Abstract
Background: Sarcomas are a rare and heterogeneous tumor group composed of a variety of histologic subtypes. Targeted next-generation sequencing (NGS) of bone and soft tissue sarcomas is a nascent field with limited evidence for its use within clinical practice. Therefore, further research is needed to validate NGS in sarcoma and assess the clinical utility of these techniques with the hope of improving treatment options.
Methods: Comprehensive molecular profiling with NGS was performed on 136 tumors (116 soft tissue, 20 bone) using two commercial vendors. Patient records were retrospectively reviewed, and the clinical impact of NGS-related findings were qualitatively analyzed to determine actionable mutations and number of changes in treatment.
Results: The median age was 55.0 years (IQR 42–67 years), and most patients were non-metastatic at presentation (80.9%, n = 110). Prior to performing NGS, 72.1% (n = 98) were treated with a mean 1.1 ± 1.2 lines of systemic chemotherapy. NGS identified 341 putative alterations with at least one mutation present in 89.7% (n = 122) of samples. In a subset of 111 patients with available TMB data, 78.7% (n = 107) had a low (<6 m/Mb) mutational burden. Among all 136 cases, 47.1% (n = 64) contained clinically actionable alterations, and 12 patients had a change in medical treatment based on NGS. Those who underwent a treatment change all had metastatic or recurrent disease; three of these patients experienced a clinical benefit.
Conclusion: Most bone and soft tissue sarcomas harbor at least one genetic alteration, and it appears a sizeable number of tumors contain mutations that are clinically actionable. While a change in treatment based off NGS-related findings occurred in 12 cases, three patients experienced a clinical benefit. Our data provide further proof-of-concept for NGS in sarcoma and suggest a clinical benefit may be observed in select patients.
Introduction
Sarcomas are a rare and heterogeneous tumor group that account for nearly 1% of adult malignancies [Citation1]. Given that sarcomas are comprised of over 60 different histologic subtypes, obtaining an accurate diagnosis is challenging. While the divergent biology of most sarcomas has been exploited by diagnostic tools that can readily identify distinct genetic patterns, for most subtypes there are limited United States Food and Drug Administration (FDA)-approved therapies that treat specific molecular targets [Citation2,Citation3]. Rather, when indicated, the treatment strategy for most patients including those with metastatic sarcoma consists of cytotoxic agents and tyrosine kinase inhibitors (e.g., pazopanib). Unfortunately, the overall response rate to cytotoxic drugs is usually low, depending on the drug or combination of drugs, and the current treatment options for patients with advanced disease are generally not curative [Citation4,Citation5].
In the era of personalized medicine, molecular profiling with next-generation sequencing (NGS) may enable expanded treatment options for sarcoma patients, especially in cases of advanced or treatment-refractory disease. Although few studies have demonstrated targeted NGS as a potentially useful tool for tailoring individualized therapies in this setting, there are limited data that address the clinical utility of this practice [Citation6–11].
The purpose of the current study was to describe the experience of our institution using NGS in non-gastrointestinal stromal tumor (GIST) sarcoma patients whose tumor samples underwent comprehensive molecular profiling. The primary goal was to document NGS-related findings and treatment changes, if applicable, and to assess outcomes among those treated based on the recommendation of NGS. To our knowledge, this study is one of the largest of its kind.
Material and methods
Patients
Following Institutional Review Board approval, our sarcoma database was retrospectively reviewed to identify all patients diagnosed with biopsy-proven, non-GIST sarcoma. Inclusion criteria for the purpose of this study were those who underwent treatment along with NGS testing at a tertiary medical center. There were no exclusion criteria based on histologic subtype. A total of 136 patients were identified (116 soft tissue, 20 bone), and comprehensive molecular profiling was performed on 136 tumor samples using commercially available platforms (Foundation Medicine, Inc. [FoundationOne, http://www.foundationone.com] or Tempus [Tempus, http://www.tempus.com]), both of which are CLIA-approved NGS testing vendors [Citation12]. This study reflects an updated cohort from our preliminary findings presented in abstract form [Citation6].
Comprehensive molecular profiling
Following the initial oncologic encounter, each case was presented to the tumor board during which the histologic diagnosis and options for treatment were discussed. Following either biopsy of initial or metastatic disease, or surgical resection, a tumor sample was prospectively assayed, and genomic alterations such as insertions/deletions, substitutions, copy number mutations, and rearrangements were recorded and made available for review in commercial reports. Within our institution, the original histologic diagnoses were finalized by a team of experienced musculoskeletal pathologists, and instances of a diagnosis change, or modification based on NGS-related findings were noted if applicable. Actionable genetic alterations were defined as any mutation found using NGS that could be directly targeted by investigational or FDA-approved drugs (on or off-label). Clinical actionability was determined both from the findings of commercial testing reports and/or at the consensus of the multidisciplinary group. Eligibility for clinical trials was also noted. Variants of unknown significance were not included in the group of identifiable alterations. Tumor mutational burden (TMB), which is an estimate of the number of mutations per Mb, was also calculated for a majority of included patients. TMB data were assessed when available.
Outcomes
Patient demographics and tumor characteristics were reviewed and collected for each case. Clinical data including systemic treatment lines employed prior to NGS were also recorded. In addition, treatment changes that occurred as a result of NGS-related findings were documented, and among this group, electronic medical records were reviewed for evidence of a subsequent clinical benefit. Clinical benefit was defined as the absence of new metastatic lesions and no significant growth in size of the primary tumor lasting six months after the change.
Results
Patients
The median patient age was 55.0 years at diagnosis (interquartile range [IQR] 42–67 years), and there was slight female predominance (52.9%, n = 72). A variety of non-GIST subtypes were included, which largely reflects the scope of our clinical practice (). For example, the most frequently recorded histologic subtypes ranged from common diagnoses such as undifferentiated pleomorphic sarcoma (UPS; 24.3%, n = 33), leiomyosarcoma (LMS; 13.2%, n = 18), and osteosarcoma (n = 5), to more rare sarcomas such as epithelioid sarcoma (n = 1) and rhabdomyosarcoma (n = 2). Most patients were non-metastatic at presentation (80.9%, n = 110), and prior to performing NGS testing, 72.1% (n = 98) were treated systemically with a mean 1.1 ± 1.2 lines of chemotherapy.
Table 1. Clinicopathologic data of included patients (n = 136).
Comprehensive molecular profiling
Two commercial vendors were used for NGS testing (Foundation Medicine, Inc.: 24.3%, n = 33; and Tempus: 75.7%, n = 103). A cumulative total of 341 genomic alterations were detected, with at least one alteration present in 89.7% (n = 122) of tumors (). The remaining 10.3% (n = 14) did not harbor a significant variant according to diagnostic reports. A median of two mutations were detected per sample (range, 0 to 9), with the most common being TP53 (30.9%, n = 38), CDKN2A/B (12.2%, n = 15), RB1 (10.6%, n = 13), and CDKN2A (9.8%, n = 12) (). Notable mutations included SSX-SS18 (synovial sarcoma) and HMGA2 (liposarcoma). In addition, each (n = 2) rhabdomyosarcoma was FOXO1 negative, and half of liposarcoma (50.0%, n = 7) harbored MDM2/CDK4 co-amplifications.
Figure 1. Frequency of the most common genetic alterations found among all included sarcoma patients undergoing next-generation sequencing (n = 136).
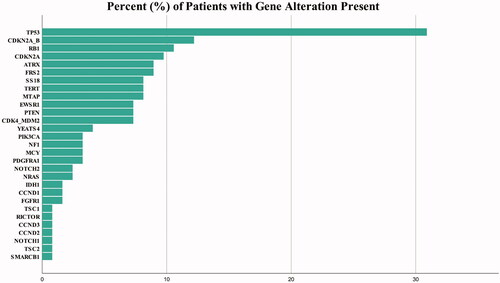
Table 2. Actionable changes and recommendations per histologic subtype.
There were 16 total PI3Kinase alterations including TSC2 (n = 1), TSC1 (n = 1), RICTOR (n = 1), PIK3Ca (n = 4), and PTEN (n = 9). There were 11 ATRX mutations which predominantly occurred in UPS (n = 3) and LMS (n = 3). Disease-defining gene alterations were detected as mentioned above, and also included IDH (dedifferentiated chondrosarcoma), EWSR1-ATF1 and EWSR1-FLI1 (Ewing sarcoma).
CDK alterations were relatively common and included 25 patients with loss of CDKN2A or CDKN2A/B, who were diagnosed with UPS (n = 5), malignant peripheral nerve sheath tumor (n = 4), myxofibrosarcoma (n = 3), chondrosarcoma (n = 2), fibrosarcoma (n = 2), Phyllodes sarcoma (n = 2), and one case each of angiosarcoma, schwannoma, rhabdomyosarcoma, liposarcoma, LMS, cystosarcoma, and Ewing sarcoma. Two CCND1 mutations were recorded (UPS, extraskeletal osteosarcoma), in addition to one mutation in CCND2 (LMS) and CCND3 (UPS), respectively. Additionally, NRAS mutations were present in three patients (LMS, osteosarcoma, malignant peripheral nerve sheath tumor), and one patient with epithelioid sarcoma harbored an SMARCB1 mutation. There were zero instances in which the NGS-related findings were felt to be diagnosis-changing or diagnosis-modifying.
Genomic instability
In a subset of 111 patients (81.6%), 96.4% (n = 107) had a low mutational burden (<6 m/Mb), while the remaining four samples were classified as ‘intermediate’ (6–20 m/Mb). Among this 111-patient subgroup, the mean mutational burden was 2.6 ± 2.1 m/Mb, and there were zero instances of microsatellite instability (MSS for each of the 89 of 111 samples with available data).
Outcomes
Sixty-four (47.1%) patients had an actionable mutation and were recommended at least one therapeutic option by commercial NGS testing reports. Furthermore, 67.6% (n = 92) of patients had an alteration that was potentially targetable by currently available clinical trial options (). NGS-related findings influenced a treatment change in 12 patients, all of whom had metastatic or recurrent disease. Some patients with actionable molecular profiles had locally advanced disease and therefore were not treated with NGS-targeted therapy. In this subset of patients with metastatic or recurrent disease, three patients had a benefit in clinical course (stable disease > 6 months) after NGS-related treatment change, with some having stable disease for longer than one year (, ).
Figure 2. Clinical course following NGS-related treatment change. All patients had progressive disease on prior treatment regimen and had metastatic or recurrent disease.
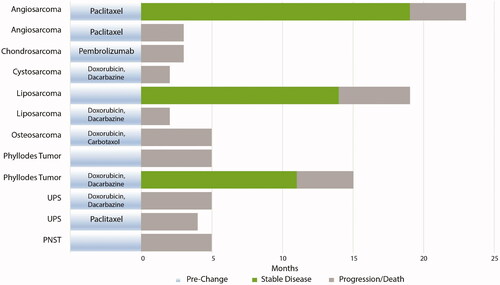
Table 3. Treatment changes based on next-generation sequencing-related findings.
Stable disease [Citation14], then progression and death [Citation5]Progression with eventual death [Citation5]
Discussion
There has been a substantial improvement in molecular therapy for patients with solid and hematologic malignancies, and the development of targeted drugs for sarcoma patients is a quickly emerging field. NGS may enable high-throughput identification of various mutations that can serve as targets for novel drugs, and NGS is becoming a common practice in sarcoma. However, there are few available data that describe NGS in this setting, with less data that describe the clinical utility of these techniques [Citation7–11]. In the current study, molecular profiling using NGS was able to identify a sizeable number of genetic alterations in nearly 90% of patients, with only about 10% lacking a genomic variant of significance. It also appears NGS-related findings influence the subsequent treatment in a small minority. Despite a treatment change occurring in only 12 patients, however, a favorable clinical response was observed in three of these patients.
Patients with metastatic sarcoma portend a poor prognosis with survival estimates of about 30% at two years [Citation13]. Therefore, improvements in treatment options in this group are much needed. Molecular profiling with NGS may offer hope for additional treatment options resulting in improved survival. While there are promising data that suggest molecular-based trial criteria for selection of early phase clinical trials in advanced sarcoma improves success rates, it would also make sense that each sarcoma, whether metastatic or not, would benefit from a more personalized treatment approach [Citation14,Citation15].
To our knowledge, the current study is one of the largest that describes comprehensive molecular profiling in non-GIST sarcoma that also attempts to identify the clinical utility of NGS-related findings. We found 47.1% of all patients harbored a clinically actionable mutation, and our results are consistent with the available published rates of about 40.0% to 60.0%. In their series of 133 sarcoma patients, Cote et al. recorded a cumulative 342 alterations among all patients, with 88.0% of tumors harboring at least one mutation [Citation8]. They suggest targeted NGS may be a useful tool with implications for clinical practice, however, they did not report outcomes. In a separate study of 114 sarcoma patients by Boddu et al., at least one alteration was identified in 96.7% of tumors, and in a similar report by Groisberg et al., 93.0% of the total cohort (n = 102) had at least one alteration [Citation7,Citation9]. Both these groups included clinical outcome data described below, and together with our findings, it appears most sarcomas harbor genetic alterations that can potentially lead to a change in treatment if the mutations are considered actionable.
Despite a relatively high rate of actionable mutations identified using NGS, however, only a minority of sarcoma patients tend to undergo a subsequent change in treatment. Our rate of a treatment change (8.8%) aligns with other recent rates of about 13.0% to 16.0%. A similar study also reported nearly 30.0% enrollment in a matched trial option after NGS testing in sarcoma [Citation11]. Of note, all patients who underwent a treatment change had metastatic or recurrent disease, while others with actionable mutations did not undergo a treatment change due to having localized disease. Although these rates are low, patients who undergo a change in treatment based on clinical genomic profiling may experience an improved clinical course. A fourth of our patients with a treatment change had a clinical benefit of stable disease for longer than six months, and these findings align with similar response rates of 26.7% to 50.0% after changes in treatment elsewhere in the literature, respectively [Citation7,Citation9]. Together with the findings of these other groups, the clinical success observed in our subset of patients highlights the importance of NGS testing to identify potentially actionable mutations that may translate into a successful clinical outcome for a select few.
Notably, all cases of phyllodes sarcoma (n = 2 of 2) and cystosarcoma (n = 1 of 1) experienced an NGS-related treatment and were started on palbociclib. These three cases all shared a CDKN2A mutation, which is a common genetic alteration observed not only in sarcoma, but all malignancies. With respect to treatment change, palbociclib has been shown to demonstrate varying levels of clinical benefit in patients with CDKN2A mutations in previous studies [Citation16,Citation17]. Considering the small number of three patients, these observed treatment changes are anecdotal in nature and further studies are required to better elucidate specific mutations and potential targeted therapies for these particular tumor types.
TMB is an interesting area of emerging research, and higher values are thought to predict greater genomic instability. Values less than 20 m/Mb are considered intermediate in nature, while values less than 6 m/Mb are considered to be of low instability [Citation18]. Given that most sarcomas are defined by translocation events (e.g., synovial sarcoma and Ewing sarcoma) which are of relatively low instability, it was unsurprising that the mean estimated m/Mb in the current study was “low” overall. This observation aligns with other reports that show similarly low estimates, and a low mean TMB in our study is likely a reflection of the frequency of subtypes seen within clinical practice, which often includes low-grade disease and a higher incidence of sarcomas with fusion translocations [Citation7,Citation8,Citation19,Citation20]. TMB data was available for 10 of the 12 patients who experienced a NGS-related treatment change, and in this group, all were noted to have low instability. This observation can likely be attributed to the overall low TMB seen in sarcomas. As NGS becomes more utilized in oncologic care, future studies should investigate the clinical implications of intermediate and high TMB (> 20 m/Mb).
As the indications for NGS testing in sarcoma expand, it is important to define interpretations of clinical actionability, and to ascertain whether NGS is truly needed to identify each mutation. In our current series, NGS testing was performed on some tumors with mutations that could have been identified without comprehensive molecular testing, such as MDM2 in liposarcoma or SSX-SS18 alterations in synovial sarcomas. Exclusion of these tumors would have given us a more concise depiction of the utility of NGS, and therefore the decision to test such patients should be discussed in a multidisciplinary setting. However, different centers may rely more heavily on a multidisciplinary decision, or conversely place greater emphasis on testing reports. In our institution, the results of NGS were reviewed with the medical team, and any alteration deemed potentially actionable by commercial reports was corroborated by the consensus of the group.
Limitations of this study include its retrospective and observational nature. Our series was comprised of a variety of histologic subtypes, and testing was performed across two laboratories over the course of the study period. NGS panels are updated quite frequently and therefore may differ from year to year. An additional consideration for the sarcoma care team is the extra cost and utilization of resources incurred with NGS testing, which often includes the need for more tissue to be obtained to sufficiently assay samples. Nonetheless, our data do further validate this novel technique for use in bone and soft tissue sarcoma and highlight the importance of NGS from a clinical standpoint.
Conclusions
Comprehensive molecular profiling was performed on 136 sarcoma patients with and without advanced disease at presentation, which simulates the context of a routine clinical practice. Among our patients, we were able to identify a sizeable number of mutations as well as a relatively high rate of alterations considered potentially actionable with drugs or clinical trials. In our cohort of 136 patients, 12 patients, all with metastatic or recurrent disease, underwent a change in treatment as a result of NGS testing. The potential for a marked benefit in select patients provides further proof-of-concept for the tailoring of individual therapy in sarcoma and expanding treatment options. We hope these data contribute to future efforts to identify a clinical benefit. Given that sarcomas are rare and encompass a variety of histologic subtypes, the implications of our findings are promising and should assist in future goal-oriented research.
Ethics approval and consent to participate
Rush University Medical Center obtained individual Institutional Review Board approval with an approved waiver of consent prior to beginning any research efforts.
Prior presentations
DOI: 10.1200/JCO.2019.37.15_suppl.e22552 Journal of Clinical Oncology 37, no. 15_suppl
Disclosure statement
ATB: (BMJ Case Reports: Editorial or governing board; Clinical Orthopedics and Related Research: Editorial or governing board; exparel/pacira: Stock or stock Options; Journal of Oncology Practice: Editorial or governing board; Journal of Surgical Oncology: ad hoc reviewer; Lancet - Oncology: Editorial or governing board; Musculoskeletal Tumor Society: Board or committee member; Onkos Surgical: Paid consultant; Pediatric Blood and Cancer: Editorial or governing board; Rare Tumors: Editorial or governing board; Rush Orthopedic Journal: Editorial or governing board; Swim Across America Cancer Research Grant: Research support); SG: (Onkos Surgical: Paid consultant; Stock or stock Options; USMI: Stock or stock Options). All other authors have no pertinent financial disclosures or pertinent conflicts of interest.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Spiguel A. Soft tissue sarcomas. Cancer Treat Res. 2014;162:203–223.
- Jo VY, Fletcher CDM. WHO classification of soft tissue tumours: an update based on the 2013 (4th) edition. Pathology (Phila). 2014;46(2):95–104.
- Mertens F, Tayebwa J. Evolving techniques for gene fusion detection in soft tissue tumours. Histopathology. 2014;64(1):151–162.
- Santoro A, Tursz T, Mouridsen H, et al. Doxorubicin versus CYVADIC versus doxorubicin plus ifosfamide in first-line treatment of advanced soft tissue sarcomas: a randomized study of the European organization for research and treatment of cancer soft tissue and bone sarcoma group. J Clin Oncol. 1995;13(7):1537–1545.
- Judson I, Verweij J, Gelderblom H, et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncol. 2014;15(4):415–423.
- Weiss MC, Blank A, Gitelis S, et al. Clinical benefit of next generation sequencing in soft tissue and bone sarcoma: Rush University medical center’s experience. J Clin Oncol. 2019; 37(15_suppl):e22552–e22552.
- Boddu S, Walko CM, Bienasz S, et al. Clinical utility of genomic profiling in the treatment of advanced sarcomas: a single-center experience. J Clin Oncol Precis Oncol. 2018;2:1–8.
- Cote GM, He J, Choy E. Next-generation sequencing for patients with sarcoma: a single center experience. Oncologist. 2018;23(2):234–242.
- Groisberg R, Hong DS, Holla V, et al. Clinical genomic profiling to identify actionable alterations for investigational therapies in patients with diverse sarcomas. Oncotarget. 2017; 8(24):39254–39267.
- Wilky BA, Villalobos VM. Emerging role for precision therapy through next-generation sequencing for sarcomas. J Clin Oncol Precis Oncol. 2018;2:1–4.
- Gounder MM, Ali SM, Robinson V, et al. Impact of next-generation sequencing (NGS) on diagnostic and therapeutic options in soft-tissue and bone sarcoma. J Clin Oncol. 2017; 35(15_suppl):11001–11001.
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–1031.
- Italiano A, Mathoulin-Pelissier S, Cesne AL, et al. Trends in survival for patients with metastatic soft-tissue sarcoma. Cancer. 2011;117(5):1049–1054.
- Schwaederle M, Zhao M, Lee JJ, et al. Impact of precision medicine in diverse cancers: a Meta-Analysis of phase II clinical trials. J Clin Oncol. 2015;33(32):3817–3825.
- Tsimberidou A-M, Wen S, Hong DS, et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res. 2014;20(18):4827–4836.
- Al Baghdadi T, Halabi S, Garrett-Mayer E, et al. Palbociclib in patients with pancreatic and biliary cancer with CDKN2A alterations: results from the targeted agent and profiling utilization registry study. J Clin Oncol Precis Oncol. 2019; 3:1–8.
- Ahn ER, Mangat PK, Garrett-Mayer E, et al. Palbociclib in patients with non–small-cell lung cancer with CDKN2A alterations: results from the targeted agent and profiling utilization registry study. J Clin Oncol Precis Oncol. 2020; 4:757–766.
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34.
- Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014; 7(1):104–112.
- Perry JA, Kiezun A, Tonzi P, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564–5573.
