To the Editor
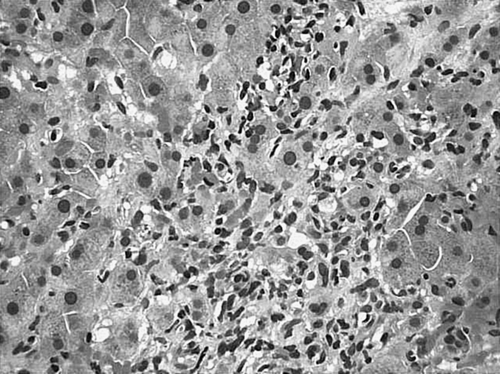
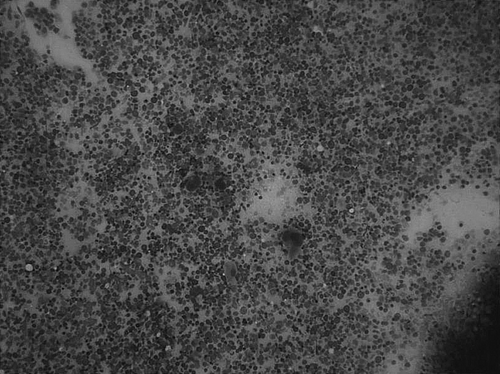
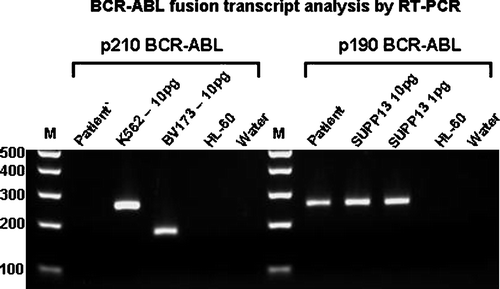
A 56-old man with past medical history of epilepsy and hypertension presented to our hospital with complaints of generalized skin rash, increasing abdominal girth, lower extremity edema and dyspnea on exertion for the past 3 months. He had had a splenectomy several months prior at an outside facility for management of refractory thrombocytopenia. The spleen weighed 1200 g and showed extramedullary hematopoiesis. Shortly after the splenectomy the patient noticed increasing abdominal girth and weight gain (50 lb over 5 months). CT-scan of the abdomen revealed moderate ascites and mild hepatomegaly. Hepatitis serology was negative, total bilirubin and liver enzymes were elevated while total protein and albumin were slightly decreased. Therapeutic paracenthesis with subsequent cytology of the ascitic fluid was non-contributory. In order to rule out liver disease, a liver biopsy was performed which revealed dense diffuse sinusoidal infiltration by immature myeloid and monocytic cells. (). These infiltrates were positive for myeloperoxidase and CD68. CBC upon admission showed mild normocytic anemia, mild leukocytosis with absolute monocytosis and moderate thrombocytopenia. Eosinophilia and basophilia were absent. Peripheral smear showed few immature myeloid and monocytic precursors and dysplastic (Pseudo-Pelger-Huet-like) neutrophils. Bone marrow was hypercellular for age (90% cellularity) with trilineage hyperplasia and dyspoiesis (). Increased monocytic precursors (blasts 2%, monoblasts 1%, promonocytes 8%) and 20% ringed sideroblasts were noted. Immunophenotypic analysis of the marrow aspirate by flow cytometry showed an abnormal monocytic population expressing CD45, bright CD14, bright CD64, HLA-DR, bright CD13, moderate CD33, dim CD15 and with an aberrant expression of dim CD56, dim CD2 and dim CD5. These morphological and immunophenotypical features are characteristic of CMML type 2 according to the WHO classification. Cytogenetic analysis revealed a normal male karyotype. Fluorescent in-situ hybridization (FISH) analysis of interphase cells using the Vysis dual fusion BCR and ABL gene probes did not detect gene fusion. Molecular analysis of the bone marrow aspirate by RT-PCR surprisingly revealed p190 BCR-ABL (e1a2) fusion transcripts (). Our patient was put on Gleevec therapy. Unfortunately, he passed away a month later due to an unrelated cause.
Discussion
The BCR-ABL hybrid gene, the product of t(9;22)(q34;q11.2) is found in leukemic clones of at least 95% of CML patients. The fusion protein encoded by this gene varies in size depending on the breakpoint in the BCR gene. Three breakpoint cluster regions have been characterized: major p210, minor p190 and micro p230 Citation[1]. The majority of CML patients express the p210 fusion transcript (b2a2/b3a2) and protein. p190 BCR-ABL (e1a2) fusion transcripts and protein have been commonly described in patients with Philadelphia (Ph) chromosome-positive acute lymphoblastic leukemia (ALL) and as an additional minor component to p210 in chronic myelogenous leukemia (CML) Citation[2], Citation[5]. Solitary expression of p190 fusion transcripts occurs very rarely in chronic myeloid leukemia (CML); it has been reported in only 18 cases so far and about half of these cases have been associated with monocytosis, resembling CMML Citation[6], Citation[7]. Melo et al. Citation[3] suggested that CML with p190 may be a specific form of CML in which the cytomorphologic characteristics are intermediate between CML and CMML. It is even rarer in other myeloproliferative diseases like essential thrombocythemia and agnogenic myeloid metaplasia in which it is usually seen as a secondary event. Roumier et al. Citation[4] described three cases of chronic myeloid disorders with a secondary cytogenetic acquisition of p190 rearrangement. Unlike the other cases described in literature, this is the first case of CMML type 2 with a p190 BCR-ABL fusion transcript detected by RT-PCR.
The failure to detect the rearrangement by cytogenetic or FISH analysis suggests that the fusion was generated by a small insertion of BCR into ABL; the length of the BCR sequence is below the level of resolution of molecular cytogenetic testing but detectable by PCR. An analogous situation exists in cases of typical CML where small insertions of BCR sequence into the ABL gene on chromosome 9 may not be detectable by molecular cytogenetic methods. The biologic and clinical significance of this finding is still to be determined. This case confirms the variability of clinical features of p190 BCR-ABL positive chronic myeloid disorders and suggests that, at least in some cases, p190 BCR-ABL rearrangement could be a primary event in the course of a myeloid disorder other than CML. The prognostic significance of this finding needs further investigation. Presence of this molecular marker may be helpful in detecting minimal residual disease. There also may be a role for tyrosine kinase inhibitors and this needs to be evaluated from the viewpoint of a therapy modification. This case suggests that p190 can not only induce proliferation of granulocytes and monocytes but may also play an initial role in the course of a myeloid disorder other than CML.
References
- Melo JV. BCR-ABL gene variants. Baillieres Clin Haematol 1997; 10: 203–22
- Ohsaka A, Shina S, Kobayashi M, et al. Philadelphia chromosome-positive chronic myeloid leukemia expressing p190 bcr-abl. Inter Med 2002; 41: 1183–7
- Melo JV, Myinth H, Galton DAG, Goldman JM. P190 bcr-abl chronic myeloid leukemia: The missing link with chronic myelomonocytic leukemia?. Leukemia 1994; 8: 208–11
- Roumier C, Daudignon A, Soenen V, et al. P190 bcr-abl rearrangement: A secondary cytogenetic event in some chronic myeloid disorders. Haematologica 1999; 84: 1075–80
- Van Rhee F, Hochhaus A, Lin F, et al. P 190 bcr-abl mRNA is expressed at low levels in p210-positive chronic myeloid ad acute lymphoblastic leukemias. Blood 1996; 87: 5213–7
- Solves P, Bolufer P, Lopez JA, et al. Chronic myeloid leukemia with expression of ALL-type bcr-abl transcript: A case report and review of the literature. Leuk Res 1999; 23: 851–4
- Ravandi F, Cortes J, Albitar M, et al. Chronic myelogenous leukemia with p185 bcr-abl expression: Characteristics and clinical significance. Br J Haematol 1999; 107: 581–6