Abstract
Background: There is conflicting evidence regarding the association between PIK3CA mutations and clinicopathological features of colorectal cancer (CRC). We performed a comprehensive meta-analysis investigating the association between PIK3CA mutations and clinicopathological features in CRC, including subgroup analysis of mutations in exons 9 and 20, to elucidate the role of PIK3CA mutations in CRC.
Materials and Methods: A detailed literature search was performed within the PubMed, Web of Science, and Embase databases, examining the associations between PIK3CA mutations and demographic characteristics, clinicopathologic parameters, and molecular features in patients with CRC. The odds ratios with 95% confidence intervals were used to estimate the effect of PIK3CA mutations on outcome parameters.
Results: Forty-four studies enrolling 17621 patients were eligible for inclusion. PIK3CA mutations were associated with proximal tumor location, mucinous differentiation, KRAS mutations, and microsatellite instability (MSI). Subgroup analysis demonstrated that PIK3CA exon 9 mutations were positively associated with proximal tumor location and KRAS mutations, and negatively associated with BRAF mutations and MSI; exon 20 mutations were associated with proximal tumor location, KRAS mutations, BRAF mutations and MSI.
Conclusions: Our findings suggest that overall or exon-specific PIK3CA mutations showed null associations with key clinicopathological parameters, including disease stage and tumor differentiation, indicating that PIK3CA mutations do not predict aggressive clinicopathological characteristics in CRC. As PIK3CA mutations were found to be closely associated with KRAS mutations, their relationship warrants further investigation. Since PIK3CA exon 9 and 20 mutations showed different tendencies with regard to BRAF mutation and MSI status, they may have distinct molecular impacts on CRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most frequent cause of cancer-related deaths worldwide, posing a serious threat to human health [Citation1]. It is known that the malignant phenotype of CRC arises from the accumulation of genetic alterations [Citation2]. Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) is one of the crucial kinases in the PI3K/AKT1/MTOR pathway, that plays an important role in cellular growth, proliferation, and survival in various solid tumors [Citation3,Citation4]. Numerous studies have indicated that various malignant tumors including CRC, harbor activated PIK3CA mutations [Citation5]. PIK3CA mutations may activate the p110a enzyme, the key catalytic subunit of PI3K, stimulating AKT signaling, and finally promoting cancer cell proliferation and migration [Citation6]. PIK3CA is one of the most frequently mutated genes in CRC, with almost 80% of mutation hotspots in the helicase and kinase domains of exon 9 and 20, respectively [Citation7,Citation8].
As PIK3CA mutations are common in CRC, their role requires further investigation. Some studies demonstrated that PIK3CA mutations conferred significantly better survival in CRC [Citation9,Citation10]; however, other studies found negative or no association [Citation11–15]. In addition, data on the relationships between PIK3CA mutations and demographic characteristics, clinicopathologic parameters, and molecular features remain unclear. In terms of disease stage, Palomba et al. reported that PIK3CA mutations were correlated with late clinical stage [Citation16], while Day et al. indicated that PIK3CA mutations were unrelated to tumor stage [Citation17]. Till date, no studies have conducted pooled analysis of associations between PIK3CA mutations and clinicopathological characteristics of CRC. A clear understanding of the influence of PIK3CA mutations on clinicopathological and molecular characteristics of CRC will aid in identifying the characteristics of patients with PIK3CA-mutated CRC. This will further aid in defining the roles of PIK3CA mutations in the progression of CRC. We therefore conducted this systematic review and meta-analysis to comprehensively analyze the association between PIK3CA mutations and clinicopathological characteristics of CRC. Since the biological effects of PIK3CA exon 9 and 20 mutations differ, we also conduced subgroup analyses based on PIK3CA exon 9 and 20 mutations in CRC [Citation18–20].
Materials and methods
Literature search strategy
We searched the PubMed, Web of Science, and Embase databases for relevant publications with the following search terms: “colorectal cancer”, “rectal cancer”, “colon cancer”, “PIK3CA”, “Phosphoinositide-3-kinase catalytic alpha polypeptide”, “PIK3 catalytic alpha polypeptide”, “mutation or mutated”. Original articles on human studies written in English and published before July 30, 2018 were included. We also manually searched the reference lists of the relevant articles.
Inclusion and exclusion criteria
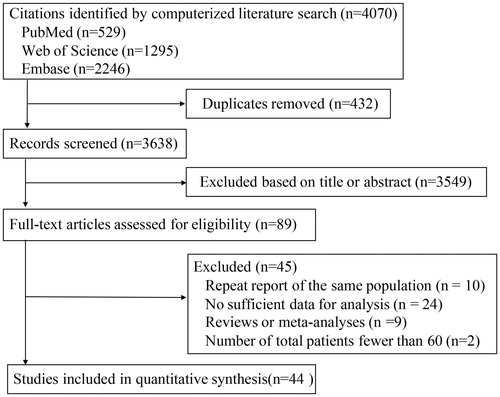
Studies published as original articles on human subjects, simultaneously evaluating PIK3CA mutation status and recording clinicopathological characteristics in CRC patients, and written in English, were included for analysis. In cases where many studies reported an association between PIK3CA mutations and similar clinicopathological characteristics was noted in the same population or overlapping populations, the study with the largest sample size was selected. To avoid the inclusion of redundant studies, we carefully checked the authors and organizations, and evaluated the accrual period and community of patients enrolled for each study. Studies having no or inappropriate data on PIK3CA mutation status or clinicopathological characteristics, studies with less than 60 patients, letters, comments, review articles or meta-analysis without original data, and abstracts published in meetings, were excluded from analysis.
Data extraction and quality assessment
Relevant data were extracted from the eligible studies, including name of the first author, publication year, country where the study was conducted, mutation detection assay, number of PIK3CA-mutated patients, number of total patients, patient demographics (age and gender), clinicopathological characteristics (including disease stage, T stage, metastasis status, tumor site, tumor differentiation and tumor histology), molecular features (KRAS mutation status, BRAF mutation status, mismatch repair capacity (MMR) and microsatellite instability status (MSI). The quality of each study was assessed using the Newcastle-Ottawa Scale (NOS) [Citation4].
Statistical analysis
Meta-analysis was performed using the STATA statistical analysis software (STATA Corp., College Station, TX) and RevMan 5.0 software (Cochrane Collaboration, Oxford, UK) packages. The discrepancies in PIK3CA mutations between groups categorized by clinicopathological factors, were presented as odds ratios (ORs) with 95% confidence intervals (CIs), which were calculated using the Mantel-Haenszel method. In the course of data pooling, statistical heterogeneity was defined using the chi-square-based Q-test. The I2 value indicated the degree of heterogeneity. A p-value < 0.05 and I2 > 50% were considered significant heterogeneity, and a random-effect model was used in these cases; in other cases, a fixed-effect model was used. Publication bias was assessed by Begg’s funnel plot and Egger’s linear regression test. Sensitivity analyses were conducted to estimate whether modification of our inclusion criteria affected the final results.
Results
Characteristics of eligible literature
The flow chart of literature search and study selection have been presented in . According to the selection criteria, 44 articles published between 2007 and 2018 were eligible for the meta-analysis, most of which were retrospective cohort studies. Among these articles, although Liao et al., McCleary et al., and Nosho et al. studied the same population, they investigated the associations of PI3KCA mutations with different clinicopathological parameters [Citation7,Citation9,Citation14,Citation21]. The articles by Chen et al. and Guo et al. also studied the same population, but provided different data [Citation22,Citation23]. A cohort of 17621 CRC patients were enrolled from multiple countries. The study sample sizes ranged from 61 to 2091. Since it was difficult to report all variables in one study, we analyzed studies reporting the association of PIK3CA mutations with certain variables of interest. The basic characteristics of the 44 eligible articles have been summarized in Supplementary Table S1. All the included studies have a NOS score of ≥5 (Supplementary Table S1).
Analysis of PIK3CA mutations in CRC
All specimens were obtained from CRC tissues, either by biopsy or surgical resection, and were tested for PIK3CA mutation status by direct sequencing, pyrosequencing, sanger sequencing, polymerase chain reaction, and next generation sequencing, among others. A total of 33 studies examined both exon 9 and 20 mutations in 10496 patients [Citation7,Citation9,Citation11,Citation12,Citation14,Citation15,Citation17,Citation21–46]; 8 studies including 1713 patients examined mutations in other exons [Citation47–53], and 3 studies with 1872 patients only examined exon 20 mutations [Citation54–56]. The overall discrepancies in PIK3CA mutation status, including mutations in exons 9, 20, and others, were evaluated between groups based on clinicopathological factors, in 41 studies with 18650 patients [Citation7,Citation9,Citation11,Citation12,Citation14,Citation15,Citation17,Citation21–53,Citation57]; 8 studies with 4906 patients reported the discrepancies in PIK3CA exon 9 mutations [Citation14,Citation15,Citation17,Citation27,Citation32,Citation45,Citation48,Citation55], and 10 studies including 6607 patients reported the discrepancies in PIK3CA exon 20 mutations [Citation14,Citation15,Citation17,Citation27,Citation32,Citation45,Citation48,Citation55,Citation56]. The rate of PIK3CA exon 9 or 20 mutations was 12.9% in our study. Notably, the frequency of PIK3CA exon 9 or 20 mutations differed according to the detection techniques used. The rate of PIK3CA exon 9 or 20 mutations was 11.3% and 14.3% based on direct and pyro-sequencing, respectively (p = 0.029) (Supplementary Table S2). The PIK3CA exon 9 and 20 mutation rate was 9.9% and 3.9%, respectively (Supplementary Table S3). PIK3CA exon 9 and 20 mutations co-existed in only 0.09% to 0.6% of CRC patients [Citation14,Citation15,Citation17,Citation45].
Correlation between PIK3CA mutations and clinicopathological characteristics
Demographic characteristics (age and gender)
A total of 7 studies (3191 patients) investigated the association between overall PIK3CA mutation status and age; the association did not reach statistical significance (OR = 1.26; 95% CI: 0.99–1.59). The results for patients with PIK3CA exon 20 mutations were similar (OR = 1.2; 95% CI: 0.51–2.84) (Supplementary Figure S1 A and ). A total of 33 studies (14976 patients) were analyzed for the association between overall PIK3CA mutations and gender. No association was noted between overall PIK3CA mutations and male gender (OR = 0.93; 95% CI: 0.84–1.02); PIK3CA exon 9 and 20 mutations also showed no association with gender (Supplementary Figure S1 B and ).
Table 1. Overall analysis of the association between PIK3CA mutation and clinicopathological features in CRC patients.
Clinical features (disease stage, T stage, distant metastasis status, tumor site tumor differentiation, and tumor histology)
Twenty studies analyzed the association between disease stage and PIK3CA overall mutations. Among 4746 and 4228 patients with stage III or IV and I or II disease, respectively, 613 (12.9%) and 556 (13.2%) patients, respectively, had overall PIK3CA mutation positivity. Poor disease stage was not associated with overall PIK3CA mutation status (OR = 1.00; 95% CI: 0.88–1.14) (Supplementary Figure S2 A and ). These results were similar to that of patients with PIK3CA exon 9 (OR = 1.01; 95% CI: 0.78–1.32) and 20 (OR = 0.77; 95% CI: 0.57–1.04) mutations. In addition, overall PIK3CA or exon 9 or 20 mutations were not associated with advanced T and metastatic stages (Supplementary Figure S2 B and C and ).
Overall, 10 studies investigated the relationship between PIK3CA mutations and tumor site. The results showed that PIK3CA mutation positivity was observed in 431 (17.7%) of 2442 patients with tumors in the proximal colons, compared with 421 (10.9%) of 3875 patients with distal colons or rectal tumors. A positive association was observed between PIK3CA mutations and proximal tumor location (OR = 1.79; 95% CI: 1.39–2.29). On subgroup analysis, the positive association also extended to PIK3CA exon 9 (OR = 1.78; 95% CI: 1.18–2.68) and 20 (OR = 1.61; 95% CI: 1.05–2.45) mutations ( and ).
Figure 2. The association of PIK3CA mutation with tumor characteristics, including tumor site (A), tumor differentiation (B) and mucinous histology (C).
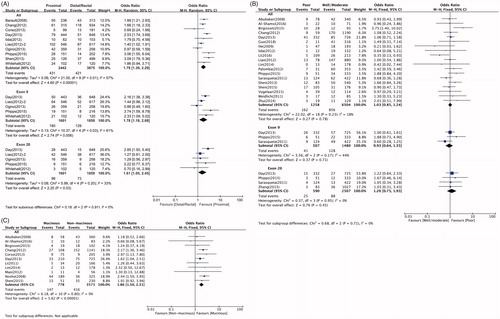
A total of 19 studies (7762 patients) investigated the association between PIK3CA mutations and tumor differentiation. No significant association was found between overall PIK3CA mutations and poor differentiation (OR = 1.03; 95% CI: 0.85–1.24). The results for exon 9 (OR = 0.93; 95% CI: 0.64–1.35) and exon 20 (OR = 1.20; 95% CI: 0.75–1.93) mutations were similar ( and ). Eleven studies were analyzed for the association between overall PIK3CA mutations and histological subtypes. Among 778 and 3571 patients with mucinous and non-mucinous types, respectively, 147 (18.9%) and 416 (11.7%) patients, respectively, were overall PIK3CA mutation positive. Therefore, overall PIK3CA mutations were associated with mucinous histology (OR = 1.86; 95% CI: 1.50–2.31). ( and ).
Molecular features (KRAS mutation status, BRAF mutation status, MSI status, and MMR capacity)
Fifteen studies analyzed the association between PIK3CA and KRAS mutation status. Among 2322 and 4126 KRAS-mutated and wild patients, respectively, 440 (18.9%) and 420 (10.2%), respectively, had PIK3CA mutations. Notably, KRAS and overall PIK3CA mutations were significantly and positively associated (OR = 2.06; 95% CI: 1.78–2.38) ( and ). For subgroup analysis, 6 studies were available for both exon 9 and 20 mutations. PIK3CA exon 9 mutations were detected in 217 (14.5%) of 1502 and 187 (6.8%) of 2753 patients, with KRAS mutations and wild-type KRAS, respectively. Exon 20 mutations were detected in 89 (5.9%) of 1502 and 110 (4.0%) of 2753 patients with KRAS mutations and wild-type KRAS, respectively. Therefore, KRAS mutation was significantly associated with both PIK3CA exon 9 (OR = 2.32; 95% CI:1.88–2.85) and exon 20 (OR = 1.51; 95% CI:1.13–2.01) mutations ( and ). The pooled results demonstrated no association between BRAF and overall PIK3CA mutations (OR = 0.92; 95% CI: 0.73–1.16). Remarkably, the association in patients with PIK3CA exon 9 and 20 mutations, showed opposing trends. Six studies were included for analysis of the association between BRAF mutations and PIK3CA exon 9 and 20 mutations. A total of 599 and 3809 patients had BRAF mutations and wild-type BRAF, respectively; among them, 43 (7.2%) and 366 (9.6%) were PIK3CA exon 9 mutation positive, respectively. Therefore, PIK3CA exon 9 mutations appeared to be negatively associated with BRAF mutations (OR = 0.69; 95% CI: 0.49–0.96). However, compared with 165 (4.3%) of 3809 BRAF-wild patients, 40 (6.7%) of 599 BRAF-mutated patients were PIK3CA exon 20 mutation positive. Therefore, PIK3CA exon 20 mutations were positively associated with BRAF mutations (OR = 1.61; 95% CI: 1.13–2.31) ( and ).
Figure 3. The association of PIK3CA mutation with molecular features, including KRAS mutation status (A), BRAF mutation status (B), MSI status (C), and MMR capacity (D).
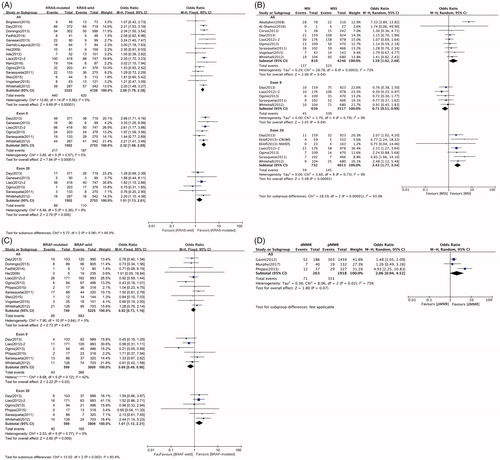
Nine studies investigated the relationship between overall PIK3CA mutations and MSI. Overall PIK3CA mutations were observed among 137 (16.9%) of 810 patients with MSI, and in 523 (12.3%) of 4240 patients with microsatellite stable (MSS). PIK3CA mutations were associated with MSI (OR = 1.59; 95% CI: 1.02–2.48). The trends were also inconsistent between PIK3CA exon 9 and 20 mutations in this case. A total of 5 and 7 studies were analyzed for exon 9 and 20 mutations, respectively. Exon 9 mutation was found in 45 (6.9%) of 650 cases with MSI and in 339 (9.6%) of 3517 patients with MSS. Exon 20 mutation was detected in 59 (8.1%) of 732 patients with MSI, and in 145 (3.6%) of the 4013 patients with MSS. In summary, MSI appeared to be negatively and positively associated with exon 9 (OR = 0.71;95% CI: 0.51– 0.99) and exon 20 (OR = 2.43;95% CI:1.77–3.34) mutations, respectively ( and ). Only three studies investigated the relationship between PIK3CA mutations and MMR capacity; 71 (27.0%) of 263 patients with deficient MMR (dMMR) capacity, and 351 (18.3%) of 1918 patients with proficient MMR (pMMR) capacity were PIK3CA mutation positive. The association between PIK3CA mutations and dMMR did not reach statistical significance (OR = 2.06; 95% CI: 0.94–4.52) ( and ).
Sensitivity analysis and publication bias
The Begg’s and Egger’s tests demonstrated no indication of publication bias among the studies included in this meta-analysis. The sensitivity analyses revealed that no individual studies unduly influenced pooled ORs and CIs significantly, suggesting that the estimates were robust.
Discussion
We conducted a systematic and comprehensive meta-analysis, assessing the associations between PIK3CA mutations and demographic characteristics, clinicopathologic parameters, and molecular features. We also compared PIK3CA exon 9 and 20 mutations to identify differences between them. In this meta-analysis, PIK3CA mutation status was not associated with the factors affecting prognosis including disease stage, distant metastasis status, and tumor differentiation. In view of the important roles of PIK3CA mutations in tumorigenesis and development, the null association between PIK3CA mutations and these key clinicopathologic characteristics was remarkable [Citation5,Citation7,Citation8]. In 2016, the meta-analysis by Mei et al. reported that the pooled hazard ratios of PIK3CA-mutated and wild-type CRC for overall- and prognosis free- survival were 0.96 (95%CI: 0.83–1.12) and 1.20 (95%CI: 0.98–1.46), respectively[Citation58]. Considering that PIK3CA mutation has no influence on clinical outcomes, our observations were rational, and strongly suggested that PIK3CA mutations showed null association with tumor progression in CRC. The null association with key clinicopathologic characteristics was probably owing to the fact that although the PIK3CA gene harbors a high mutation rate in CRC, these mutations may not be the crucial molecular alteration, as opposed to other gene mutations [Citation58,Citation59]. Experimental in vitro evidence has suggested that the influence of PIK3CA mutations on cancer cell growth may not be sufficiently potent, and that these mutations needed to cooperate with other mutations of PI3K pathway kinases to effectively influence tumor behavior [Citation3,Citation60]. In addition, CRC has high heterogeneity, with distinct molecular backgrounds [Citation61]. Smeby et al. concluded that the role of genetic alterations may depend on molecular backgrounds; it may therefore be concluded that the association of PIK3CA mutations with clinicopathologic characteristics depends on distinct molecular contexts [Citation62]. Since numerous studies have demonstrated the prognostic and predictive value of PIK3CA mutations in KRAS-wild CRC, the association between PIK3CA mutations and clinicopathologic features in wild-type KRAS may differ from unselected patients [Citation6,Citation59,Citation63].
Our findings indicated that tumors located in the proximal colon had higher PIK3CA mutation rates than those of the distal colon and rectum, irrespective of the exon mutated. This highlighted the difference in molecular features between proximal and distal colorectal tumors [Citation64]. In addition, overall PIK3CA mutations were closely associated with mucinous differentiation, an unfavorable subtype of CRC [Citation65]; the cause of this potential association remains obscure. Our results demonstrated that PIK3CA mutations were associated with KRAS mutations and MSI; this suggests significant differences between PIK3CA-mutated and wild-type patients at the molecular level. However, molecular events associated with PIK3CA mutations are not able to induce considerable alterations in the clinicopathologic phenotypes.
Studies in vitro have demonstrated that mutations in exons 9 and 20 transform cancer cells in different and independent pathways [Citation18]. In our study, significant differences were noted between overall and exon-specific PIK3CA mutations in several molecular parameters. De Roock et al. reported that in metastatic CRC, PIK3CA exon 9 mutations were strongly associated with KRAS mutations, whereas exon 20 mutations were independent [Citation59]. Interestingly, in our study, the results showed that although both exon 9 and 20 mutations were associated with KRAS mutations, exon 9 mutations had greater statistical power than both overall PIK3CA and exon 20 mutations. Additionally, although the association between overall PIK3CA and BRAF mutations did not reach statistical significance, BRAF mutations were negatively and positively associated with PIK3CA exon 9 and 20 mutations, respectively. The association between overall PKI3CA and BRAF mutations reflected the mean of exon 9 and 20 mutations. Conclusively, it appears prudent to consider the effect of PIK3CA exon 9 and 20 mutations separately.
Although our meta-analysis is systematic and detailed, there are some limitations. Firstly, most of the involved studies were retrospective, and obtained tissues from tissue specimen databases; this may have led to a bias in patient selection. Secondly, the number of studies examining the relationship between PIK3CA mutations and several features was limited, particularly in terms of subgroup analysis of exon 9 and 20 mutations; this reduces the credibility of pooled results. In addition, the sample sizes of the studies varied considerably among different studies. Small studies are likely to introduce unstable results, that are related to publication bias. Thirdly, the mutation detection assays were different among the included studies; this also affects the accuracy and precision of the pooled estimates [Citation5].
In conclusion, overall or exon-specific PIK3CA mutation status was found to have no association with most clinicopathologic parameters. Overall PIK3CA mutations were associated with proximal locations, mucinous differentiation, KRAS mutations, and MSI. Subgroup analysis showed PIK3CA exon 9 and 20 mutations had opposing trends with regard to BRAF mutation and MSI status; this needs further confirmation in future large-scale studies. The clinical implications of PIK3CA mutations, combined with other factors such as KRAS and BRAF mutations, require further investigation in future studies. Independent evaluation of PIK3CA exon 9 and 20 mutations is warranted.
Supplemental Material
Download (4.7 MB)Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Siegel RL, Miller KD, Fedewa SA. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193.
- Cathomas G. PIK3CA in colorectal cancer. Front Oncol. 2014;4:35
- Samuels Y, Diaz LA, Jr Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–573.
- Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441(7092):424
- Ogino S, Lochhead P, Giovannucci E, et al. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene. 2014;33(23):2949.
- Mao C, Yang Z, Hu X, et al. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2012;23(6):1518–1525.
- Nosho K, Kawasaki T, Longtine JA, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10(6):534–541.
- Velho S, Oliveira C, Ferreira A, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41(11):1649–1654.
- Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–1606.
- Manceau G, Marisa L, Boige V, et al. PIK3CA mutations predict recurrence in localized microsatellite stable colon cancer. Cancer Med. 2015;4(3):371–382.
- Zhu K, Yan H, Wang R, et al. Mutations of KRAS and PIK3CA as independent predictors of distant metastases in colorectal cancer. Med Oncol. 2014;31(7):16.
- He Y, Van't Veer LJ, Mikolajewska-Hanclich I, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009;15(22):6956–6962. CCR-09-1165.
- Phipps AI, Makar KW, Newcomb PA. Descriptive profile of PIK3CA-mutated colorectal cancer in postmenopausal women. Int J Colorectal Dis. 2013;28(12):1637–1642.
- Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18(8):2257–2268.
- Ogino S, Liao X, Imamura Y, et al. Predictive and prognostic analysis of PIK3CA mutation in stage III colon cancer intergroup trial. J Natl Cancer Inst. 2013;105(23):1789–1798.
- Wang Q, Shi Y-L, Zhou K, et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018;9(7):739.
- Day FL, Jorissen RN, Lipton L, et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathological and molecular associations in colorectal cancer. Clin Cancer Res. 2013;19(12):3285. clincanres. 3614.2012.
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110α of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. P Natl A Sci USA. 2008;105(7):2652–2657.
- Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15(16):5049–5059. CCR-09-0632.
- Lai Y-L, Mau B-L, Cheng W-H, et al. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15(4):1064–1069.
- McCleary NJ, Sato K, Nishihara R, et al. Prognostic utility of molecular factors by age at diagnosis of colorectal cancer. Clin Cancer Res. 2016;22(6):1489–1498.
- Chen J, Guo F, Shi X, et al. BRAF V600E mutation and KRAS codon 13 mutations predict poor survival in Chinese colorectal cancer patients. BMC Cancer. 2014;14(1):802.
- Guo F, Gong H, Zhao H, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep. 2018;8(1):6076.
- Weidlich S, Walsh K, Crowther D, et al. Pyrosequencing-based methods reveal marked inter-individual differences in oncogene mutation burden in human colorectal tumours. Br J Cancer. 2011;105(2):246.
- Mao C, Zhou J, Yang Z, et al. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PloS one. 2012;7(5):e36653
- Birgisson H, Edlund K, Wallin U, et al. Microsatellite instability and mutations in BRAF and KRAS are significant predictors of disseminated disease in colon cancer. BMC Cancer. 2015;15(1):125.
- Sarasqueta AF, Zeestraten EC, van Wezel T, et al. PIK3CA kinase domain mutation identifies a subgroup of stage III colon cancer patients with poor prognosis. Cell Oncol. 2011;34(6):523–531.
- Garrido-Laguna I, Hong DS, Janku F, et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One. 2012;7(5):e38033.
- Vogelaar FJ, van Erning FN, Reimers MS, et al. The prognostic value of Microsatellite Instability, KRAS, BRAF and PIK3CA mutations in stage II colon cancer patients. Mol Med. 2015;21(1):1038.
- Stec R, Semeniuk-Wojtaś A, Charkiewicz R, et al. Mutation of the PIK3CA gene as a prognostic factor in patients with colorectal cancer. Oncology letters. 2015;10(3):1423–1429.
- Shen Y, Wang J, Han X, et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PloS one. 2013;8(12):e81628.
- Phipps AI, Ahnen DJ, Cheng I, et al. PIK3CA somatic mutation status in relation to patient and tumor factors in racial/ethnic minorities with colorectal cancer. Cancer Epidemiol Biomar. 2015;24(7):1046–1051.
- Palomba G, Colombino M, Contu A, et al. Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. J Transl Med. 2012;10(1):178.
- Murphy C, Turner N, Wong HL, et al. Examining the impact of regular aspirin use and PIK3CA mutations on survival in stage 2 colon cancer. Intern Med J. 2017;47(1):88–98.
- Lin J-K, Lin P-C, Lin C-H, et al. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Ann Surg Oncol. 2014;21(S4):680–686.
- Li Z-Z, Wang F, Zhang Z-C, et al. Mutation profiling in chinese patients with metastatic colorectal cancer and its correlation with clinicopathological features and anti-EGFR treatment response. Oncotarget. 2016;7(19):28356.
- Li H-T, Lu Y-Y, An Y-X, et al. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25(6):1691–1697.
- Kato S, Iida S, Higuchi T, et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer. 2007;121(8):1771–1778.
- Iida S, Kato S, Ishiguro M, et al. PIK3CA mutation and methylation influences the outcome of colorectal cancer. Oncol Lett. 2012;3(3):565–570.
- Gavin P, Colangelo LH, Fumagalli D, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clin Cancer Res. 2012;18(23):6531. clincanres. 0605.2012.
- Domingo E, Church DN, Sieber O, et al. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. J Clin Oncol. 2013;31(34):4297–4305.
- Corso G, Pascale V, Flauti G, et al. Oncogenic mutations and microsatellite instability phenotype predict specific anatomical subsite in colorectal cancer patients. Eur J Hum Genet. 2013;21(12):1383.
- Christensen TD, Palshof JA, Larsen FO, et al. Associations between primary tumor RAS, BRAF and PIK3CA mutation status and metastatic site in patients with chemo-resistant metastatic colorectal cancer. Acta Oncologica. 2018;57(8):1057–1062.
- Chang S-C, Lin P-C, Lin J-K, et al. Mutation spectra of common cancer-associated genes in different phenotypes of colorectal carcinoma without distant metastasis. Ann Surg Oncol. 2016;23(3):849–855.
- Whitehall VL, Rickman C, Bond CE, et al. Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J Cancer. 2012;131(4):813–820.
- Abubaker J, Bavi P, Al-Harbi S, et al. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene. 2008;27(25):3539.
- Barault L, Veyrie N, Jooste V, et al. Mutations in the RAS‐MAPK, PI (3) K (phosphatidylinositol‐3‐OH kinase) signaling network correlate with poor survival in a population‐based series of colon cancers. Int J Cancer. 2008;122(10):2255–2259.
- Ganesan P, Janku F, Naing A, et al. Target-based therapeutic matching in early-phase clinical trials in patients with advanced colorectal cancer and PIK3CA mutations. Mol Cancer Ther. 2013;12(12):2857.
- Nam SK, Yun S, Koh J, et al. BRAF, PIK3CA, and HER2 oncogenic alterations according to KRAS mutation status in advanced colorectal cancers with distant metastasis. PloS one. 2016;11(3):e0151865.
- Al-Shamsi HO, Jones J, Fahmawi Y, et al. Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern. J Gastrointest Oncol. 2016;7(6):882.
- Balc’h EL, Grandin N, Demattei M-V, et al. Measurement of telomere length in colorectal cancers for improved molecular diagnosis. IJMS. 2017;18(9):1871.
- Chiu JW, Krzyzanowska MK, Serra S, et al. Molecular profiling of patients with advanced colorectal cancer: princess margaret cancer centre experience. Clin Colorectal Canc. 2018;17(1):73–79.
- Russo AL, Borger DR, Szymonifka J, et al. Mutational analysis and clinical correlation of metastatic colorectal cancer. Cancer. 2014;120(10):1482–1490.
- Eklöf V, Wikberg ML, Edin S, et al. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108(10):2153.
- Neumann J, Wehweck L, Maatz S, et al. Alterations in the EGFR pathway coincide in colorectal cancer and impact on prognosis. Virchows Arch. 2013;463(4):509–523.
- Zhang J, Zheng J, Yang Y, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep. 2015;5(1):18678.
- Fadhil W, Kindle K, Jackson D, et al. DNA content analysis of colorectal cancer defines a distinct ‘microsatellite and chromosome stable’group but does not predict response to radiotherapy. Int J Exp Path. 2014;95(1):16–23.
- Mei Z, Duan C, Li C, et al. Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2016;27(10):1836–1848.
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762.
- Oda K, Okada J, Timmerman L, et al. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008;68(19):8127–8136.
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350.
- Smeby J, Sveen A, Merok M, et al. CMS-dependent prognostic impact of KRAS and BRAF V600E mutations in primary colorectal cancer. Ann Oncol. 2018;29(5):1227–1234.
- Mohamed A, Twardy B, AbdAllah N, et al. Clinical impact of PI3K/BRAF mutations in RAS wild metastatic colorectal cancer: Meta-analysis results. J Gastrointest Canc. 2019;50(2):269–275.
- Maus MK, Hanna DL, Stephens CL, et al. Distinct gene expression profiles of proximal and distal colorectal cancer: implications for cytotoxic and targeted therapy. Pharmacogenomics J. 2015;15(4):354.
- Hugen N, Brown G, Glynne-Jones R, et al. Advances in the care of patients with mucinous colorectal cancer. Nat Rev Clin Oncol. 2016;13(6):361.