Background
In oligometastatic colorectal cancer (OCRC), there can be a curative potential from treating metastases [Citation1,Citation2]. Some patients obtain long-term survival whereas others show aggressive biology with early recurrence and systemic dissemination [Citation1]. No current parameters have shown a clinically relevant potential to predict the course of the disease nor the optimal selection of post-treatment chemotherapy.
In various solid tumours, circulating tumour DNA (ctDNA) has gained considerable interest as a prognostic and predictive marker in both localised and metastatic disease [Citation3,Citation4]. In a pilot study, we have shown that patients with detectable ctDNA two weeks after treatment of OCRC have a high risk of recurrence 2 years post-ablation [Citation5]. Furthermore, we observed a temporal clearance of ctDNA by adjuvant chemotherapy, but also ctDNA recurrence after the termination of chemotherapy indicating that patients with ctDNA positive status may need more intensified adjuvant chemotherapy than the standard of care (SOC) [Citation5].
Consequently, it is highly relevant to initiate a clinical trial of ctDNA-guided adjuvant treatment. Such a strategy must be evaluated against current SOC to investigate the true clinical utility, and for that purpose, feasibility measures need to be established prospectively. The current study will initiate with a run-in phase investigating feasibility. This will progress into a multicentre, phase II randomised study, investigating the clinical utility of ctDNA-guided treatment.
Hypotheses
Patients with detectable ctDNA post-treatment of OCRC have microscopic residual disease and consequently a high risk of recurrence, which can be prevented with intensified adjuvant chemotherapy. Patients without detectable ctDNA have undergone successful radical treatment and do not benefit from adjuvant chemotherapy. Hence, ctDNA-guided adjuvant treatment may improve disease-free survival and minimise chemotherapy-related toxicity.
Aim
The aim of the run-in phase is the testing of feasibility parameters. The primary aim of the main study is to investigate the benefit of ctDNA-guided adjuvant treatment compared to SOC in OCRC. Secondary aims include investigating the molecular biological response to chemotherapy, cost-effectiveness, and quality of life (QoL) in patients undergoing ctDNA-guided adjuvant treatment.
Material and methods
Study design
OPTIMISE is an open-label 1:1 randomised phase II study. The protocol is available in supplementary.
Eligibility criteria
Inclusion criteria:
Radical intended treatment for metastatic spread from CRC by resection, radiofrequency ablation, stereotactic body radiation therapy, or other experimental local treatments.
No evidence of further disease based on pre-treatment workup according to SOC.
Age ≥ 18 years.
ECOG performance status 0–2.
Clinically eligible for adjuvant triple chemotherapy at investigators decision.
Adequate bone marrow function allowing systemic chemotherapy.
Anticonception for fertile women and for the male patients with a fertile partner.
Written and verbally informed consent.
Exclusion criteria:
Neuropathy National Cancer Institute CTCAE grade > 1.
Other malignant tumour within 5 years except for non-melanoma skin cancer or carcinoma in situ cervicis uteri.
Pregnant or breastfeeding women.
Intolerance or allergy to 5-FU, leucovorin, oxaliplatin, irinotecan or capecitabin.
Recruitment
Patients who meet the eligibility criteria are approached at the treating department of oncology. Patients are included based on both verbal and written informed consent. Patients are informed that they may withdraw their consent at any time.
Randomisation
Randomisation is blinded and automated. Randomisation is performed using a predefined electronic randomisation tool module in the Research Electronic Data Capture (REDCap).
ctDNA analysis
Circulating free DNA will be extracted from 4 ml of plasma with the QiaSymphony (Qiagen) or a Chemagic 360 (PerkinElmer) purification system for further analyses.
Detection and quantification of ctDNA will be done using two different approaches:
Digital droplet PCR (ddPCR) panel detecting CRC specific mutations in KRAS, NRAS and BRAF.
Tumour specific methylation assay detecting methylation of the NPY gene.
The samples are analysed for known CRC specific mutations in KRAS, NRAS and BRAF by a ddPCR panel and for tumour DNA by a methylation assay [Citation5–11]. Methylated ctDNA is analysed as DNA with methylation of the NPY gene. The purified DNA is bisulphite converted and analysed by NPY/ALB duplex ddPCR using the QX200 system from BioRad.
The mutational analysis has a limit of blank of 0.025–0.28% (depending on the mutation analysed) and a limit of detection of 0.05–0.3% (depending on the mutation analysed), whereas the methylation analysis has a limit of blank of 0.03% and a limit of detection of 0.04%.
Interventions
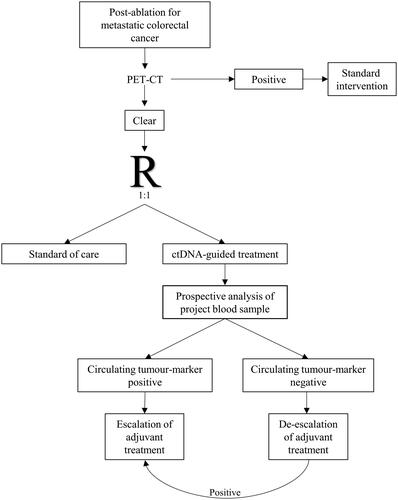
The study flow is illustrated in . Before randomisation, all study participants will undergo a PET-CT scan to exclude metabolically detectable metastatic lesions or another unforeseen concurrent disease.
All study participants will have the first project blood samples drawn 2–4 weeks post-ablation. Hereafter project blood samples will be drawn at follow-up visits. Follow-up with imaging will be in accordance with the current Danish SOC. All study participants will complete QoL questionnaires at baseline and at 12, 24, 36, 48, and 60 months.
The control group (SOC): Patients will be offered adjuvant chemotherapy according to SOC. Blood samples will be analysed retrospectively to evaluate ctDNA status.
The experimental group (ctDNA-guided strategy): Blood samples will be analysed prospectively. Circulating tumour-marker positivity () will lead to escalated adjuvant treatment (4 months of FOLFOXIRI followed by 2 months of 5-FU monotherapy). Circulating tumour-marker negativity () opens the option of observation without further treatment. The advice to observation, single-agent chemotherapy, or combination chemotherapy will be based on shared decision-making. Conversion from circulating tumour-marker negativity to positivity, without radiological evidence of disease, will lead to rescue therapy (4 months of FOLFOXIRI followed by 2 months of 5-FU as monotherapy). Detection of visible metastases will lead to a referral for SOC.
Table 1. Overview of conclusion on circulating tumour-marker status based on molecular analysis.
Trial status
The inclusion of patients in the run-in phase began in October 2021. Two centres are currently including. The trial has been registered in the Clinical Trials database: NCT04680260.
Biobank
At end of the study, any excess biological material will be transferred to a biobank for the purpose of future research. The biobank will be located in the Department of Experimental Clinical Oncology at Aarhus University Hospital. Analyses not specifically related to the current study can only be initiated after a new application and approval by the ethics committee.
Ethical considerations
The triplet chemotherapy regimen is more time-consuming and toxic compared to SOC doublet adjuvant treatment and requires a central venous catheter. The intensified adjuvant treatment is solely given to patients with a known very high risk of recurrence (tumour-marker positive).
Circulating DNA investigations are limited to tumour-specific genomic changes, and do not analyse for known genetic variants related to inherited disease.
One additional PET-CT will be acquired for trial participants. Ionising radiation is a known carcinogenic and increases the risk of cancer. We state that the potential benefit from identifying recurrence at an early stage with possible curative treatment outweighs the risk of radiation-induced secondary cancer.
Project blood samples are primarily drawn as part of routine management before radiological evaluations, and the extra amount of blood is regarded negligible.
Definition of endpoints
Feasibility measures for the run-in phase:
Inclusion of 30 patients over 12 months in two clinical centres
Compliance with randomisation >80%
Rate of unexpected PET-CT positive cases <20%
Circulating tumour-marker positivity rate >20%
Eligibility for triple chemotherapy >80%
The feasibility measures will be evaluated at 12 months
The primary endpoint is
Rate of patients free from recurrent CRC at two years post-ablation
Secondary endpoints include:
Rate of CTCAE grade 3–5 toxicity 6 months post-ablation
Rate of patients with tumour-marker negativity 6 months after adjuvant treatment
Rate of patients with tumour-marker negativity 1 year after inclusion
Time to molecular biological recurrence: Calculated from the first time of tumour-marker negativity until tumour-marker positivity
Time to radiological recurrence: Calculated from inclusion until radiological evidence of disease recurrence
Rate of patients with local and distant relapse
Overall survival: Time from inclusion until death from any cause
QoL
Cost-effectiveness (explorative)
Data management
The study complies with the Data Protection Act and the General Data Protection Regulation. The project has been reported to the Central Denmark Region. Data are entered into electronic case report forms (eCRF) established using the RedCap framework at Aarhus University.
Monitoring
The trial sites will be visited by the Clinical Trial Manager (monitor) at times agreed with the investigator. The monitor has to ascertain protocol compliance and that the trial conduction conforms to applicable regulatory requirements and established rules for Good Clinical Practice. The monitor will review and verify the data collected in the eCRFs against the source documents and address any discrepancies. All corrections will be documented in the audit file.
Statistical analysis and sample size
All data will be presented using descriptive statistics and analysed as intention-to-treat. Kaplan-Meier estimates will be used to estimate median times to recurrence or death. Endpoints will be assessed using the log-rank test or a Cox regression model with time to event as response variable. QoL analysis will be done using the validated Danish versions of the EORTC QLQ-C30 and –CR29. A plan for explorative cost-effectiveness analyses will be drawn based on the results from the run-in phase.
The primary endpoint is the fraction of patients alive without recurrence 2 years after inclusion. A fraction of 35% is expected by standard treatment. The study is interesting for a further phase III trial if the fraction is increased to 50%. We expect that a total sample size of 350 patients is needed to full fill this. This must be revised based on the findings in the run-in phase.
Dissemination
The results are planned for publication in a peer-reviewed scientific journal
Discussion
Treatment of OCRC continues to gain ground, but the vast majority of the treated patients will relapse [Citation12,Citation13]. The use of adjuvant chemotherapy in this setting is still debateable, and predictive and prognostic markers are lacking. In this study, we evaluate ctDNA-guided adjuvant treatment.
The ultimate goal, when validating a new intervention is to test the clinical utility, which means a direct comparison with SOC. In this case, the SOC in terms of adjuvant chemotherapy after treatment for OCRC is based on limited evidence [Citation14–16]. The frequency of imaging during follow-up is also debateable, based on limited evidence. Hence, sample size calculations are based on rough assumptions. The run-in phase with an evaluation of feasibility parameters and revision of sample size calculation is crucial to be able to continue with a solid large-scale multicentre study.
A PET/CT scan will be performed before randomisation. This can compromise the external validity of the study since this is not SOC. However, an ongoing study in colorectal cancer suggests a high rate of positive PET/CT scans detecting residual disease or new cancers, which undermines the immediate use of adjuvant treatment. It is essential to exclude study participants with detectable residual disease when evaluating the effect of adjuvant treatment on minimal residual disease (MRD). Based on the results from the feasibility study, we will reconsider whether the PET/CT scan should continue to be a mandatory part of the study.
The timing of the post-ablation blood sample can be discussed. The elimination half-life of ctDNA is less than a few hours [Citation17,Citation18]. Therefore, clearance from the bloodstream is expected within a day following a curative removal of tumour tissue. However, a blood sample following major surgery can be false negative, since tissue trauma can lead to an elevated level of cell-free DNA and thereby dilute ctDNA to a concentration below the detection level [Citation19]. Therefore, we have chosen a minimum interval of 2–3 weeks from ablation to blood sampling. In patients treated with SBRT, a later time point of 4 weeks was chosen to allow for the deferred tumour reduction after radiotherapy. This time point is chosen to allow for a reasonable time for stress response to have settled but also for timely initiation of adjuvant chemotherapy if needed.
This trial is designed with randomisation against SOC, which can be discussed. Available data show that ctDNA positivity indicates a very high risk of recurrence, and the ethical dilemma of randomisation can therefore be questioned. However, it is still unknown if the risk of recurrence can be reduced by intensified chemotherapy. Furthermore, randomisation is needed to evaluate the differences in treatment-related toxicity and QoL, and hereby the overall benefits or harm from the ctDNA guided approach, although treatment regimens are well known. Feasibility for randomisation is thus a part of the run-in phase.
To the best of our knowledge, OPTIMISE is the first ongoing randomised trial to evaluate ctDNA-guided adjuvant treatment in patients treated for OCRC. Hopefully, the results will optimise the adjuvant treatment of patients treated for OCRC prolonging survival and reducing chemotherapy-related toxicity.
Regulatory approvals and consent to participate
The study is approved by the Central Denmark Region Committees on Health Research Ethics (j. no. 1-10-72-249-20) and by the Danish Medicines Agency (EudraCT no: 2020-004524-41). Informed consent is given voluntary and is obtained prior to any study-related procedure. The consent may be withdrawn at any time without consequence. The study will not present any personal data in any way and will not require consent for publication by any participants
Author contributions
LC drafted the manuscript. KS, TH, AJ, and HJ designed the study. All authors drafted the full protocol. All authors reviewed and approved the final manuscript.
Acknowledgment
The authors want to thank the OPTIMISE-study-group and the participating departments for their corporation in this study.
Disclosure statement
The authors have no affiliations or financial involvement with any organisation or entity with a financial interest or financial conflict with the study subject or materials discussed in the manuscript.
Data availability statement
The datasets used and/or analysed during the current study will be available from the corresponding author on reasonable request.
Additional information
Funding
References
- Boysen AK, Spindler KLG, Høyer M, et al. Metastasis directed therapy for liver and lung metastases from colorectal cancer – a population-based study. Int J Cancer. 2018;143(12):3218–3226.
- Nosher JL, Ahmed I, Patel AN, et al. Non-operative therapies for colorectal liver metastases. J Gastrointest Oncol. 2015;6(2):224–240.
- Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379(18):1754–1765.
- Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J Clin Oncol. 2018;36(16):1631–1641.
- Boysen AK, Pallisgaard N, Andersen CSA, et al. Circulating tumor DNA as a marker of minimal residual disease following local treatment of metastases from colorectal cancer. Acta Oncol. 2020;59(12):1424–1429.
- Hamfjord J, Guren TK, Glimelius B, et al. Clinicopathological factors associated with tumour-specific mutation detection in plasma of patients with RAS-mutated or BRAF-mutated metastatic colorectal cancer. Int J Cancer. 2021;149(6):1385–1397.
- Spindler KLG, Pallisgaard N, Vogelius I, et al. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18(4):1177–1185.
- Thomsen CB, Andersen RF, Lindebjerg J, et al. Correlation between tumor-specific mutated and methylated DNA in colorectal cancer. J Clin Oncol Precis Oncol. 2019;3:1–8.
- Thomsen CB, Hansen TF, Andersen RF, et al. Early identification of treatment benefit by methylated circulating tumor DNA in metastatic colorectal cancer. Ther Adv Med Oncol. 2020;12:1758835920918472.
- Roperch JP, Incitti R, Forbin S, et al. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566.
- Garrigou S, Perkins G, Garlan F, et al. A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker. Clin Chem. 2016;62(8):1129–1139.
- Mise Y, Imamura H, Hashimoto T, et al. Cohort study of the survival benefit of resection for recurrent hepatic and/or pulmonary metastases after primary hepatectomy for colorectal metastases. Ann Surg. 2010;251(5):902–909.
- Serrano PE, Gu CS, Husien M, et al. Risk factors for survival following recurrence after first liver resection for colorectal cancer liver metastases. J Surg Oncol. 2019;120(8):1420–1426.
- Mitry E, Fields ALA, Bleiberg H, et al. Adjuvant chemotherapy after potentially curative resection of metastases from colorectal cancer: a pooled analysis of two randomized trials. J Clin Oncol. 2008;26(30):4906–4911.
- Langer B, Bleiberg H, Labianca R, et al. Fluorouracil (FU) plus l-leucovorin (l-LV) versus observation after potentially curative resection of liver or lung metastases from colorectal cancer (CRC): result of the ENG (EORTC/NCIC CTG/GIVIO). Proc Am Soc Clin Oncol. 2002;21:149.
- Portier G, Elias D, Bouche O, et al. Multicenter randomized trial of adjuvant fluorouracil and folinic acid compared with surgery alone after resection of colorectal liver metastases: FFCD ACHBTH AURC 9002 trial. J Clin Oncol. 2006;24(31):4976–4982.
- Lo YM, Zhang J, Leung TN, et al. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999;64(1):218–224.
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990.
- Henriksen TV, Reinert T, Christensen E, et al. The effect of surgical trauma on circulating free DNA levels in cancer patients—implications for studies of circulating tumor DNA. Mol Oncol. 2020;14(8):1670–1679.