Abstract
Background
Local treatment of liver and/or lung metastases from colorectal cancer (CRC) is increasingly used in daily practice and comprises resection, radiofrequency ablation (RFA) and stereotactic radiotherapy (SBRT). The need for prognostic markers for patients undergoing such treatment is currently unmet. We investigated post-treatment circulating tumor-specific DNA (ctDNA) analysis and address a possible prognostic value in a pilot study.
Materials
From July 2015 to September 2017, patients undergoing standard of care local treatment of liver and/or lung metastases were included in a prospective translational study. Blood samples were drawn 2 weeks after local treatment and during follow-up. CtDNA was detected by ddPCR and a mass spectrometry-based platform MassARRAY®.
Results
Post treatment blood samples were available for 35 patients including five with detectable ctDNA (KRAS mutation, n = 2; NRAS mutation, n = 2; BRAF mutation, n = 1) by ddPCR. 17 out of 35 patients (49%) developed recurrence within a median of 273 days (95%CI 95–NA) among patients positive for ctDNA, while the median time to recurrence was not reached for the group of patients negative for ctDNA (p = .03).
Conclusion
The presence of ctDNA following local treatment of metastatic CRC is associated with an increased risk of recurrence and a short time to failure.
Introduction
Globally, more than 1.3 million patients are yearly diagnosed with colorectal cancer (CRC), which hold a poor prognosis [Citation1]. Across the past decades, there has been an increased use of local therapy modalities for patients with a limited extent of metastatic lesions [Citation2,Citation3]. These include surgical resection, radiofrequency ablation (RFA), and stereotactic radiotherapy (SBRT). The reported median 5-year survival ranges from 30–50% indicating that only a subset of patients become long term survivors [Citation4–6]. Selecting the right candidates for local therapies and making decisions of perioperative chemotherapy are major challenges due to unclear data regarding a potential benefit in this setting [Citation7,Citation8].
Active surveillance following local treatment of metastases is mandatory since close to 80% of patients who undergo resection of colorectal cancer liver metastases (CRCLM) eventually develop new metastatic lesions [Citation9]. Today, clinical examination, imaging, and measurement of carcinoembryonic antigen (CEA) have limitations in terms of individualizing the approach, both for early detection of recurrence and optimal selection of post-treatment chemotherapy.
Circulating tumor DNA (ctDNA) represents small DNA fragments in the bloodstream with tumor-specific mutations comprising a minor part of the total cell-free DNA (cfDNA). Across a variety of solid tumors, this has gained considerable interest as a potential biomarker in both localized and metastatic disease [Citation10,Citation11]. Following resection of primary colorectal cancer, emerging evidence supports a correlation between the presence of ctDNA in the plasma post-treatment and the existence of microscopic residual disease and a subsequent poor outcome [Citation12,Citation13].
In this study, we analyzed the biological nature of total cfDNA and ctDNA following local treatment of metastases from CRC including a comparison of ddPCR genotyping with a mass spectrometric based multiplexed platform, MassARRAY® (MA). The primary aim of this study was to examine the feasibility of ctDNA detection after local treatment for metastases and describe the potential clinical impact of ctDNA post-treatment.
Material and methods
Patients and treatment
Patients with metastatic colorectal adenocarcinoma, age >18 years, and no history of other malignancies were included in a single armed phase II observational study at Aarhus University Hospital between July 2015 and September 2017. Treatment consisted of standard local therapy for liver and/or lung metastases following multidisciplinary evaluation in accordance with institutional guidelines. Patients could undergo surgical resection, open or laparoscopic RFA, or SBRT with a dosage ranging from 56.25 Gy to 67.5 Gy in 3(–6) fractions depending on the size and location of the metastasis. A combination of modalities was allowed and chemotherapy was given at the discretion of the treating oncologist.
Patients underwent post-treatment Computed Tomography scan (CT) every 3 months during the first year, at 18 and 24 months during the second year, then yearly up to 5 years. Corresponding blood samples were drawn. Upon recurrence all standard of care modalities were available.
Both local and distant recurrences alongside death were considered as an event in the time to recurrence analysis.
Sampling and pre-analytical procedures
The post-treatment blood sample for translational analysis was drawn 2 weeks after the local procedure. Further samples were taken, if available, before treatment and at a fixed schedule during follow up. Thirty mL of blood was collected in EDTA tubes and plasma isolated by centrifugation at 2000 g for 10 min within 2 h and stored at −80 °C until further analysis. For patients receiving adjuvant chemotherapy, blood samples were drawn during treatment. The number of samples per patient ranged from 1–8 plasma samples.
Circulating DNA analysis
For detection and quantification of ctDNA 4 mL of plasma was used. Prior to DNA purification, an in vitro generated 191 bp CPP1 DNA fragment was spiked into the plasma [Citation14]. DNA was purified on a Chemagen 360 purification robot (PerkinElmer, Waltham, MA) using a CMG-1104 kit (PerkinElmer) according to the manufacturer's recommendations and eluted in 100 µL. Potential contamination of the purified DNA with white blood cell DNA was evaluated using an immunoglobulin gene-specific assay (PBC) and loss of DNA during the purification and handling was assessed by measuring the amount of spike in the fragment as previously reported by a digital droplet PCR [Citation14]. Depending on how well the observed mutations in the tissue had been characterized, ctDNA was analyzed either by singleplex or multiplex ddPCR assays, which had a limit of detection (LoD) of 0.1% or better.
The total level of cfDNA was analyzed by ddPCR targeting the housekeeping gene Beta-2-Microglobulin (B2M) as previously described [Citation14].
The plasma DNA was additionally analyzed by a mass spectrometric based multiplexed platform (MassARRAY® (MA) Agena Bioscience). In brief, MA genotyping utilizes PCR reactions, DNA extension, and mass spectrometric in a 96-well format. The UltraSEEK MA Colon Panel is a high-throughput assay with a capacity to screen for more than 100 somatic mutations in the 5 pivotal oncogenes KRAS, NRAS, BRAF, EGFR and PIK3CA with an LoD of 0.1%. All mutations targeted by the ddPCR were included in the MA gene panel. The technology is based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF).
PCR was performed according to the manufacturer’s instructions using an input of 10 ng cfDNA. After amplification, the PCR products were treated with shrimp alkaline phosphatase and a single-base extension was performed. Amplicons were purified using streptavidin-coated magnetic beads. Before analysis, the product was conditioned with resin and dispensed onto a SpectroCHIP using the MA® Nanodispenser. The SpectroCHIP was loaded on the MA Analyzer 4 and data acquired using TyperAnalyzer software version 4.1.83.
Statistics
Data for ctDNA is reported as a binary categorical variable and as a continuous variable as a percentage of total DNA, when applicable, and the total cfDNA as alleles/mL with median values and 95% Confidence Intervals (CI). Non-parametric statistic, including Mann Whitney u test, was applied describing any correlation between cfDNA and the clinical parameters.
We analyzed the time to recurrence by the Kaplan-Meier method and log-rank test for equality between groups. Time to recurrence was reported as the median value and patients were followed until recurrence, death, or end of follow-up (September 2018). Hazard Ratios (HR) for recurrence was calculated by the Cox model for both uni- and multivariate analysis.
Data management and statistical analyses were performed using STATA version 14.1 (StataCorp, College Station, TX).
Ethics
This protocol was approved by the Regional Ethics Committee (# 1-16-02-178-15) and the Data Protection Agency. All patients signed a written consent form.
Results
Patient characteristics and outcome
A total of 35 patients were included. The male/female ratio was 18/17 with a median age of 70.5 years (range 36–81 years). Colon cancer comprised 16 patients while 19 had a rectal primary. The applied standard modalities were liver resection (n = 12), liver RFA (n = 10), liver SBRT (n = 4), lung resection (n = 5), lung RFA (n = 1) and lung SBRT (n = 3). A total of 23 patients were treated for a solitary metastasis, 4 patients received treatment for 2 metastases, and 8 patients for three metastases or more comprising a total of 61 metastatic lesions treated. Details of patient characteristics are displayed in . With a median follow up of 21 months 17 patients had to develop recurrent disease. Two cancer-related deaths had occurred. A flowchart depicting the distribution of recurrences and presence of ctDNA is shown in .
Figure 1. Flowchart of study patients depicting the mutational status for tissue, plasma and the number of patients with recurrent disease. Pos ctDNA positive samples for ctDNA, neg ctDNA negative samples for ctDNA.
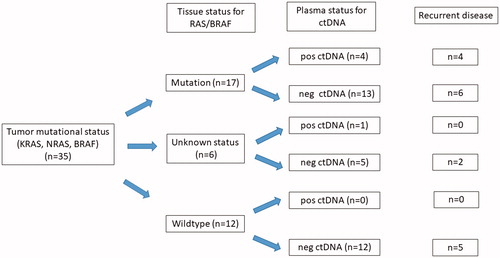
Table 1. Patient characteristics.
Tumor mutation status
Archival tumor mutational status was performed as routine. Wildtype status was found in 12 patients whereas primary tissue mutations were detected in 17, comprising 15 patients with a mutation in KRAS/NRAS and 2 BRAF mutations.
Total cfDNA and tumor DNA levels in plasma samples by ddPCR
In total, 84 plasma samples were analyzed. Post-treatment blood samples were available for 35 patients, and follow-up samples were analyzed in all patients harboring ctDNA in the primary post-treatment sample, and in patients who had developed recurrence during the observation period.
The median level of total DNA in the post-treatment samples was 32,800 alleles/mL (95%CI 19,934–133,273) with a range from 5600 to 1,560,000 alleles/mL by ddPCR, and a positive pb score in 33 (out of a total of 84 samples), suggesting potential contamination of lymphocytes falsely elevating the total level of cfDNA [Citation13]. There were no statistically significant differences between cfDNA levels according to pretreatment clinical or pathological parameters. However, correction for potential contamination of cfDNA from normal lymphocytes was not performed for the primary analysis.
Five patients (5/35; 14%) had detectable mutations in post-treatment plasma samples, 2 had KRAS, 1 NRAS, and 1 BRAF mutations (). Among patients with known tissue mutation (n = 17) the detection rate of ctDNA was 24% (4/17). A total of 15 mutations were detected in all plasma samples. Of the five patients who had detectable ctDNA post-treatment, two patients had undergone RFA, two resections, and one SBRT for liver metastases. The median level of mutated ctDNA in the post-treatment samples (n = 5) was 0.05% (95%CI 0.01–0.26%) by ddPCR.
Table 2. Overview of 5 patients with detectable tumor DNA in plasma following a local treatment of metastatic colorectal cancer.
Explorative analysis by MA technique
To increase the potential ctDNA detection rate a new commercially available method for identification of a broader panel of tumor-specific mutations by MA was validated. We aimed to compare the sensitivity for mutation detection in patients with limited ctDNA levels by MA and ddPCR, to test the performance and potential clinical value. We analyzed a total of 47 blood samples by both ddPCR and MA. Two of the MA samples were inconclusive due to the low volume plasma sample. The overall concordance rate was 87%, between ddPCR and MA. In one sample co-existing KRAS and PIK3CA mutation was detected by the MA. Notably, the ddPCR was not designed to analyze for PIK3CA mutations. Only one post-treatment sample was found positive by MA compared to 5 by ddPCR. In the sample with ctDNA positivity by both methods, the ctDNA/cfDNA fraction was 0.3% by MA compared to 0.14% by ddPCR, indicating a higher sensitivity by the ddPCR. In samples with an overall low total DNA yield, the MA did not provide supplementary information and seemed inferior to ddPCR methods.
Circulating tumor DNA and risk of recurrence
In the overall cohort (n = 35) ctDNA positive patients (n = 5) had a significantly shorter median time to recurrence of 273 days (95%CI 95-NA) compared to ctDNA negative patients (n = 30), where the median time to recurrence was not reached at end of follow up (p = .03). The time to recurrence by the presence of ctDNA is displayed in .
Figure 2. Time to recurrence from date of local treatment for all patients with presence (red line, n = 5) and absence (blue line, n = 30) of ctDNA post-treatment (p = .03). Pos ctDNA positive samples for ctDNA, neg ctDNA negative samples for ctDNA.
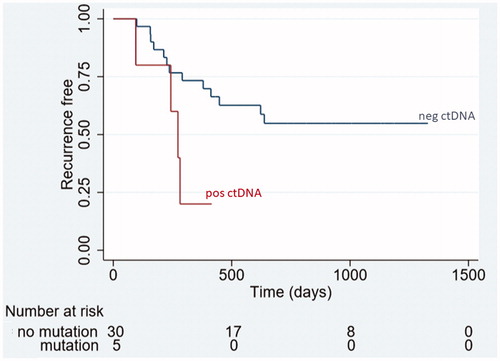
In Cox regression analysis including the parameters age, gender, N stage, site of primary (colon/rectum), K/NRAS mutational status in tissue and number of metastases the HR for recurrence if ctDNA positive with reference to ctDNA negative was 3.36 (95%CI 1.03–10.94, p = .03) in univariate and 7.48 (95%CI 1.47–38.36), p = .02) in multivariate analysis. The presence of ctDNA was the only variable independently associated with an increased HR for recurrence.
Dynamics of circulating DNA during follow-up
The clinical courses, dynamics of ctDNA, and total cfDNA are displayed in for the four patients with ctDNA presence post-treatment with sequential samples (one patient (A) only had the post-treatment sample taken, so only four curves are presented). All received adjuvant chemotherapy (single-agent capecitabine, n = 1; oxaliplatin doublet, n = 3). As demonstrated all four patients had declining ctDNA levels during adjuvant chemotherapy, indicating potential ctDNA clearance by therapy.
Figure 3. Clinical courses and dynamics of the total level of cfDNA (upper row) and the tumor specific DNA (lower row) for the four patients with detected ctDNA post-treatment with available serial measurements of circulating DNA after a radical local treatment of metastases from colorectal cancer (patient A is not presented, only one available sample).
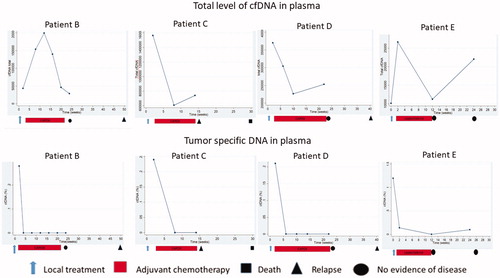
One patient (C) with a BRAF (V600E) mutated colon cancer had detectable BRAF mutation in plasma following resection of liver metastases. Despite adjuvant CAPOX, the patient developed an early recurrence and died 7 months after the liver resection. The ctDNA was undetectable during follow-up, but the total cfDNA increased at the time of relapse. A similar pattern of cfDNA mirroring the disease course was seen for another patient (, patient D).
Patient E underwent SBRT for a metachronous liver metastasis one year after the resection of a low-risk colonic cancer. This patient had a rapid decline in ctDNA post-SBRT and during adjuvant chemotherapy (single-agent capecitabine omitted due to toxicity after 6 cycles). After 6 months, emerging of the same NRAS mutation (0.11%) was detected and all imaging during the study period was without recurrence. Nine months post-study the patient developed recurrence.
Two patients with known tumor KRAS mutations had emerging ctDNA (KRAS) in relation to the occurrence of relapse. One patient developed a detectable KRAS mutation (6.3%, KRAS G12S) in plasma one month prior to the diagnosis of a multifocal recurrence. The second patient, at the time of recognition of a hepatic recurrence.
Discussion
The literature on liquid biopsies is broad, heterogeneous and rapidly evolving [Citation15]. We have tested ctDNA from plasma samples by a highly sensitive and feasible ddPCR also used in a prospective utility study in the palliative setting by our group (optipal II clinical trials.gov). In addition, we tested the potential methodological advantage of a MA platform, using a panel for more than 100 somatic mutations in 5 characteristic oncogenes (KRAS, NRAS, PIK3CA, EGFR and BRAF). With a detection limit of 0.1%, the MA showed limited value as a supplementary tool for a more sensitive ddPCR in our study, yielding a concordance rate of 87% for the 47 samples analyzed by both ddPCR and MA. While ddPCR identifies a specific mutation, known beforehand, MA might serve as a screening tool identifying mutations in KRAS, NRAS, PIK3CA, EGFR and BRAF given the mutational load is not too low given an expected detection limit of MA of around 5%. Further optimizing steps, including available plasma volume, might improve the performance of MA.
In this study, we have analyzed plasma samples for 35 patients following treatment for liver and/or lung metastases, and demonstrate a clinically relevant association between the presence of ctDNA and risk of recurrence. The calculated HR of 7.48 for recurrence by cox regression should be interpreted with caution due to the low sample size and a number of variables included, also reflected in the wide confidence intervals. However, this analysis signals a potential prognostic value of ctDNA post-treatment.
To the best of our knowledge, only 3 published studies have [Citation13,Citation16,Citation17] addressed ctDNA and resection for colorectal cancer liver metastases, all supporting a strong negative prognostic impact of ctDNA. Schøler et al. [Citation13] examined 23 patients undergoing resection for CRCLM and found the presence of ctDNA 3 months post-treatment to be associated with an increased risk of recurrence (HR 4.9 (95%CI 1.5–15.7)). Shin and colleagues [Citation16] report data from 103 patients with mCRC receiving a curatively intended treatment for all metastatic lesions. Blood samples for ctDNA analysis were drawn pretreatment, and the authors report an inferior survival for patients with detectable ctDNA. However, no post-treatment samples were available so the issue of ctDNA as a marker of residual disease could not be addressed. Finally, in the paper by Benešová [Citation17] all patients (n = 22) who developed recurrence after resection of liver metastases had a positive plasma sample for ctDNA post-treatment. Notably, in this study patients were selected for inclusion only if a pretreatment sample was positive for ctDNA, possibly explaining the high level of detection of ctDNA in this study.
Our study is the first to report this proof of principle for a mixed group of oligometastatic patients, who have undergone a variety of local treatment of metastatic disease, and our data confirm the observations from the primary setting of a strong detrimental effect of postoperative ctDNA measurement.
We noted a tendency for the increasing total level of cfDNA (patients C and D) prior to the detection of recurrent disease, despite non-detectable ctDNA. The total cfDNA level is considered a non-tumor specific biomarker known to be affected by other medical conditions and co-morbidity [Citation18]. In this descriptive study, it appears the level of total cfDNA could supplement the use of tumor-specific DNA as biomarkers.
The current literature supporting adjuvant chemotherapy in the oligometastatic setting is sparse [Citation19]. We observed ctDNA positive patients had a high risk of recurrence, despite all receiving standard adjuvant chemotherapy (capecitabine ± oxaliplatin), stressing the challenge for optimal treatment of these patients. The potential gain from intensified adjuvant chemotherapy in these high-risk patients is still unknown and needs to be prospectively investigated. Furthermore, ctDNA guidance of imaging and follow-up schedules for this heterogeneous group of patients might contribute to a more personalized approach needing further attention in future trials.
As a pilot study, there are limitations. We included all standard of care modalities of both liver and lung metastasis directed treatment, to have our study population as similar as possible to daily practice. As a consequence, this leads to heterogeneity for a small sample size (n = 35). Furthermore, only 17 out of 35 patients (49%) had a known mutation in tissue to target.
We applied a highly sensitive ddPCR assay to detect the presence of ctDNA in plasma. This platform is based on testing for the common CRC mutations in the KRAS, NRAS, and BRAF oncogenes, thus any potential tumor DNA with a different mutational profile (e.g., APC or TP53) would not be quantified by our assay. With these limitations and methodology in mind, our data primarily support the utility of ctDNA for patients with beforehand known mutations in K/NRAS oncogenes, while patients without mutations in K/NRAS might be best examined by a different approach.
In conclusion, these observational data suggest clinically relevant information from ctDNA analysis as a marker of both minimal residual disease post-treatment. Clinical trials must be designed to prospectively address the true clinical utility.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386.
- Bartlett E, Simmons K, Wachtel H, et al. The rise in metastasectomy across cancer types over the past decade. Cancer. 2015;121(5):747–757.
- Boysen AK, Spindler KL, Høyer M, et al. Metastasis directed therapy for liver and lung metastases from colorectal cancer-a population-based study. Int J Cancer. 2018;143(12):3218–3226.
- Gillams AR, Lees WR. Five-year survival in 309 patients with colorectal liver metastases treated with radiofrequency ablation. Eur Radiol. 2009;19(5):1206–1213.
- Guerrera F, Mossetti C, Ceccarelli M, et al. Surgery of colorectal cancer lung metastases: analysis of survival, recurrence and re-surgery. J Thorac Dis. 2016; 8(7):1764–1771.
- Fong Y, Fortner J, Sun R, et al. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer. Annal Surg. 1999;230(3):309–321.
- Mitry E, Fields AL, Bleiberg H, et al. Adjuvant chemotherapy after potentially curative resection of metastases from colorectal cancer: a pooled analysis of two randomized trials. J Clin Oncol. 2008;26(30):4906–4911.
- Nordlinger B, Sørbye H, Glimelius B, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14(12):1208–1215.
- Tomlinson JS, Jarnagin WR, DeMatteo RP, et al. Actual 10-year survival after resection of colorectal liver metastases defines cure. J Clin Oncol. 2007;25(29):4575–4580.
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990.
- Spindler KG, Appelt A, Pallisgaard N, et al. KRAS mutated Plasma DNA as predictor of outcome from irinotecan monotherapy in metastatic colorectal cancer. Br J Cancer. 2013;109(12):3067–3072.
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346.
- Schøler LV, Reinert T, Ørntoft MW, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–5445.
- Pallisgaard N, Spindler KL, Andersen RF, et al. Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta. 2015;446:141–146.
- Spindler KL. Methodological, biological and clinical aspects of circulating free DNA in metastatic colorectal cancer. Acta Oncol. 2017;56(1):7–16.
- Shin SJ, Chun SM, Kim TI, et al. Feasibility of multiplexed gene mutation detection in plasma samples of colorectal cancer patients by mass spectrometric genotyping. PLoS One. 2017;12(5):e0176340.
- Benešová L, Hálková T, Ptáčková R, et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J Gastroenterol. 2019;25(48):6939–6948.
- Spindler KL, Appelt A, Pallisgaard N, et al. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int J Cancer. 2014;135(12):2984–2991.
- Khoo E, O’Neill S, Brown E, et al. Systematic review of systemic adjuvant, neoadjuvant and perioperative chemotherapy for resectable colorectal-liver metastases. HPB. 2016;18(6):485–493.
