Occup. Hyg. , 5 : 143 – 151 .
- Butcher , S. J. , Grimes , J. M. , Makeyev , E. V. , Bamford , D. H. and Stuart , D. I. 2001 . A Mechanism for Initiating RNA-Dependent RNA Polymerization . Nature. , 410 : 235 – 240 .
- Davis , V. S. and Boylet , J. A. 1990 . Adapting the Polymerase Chain Reaction to a Double-Stranded RNA Genome . Analytical Biochemistry. , 189 : 30 – 34 .
- Ellis , L. F. and Schlegel , R. A. 1974 . Electron Microscopy of Pseudomonas φ 6 Bacteriophage . J. Virology. , 14 : 1547 – 1551 .
- Eninger , R. M. , Hogan , C. J. , Biswas , P. , Adhikari , A. , Reponen , T. and Grinshpun , S. A. 2009 . Electrospray vs. Nebulization for Aerosolization and Filter Testing with Bacteriophage Particles . Aerosol Sci. Technol. , 43 : 298 – 304 .
- Etten , J. L. V. , Vidaver , A. K. , Koski , R. K. and Burnett , J. P. 1974 . Base Composition and Hybridization Studies of the Three Double-Stranded RNA Segments of Bacteriophage φ 6 . J. Virology. , 13 : 1254 – 1262 .
- Fabian , P. , McDevitt , J. J. , Lee , W. M. , Houseman , E. A. and Milton , D. K. 2009 . An Optimized Method to Detect Influenza Virus and Human Rhinovirus from Exhaled Breath and the Airborne Environment . J. Environ. Monitoring. , 11 : 314 – 317 .
- Golmohammadi , R. , ValegÂrd , K. , Fridborg , K. and Liljas , L. 1993 . The Refined Structure of Bacteriophage MS2 at 2.8 A Resolution . J. Molecular Biology. , 234 : 620 – 639 .
- Gottlieb , P. , Metzger , S. , Romantschuk , M. , Carton , J. , Strassman , J. , Bamford , D. H. , Kalkkinen , N. and Mindich , L. 1988 . Nucleotide Sequence of the Middle dsRNA Segment of Bacteriophage Phi 6: Placement of the Genes of Membrane-Associated Proteins . Virology. , 163 : 183 – 190 .
- Grinshpun , S. A. , Adhikari , A. , Honda , T. , Kim , K. Y. , Toivola , M. , Rao , K. S. R. and Reponen , T. 2007 . Control of Aerosol Contaminants in Indoor Air: Combining the Particle Concentration Reduction with Microbial Inactivation . Environ. Sci. Technol. , 41 : 606 – 612 .
- Hermann , J. R. , Hoff , S. J. , Yoon , K. J. , Burkhardt , A. C. , Evans , R. B. and Zimmerman , J. J. 2006 . Optimization of a Sampling System for Recovery and Detection of Airborne Porcine Reproductive and Respiratory Syndrome Virus and Swine Influenza Virus . Applied and Environ. Microbiol. , 72 : 4811 – 4818 .
- Hogan , C. J. , Kettleson , E. M. , Lee , M.-H. , Ramaswami , B. , Angenent , L. T. and Biswas , P. 2005 . Sampling Methodologies and Dosage Assessment Techniques for Submicrometre and Ultrafine Virus Aerosol Particles . J. Applied Microbiol. , 99 : 1422 – 1434 .
- Hogan , C. J. , Kettleson , E. M. , Ramaswami , B. , Chen , D. R. and Biswas , P. 2006 . Charge Reduced Electrospray Size Spectrometry of Mega- and Gigadalton Complexes: Whole Viruses and Virus Fragments . Anal. Chem. , 78 : 844 – 852 .
- Hogan , C. J. , Lee , M. H. and Biswas , P. 2004 . Capture of Viral Particles in Soft X-Ray-Enhanced Corona Systems: Charge Distribution and Transport Characteristics . Aerosol Sci. Technol. , 38 : 475 – 486 .
- Holton , J. and Webb , A. R. 1994 . An Evaluation of the Microbiol Retention Performance of 3 Venilator-Circuit Filters . Intensive Care Medicine. , 20 : 233 – 237 .
- Kettleson , E. M. , Ramaswami , B. , Hogan , C. J. , Lee , M. H. , Statyukha , G. A. , Biswas , P. and Angenent , L. T. 2009 . Airborne Virus Capture and Inactivation by an Electrostatic Particle Collector . Environ. Sci. Technol. , 43 : 5940 – 5946 .
- Li , H.-W. , Wu , C.-Y. , Tepper , F. , Lee , J.-H. and Lee , C. N. 2009 . Removal and Retention of Viral Aerosols by a Novel Alumina Nanofiber Filter . J. Aerosol Sci. , 40 : 65 – 71 .
- McCluskey , R. , Sandin , R. and Greeneb , J. 1996 . Detection of Airborne Cytomegalovirus in Hospital Rooms of Immuncompromised Patients . J. Virological Methods. , 56 : 115 – 118 .
- McGraw , T. , Mindich , L. and Frangione , B. 1986 . Nucleotide Sequence of the Small Double-Stranded RNA Segment of Bacteriophage φ 6: Novel Mechanism of Natural Translational Control . J. Virology. , 58 : 142 – 151 .
- Mindich , L. , Nemhauser , I. , Gottlieb , P. , Romantschuk , M. , Carton , J. , Frucht , S. , Strassman , J. , Bamford , D. H. and Kalkkinen , N. 1988 . Nucleotide Sequence of the Large Double-Stranded RNA Segment of Bacteriophage Phi 6: Genes Specifying the Viral Replicase and Transcriptase . J. Virology. , 62 : 1180 – 1185 .
- Mittal , D. , Jindal , N. , Gupta , S. L. , Kataria , R. S. and Tiwari , A. K. 2005 . Detection of Infectious Bursal Disease Virus in Field Outbreaks in Broiler Chickens by Reverse Transcription-Polymerase Chain Reaction . International J. Poultry Sci. , 4 : 239 – 243 .
- Myatt , T. A. , Johnston , S. L. , Rudnick , S. and Milton , D. K. 2003 . Airborne Rhinovirus Detection and Effect of Ultraviolet Irradiation on Detection by a Semi-Nested RT-PCR Assay . BMC Public Health , 3
- Ojala , P. M. , Romantschuk , M. and Bamford , D. H. 1990 . Purified Phi 6 Nucleocapsids are Capable of Productive Infection of Host Cells with Partially Disrupted Outer Membranes . Virology. , 178 : 364 – 372 .
- Palmgren , U. , Strom , G. , Blomquist , G. and Malmberg , P. 1986 . Collection of Airborne Microorganisms on Nuclepore Filters, Estimation and Analysis—Camnea Method . J. Applied Bacteriology. , 61 : 401 – 406 .
- Perrott , P. , Smith , G. , Ristovski , Z. , Harding , R. and Hargreaves , M. 2009 . A Nested Real-Time PCR Assay has an Increased Sensitivity Suitable for Detection of Viruses in Aerosol Studies . J. Applied Microbiol. , 106 : 1438 – 1447 .
- Poranen , M. M. and Tuma , R. 2004 . Self-Assembly of Double-Stranded RNA Bacteriophages . Virus Res. , 101 : 93 – 100 .
- Reitz , V. 2005 . A Few Guidelines for Selecting Filters . Medical Design. , http://medicaldesign.com/iv-components/few_guidelines_selecting/(Accessed 20 July, 2010)
- Rimstad , E. , Hornes , E. , Olsviki , O. and Hyllseth , B. 1990 . Identification of a Double-Stranded RNA Virus by Using Polymerase Chain Reaction and Magnetic Separation of the Synthesized DNA Segments . J. Clinical Microbiol. , 28 : 2275 – 2278 .
- Steger , G. , Mçller , H. and Riesner , D. 1980 . Helix-Coil Transitions in ouble-Stranded Viral RNA:. Fine Resolution Melting and Ionic Strength Dependence . Biochimica et Biophysica Acta (BBA)—Nucleic Acids and Protein Synthesis. , 606 : 274 – 284 .
- Tsai , Y.-H. , Wan , G.-H. , Wu , Y.-K. and Tsao , K.-C. 2006 . Airborne Severe Acute Respiratory Syndrome Coronavirus Concentrations in a Negative-Pressure Isolation Room . Infect Control Hosp Epidemiol. , 27 : 523 – 525 .
- Tseng , C.-C. and Li , C.-S. 2005 . Collection Efficiencies of Aerosol Samplers for Virus-Containing Aerosols . J. Aerosol Sci. , 36 : 593 – 607 .
- Tseng , C.-C. and Li , C.-S. 2006 . Ozone for Inactivation of Aerosolized Bacteriophages . Aerosol Sci. Technol. , 40 : 683 – 689 .
- Verreault , D. , Moineau , S. and Duchaine , C. 2008 . Methods for Sampling of Airborne Viruses . Microbiology and Molecular Biology Reviews. , 72 : 413 – 444 .
- Verreault , D. , Rousseau , G. M. , Gendron , L. , Massé , D. , Moineau , S. and Duchaine , C. 2010 . Comparison of Polycarbonate and Polytetrafluoroethylene Filters for Sampling of Airborne Bacteriophages . Aerosol Sci. Technol. , 44 : 203 – 207 .
- Walker , C. M. and Ko , G. 2007 . Effect of Ultraviolet Germicidal Irradiation on Viral Aerosols . Environ. Sci. Technol. , 41 : 5460 – 5465 .
- Wang , M. and Brion , G. 2007 . Effects of RH on Glass Microfiber Filtration Efficiency for Airborne Bacteria and Bacteriophage Over Time . Aerosol Sci. Technol. , 41 : 775 – 785 .
- WHO . 2009 . Acute Respiratory Infections (Update February 2009) . World Health Organization ,