Abstract
Background
We aimed to evaluate the correlation of apoprotein E (APOE) transcription and its methylation with immune microenvironment in HCC patients.
Material and methods
The expression profiles of APOE transcription, APOE methylation, and APOE protein were investigated via comprehensive bioinformatic analyses. After that, the association between the immune activation of HCC and APOE transcription and methylation were analyzed. Finally, the prognostic role and immune correlation of the APOE protein in 92 HCC individuals was determined.
Results
Based on data from TCGA, GEO, and ICGC datasets, the APOE mRNA was differentially expressed in HCC tissues compared with normal liver tissues. Further, APOE methylation was down-regulated in HCC tissues compared to normal liver tissues. APOE methylation was negatively correlated with APOE transcription in HCC (r=–0.52, p < 0.0001). Based on APOE methylation, the HCC patients were stratified into hypermethylation and hypomethylation subgroups as they exhibited different immune activation statuses. Further, HCC individuals with APOE hypermethylation had a closer immune correlation than those with hypomethylation. Notably, APOE transcription was associated with weak immune infiltrates and activation. Finally, over-expression of the APOE protein was correlated with better survival outcomes, but not correlated with PD-1 or CTLA4 protein in HCC revealed by immunohistochemistry.
Conclusion
APOE methylation had a closer correlation with immune cells than APOE mRNA, indicating that APOE methylation might play an important role in immune regulation in HCC.
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, and usually occurs on the basis of preexistent liver diseases [Citation1]. HCC is reported to be the third most deadly tumor in the world [Citation2]. The exact pathogenesis of HCC still remains obscure, but viral hepatitis, excessive alcohol consumption, liver cirrhosis, genetic factors and immune imbalance are all involved in the occurrence of HCC [Citation3]. Although some advances in the treatment of HCC are acknowledged, the clinical outcomes of HCC patients remain unsatisfactory, specifically those with advanced TNM stage of HCC. Most individuals with HCC are diagnosed only when the disease had progressed to advanced stage, and thus lost the opportunity of radical resection. Currently, immunotherapy with immune checkpoint inhibitors (ICIs), shows promising future for advanced HCC individuals. However, some patients with advanced HCC are failed to benefit from immunotherapy with ICIs [Citation4]. An insight into the understanding of the heterogeneity of HCC immune microenvironment could aid in offering personalized immunotherapy management, and thus to improve the survival outcomes of HCC patients.
The mounting recognition of the complexity of the tumor microenvironment has inspired quite a few investigations of key genes in the regulation of the immune infiltration in HCC. Apoprotein E (APOE), a multifunctional protein, is encoded by APOE gene. Apoprotein E serves as a ligand for low-density lipoprotein receptor family members to clear certain kinds of lipoproteins [Citation5]. Thus, APOE has demonstrated to play an important role in the cardiovascular disease [Citation6] and Alzheimer’s disease [Citation7]. However, recent studies have showed that APOE was implicated in the occurrence and progression of malignant tumors [Citation8–10]. Kemp and his coworkers [Citation11] clarified that increased APOE mediates immune suppression via the regulation of NF-κB in pancreatic cancer, and high levels of serum APOE signified unfavorable patient survival. Zhang et al. [Citation12] showed that the exosome-mediated transfer of APOE protein from tumor-associated macrophages to the cancer cells could contribute to the migration of gastric tumor cells. Tavazoie et al. [Citation13] demonstrated that LXR targets APOE, which could mediate enhanced T cell activation and elicit extensive anti-tumor responses in mice. However, no study has expounded the correlation between APOE and immune microenvironment in HCC.
In the present study, we initially compared the expression profiles of APOE mRNA, APOE methylation and APOE protein between HCC tissues and normal liver tissues. Then, we explored the prognostic significance of APOE mRNA and APOE methylation in HCC though survival analysis. Subsequently, the relationship between APOE expression as well as APOE methylation and immune activation was analyzed. Finally, we performed immunohistochemistry assay to clarify the prognostic role of APOE protein in HCC patients, and its correlation with PDL1 and CTLA4.
Methods
Integrative bioinformatic analysis
RNA-seq data, DNA methylation levels of APOE and clinical information of the LIHC cohort were downloaded from UCSC website (https://xenabrowser.net/). HCCDB database (http://lifeome.net/database/hccdb/search.html), was searched to make a thorough inquiry about APOE mRNA expression in various HCC datasets. Xena Shiny (https://shiny.hiplot.com.cn/ucsc-xena-shiny/) was mined to explore differential expressions of APOE mRNA and APOE methylation in the pan-cancer datasets from The Cancer Genome Atlas (TCGA). Xena Shiny was also mined to study the prognostic role of APOE mRNA in HCC patients. Kaplan Meier Plotter database (http://kmplot.com/analysis/) is an online tool for survival analysis, which contains 371 HCC patients from the TCGA-LIHC dataset. MethSurv (https://biit.cs.ut.ee/methsurv/) was searched to determine the associations between APOE DNA methylation and clinical features. As for clinical information of HCC individuals from TCGA-LIHC data set, the mean age of the HCC patients is 59.36 ± 13.68 years. Two hundred and fifty one male individuals and 120 female HCC patients. Among them, 117 patients were infected with hepatitis virus. One hundred and eight four individuals are white people, 158 patients are Asians and 18 patients are black or Africa Americans. 257 HCC individuals are staged I-II, and 90 HCC individuals are staged III-IV. Thirty individuals with HCC received solafeini and 8 individuals received systemic radiation
Immune cells and immune activation
The single sample gene set enrichment analysis was adopted to calculate the enrichment score for each HCC individual from the TCGA LIHC data set. We defined the HCC individuals into low APOE group and high APOE group based on the median value of APOE expression in the TCGA-LIHC dataset. Similarly, HCC individuals were divided into APOE hypermethylation group and hypomethylation group based on the median value of APOE DNA methylation in the TCGA LIHC data set. Spearman correlation analysis was utilized to measure the correlations between 28 types of immune cells and APOE mRNA/methylation expression in HCC patients. We also investigated the associations between APOE mRNA/methylation expression and immune status which included 25 kinds of innate and adaptive responses. We compared the abundance of 24 immune cells, including 18 T cell subtypes and 6 other immune cell types, between the two groups based on data from Immune Cells Abundance Identifier database [Citation14]. The HCC immunity cycle stands for the anticancer immune response and thus comprises 7 critical steps [Citation15].
Collection of HCC tissues
We obtained tissue microarray chip (No. HLivH180Su17 010) from Shanghai Outdo Biotech Company (Shanghai, China) (http://www.superchip.com.cn/biology/tissue.html). This tissue microarray chip consists of 92 HCC tissue dots and 88 para-cancerous tissue dots. The clinical stages of 92 cases of HCC individuals were from stage TNM I to stage TNM III. The follow-up time ranged from 3.4 to 4.7 years, and last follow up time was up to 2016-6. Our study plan was reviewed and approved by the Ethics Committee of Renmin Hospital of Wuhan University (No. WDRY-2019-K104).
Immunohistochemical assay
Immunohistochemistry (IHC) assay was conducted to determine the expression of APOE protein in HCC tissues and para-cancerous tissue as previously reported [Citation16]. Briefly, tissue microarray was deparaffinized and rehydrated in ethanol solutions with different concentration. Then, the deparaffinized tissue was boiled in citrate buffer (pH = 6.0) for antigen retrieval for 15 min. Subsequently, 3% H2O2 solution was utilized to block the endogenous peroxidase activity of tissue microarray. The tissue microarray was incubated with the anti-APOE protein antibody (ab109117, Abcam, UK), PD-1 protein antibody (GT2280, Gene Tech, Shanghai, China) and CTLA4 protein antibody (ab19792, Abcam, UK). Then, the tissue microarray was incubated with the secondary antibody (biotinylated goat anti-mouse serum IgG). Finally, the tissue microarray was stained with diaminobenzidine and counter stained with hematoxylin. PBS resolution was added as a negative control in replace of the primary antibody.
Immunohistochemical scoring
Semi-quantitative scale on the basis of staining intensity and staining scope was applied in our analysis. The positive staining of APOE protein was defined as cytoplasmic staining in hepatocytes. The intensity of APOE protein staining was divided into four classes, including negative group (0 point) (negative), weak group (1 point), moderate group (2 points) and strong group (3 points). The assessment of IHC staining was completed by Jiao Li who is blinded to the patients’ clinical information. The final IHC scores of each tissue were multiplication of staining scope and intensity scores. X-tile (version 3.6) was employed to determine the cutoff IHC score.
Statistical analysis
Our analysis was finished with SPSS software (version 21) and R software (version 3.5.1). For comparisons of APOE IHC scores between HCC tissues and adjacent tissues, the statistical significance was detected by Student’s t-tests. Chi-square or Fisher’s exact test was used to determine correlations between the APOE protein expression and clinical metrics of HCC. To ensure whether expression of APOE mRNA was higher in HCC than that in correspondingly normal tissues, meta-analysis was performed via STATA software (version 12.0). The Spearman’s correlation analysis was utilized to measure the linear association between APOE mRNA expression and APOE DNA methylation. Associations between survival outcomes and APOE mRNA and protein expression were presented using Kaplan–Meier survival analysis, and the survival difference between two groups was measured by log-rank test. P value less than 0.05 indicated statistical significance.
Results
Differential expression of APOE mRNA in different HCC datasets
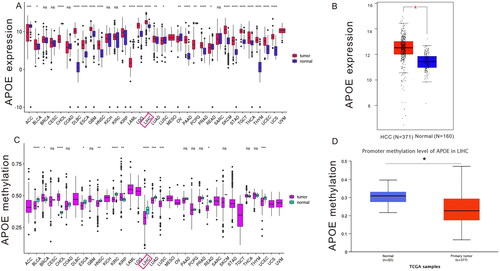
First, we explored the expression profiles of APOE transcription across various cancers using Xena Shiny (). The APOE mRNA was found to be upregulated in most malignant tumors, including HCC. In contrast, the APOE mRNA was down-regulated in lung squamous cell carcinoma and ovarian cancer. Next, we quantitatively investigated the transcription levels of APOE between HCC tissues (N = 371) from the TCGA-LIHC dataset and normal tissues (N = 160) from the GTEx database. Based on our findings, expression of APOE mRNA was up-regulated in HCC tissues compared to normal tissues (). After that, Xena Shiny was employed to investigate the methylation status of APOE across different malignant tumors. shows abnormal APOE methylation between cancerous specimens and normal tissues, including HCC ().
Figure 1. Profiles of APOE transcription (A-B), methylation (C-D) in paracancerous and HCC tissues. LIHC stands for liver hepatocellular carcinoma.
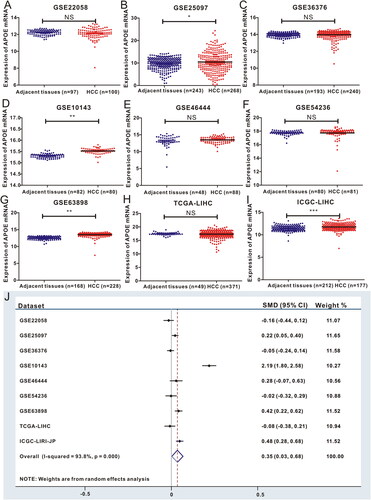
To validate the high expression of APOE mRNA in HCC tissues, we evaluated the expression levels of APOE mRNA in HCC tissues and normal specimens based on 9 different HCC datasets from TCGA, GEO and ICGC datasets. After the transformation of sequencing data from different datasets, we found that APOE mRNA was abnormally expressed in HCC (). Hence, we utilized meta-analysis to ensure whether the expression of APOE mRNA in HCC was significantly up-regulated compared to in normal tissues, and the forest plot showed the positive result (SMD = 0.35, 95%CI: 0.3–0.68, p < 0.0001, ). In brief, HCC samples displayed significantly higher expression of APOE mRNA in comparison to the expression in normal liver tissues, indicating that APOE might play a key role in the neoplastic transformation of HCC.
Survival analysis of APOE mRNA in HCC
Based on the 371 HCC individuals, the survival analysis revealed that HCC individuals with high expression of APOE transcription experienced more favorable overall survival (OS) than those with low expression of APOE transcription (HR = 0.59, 95%CI: 0.42–0.83, p = 0.0026, Supplementary Figure S1(A)). Similarly, HCC patients with high expression of APOE transcription displayed more favorable disease-free survival (DFS) than those with low expression of APOE transcription (HR = 0.58, 95%CI: 0.36–0.92, p = 0.018, Supplementary Figure S1(B)). To verify the prognostic role of APOE transcription in HCC, we also searched the Xena shiny database. Unsurprisingly, the survival curves (Supplementary Figure S1(C and D)) also indicated the similar conclusion. In a word, over-expression of APOE transcription was linked to relatively better survival outcomes of HCC patients.
Epigenetic regulation of APOE in HCC
Epigenetic regulation is one of the most common reasons for the aberrant expression of targeted gene, so we explored the DNA methylation of APOE in HCC. We browsed the UCSC Xena database to determine the epigenetic regulation of APOE in HCC. As shown in Supplementary Figure S2(A), we observed that levels of methylation of APOE promoter were decreased with increase of APOE transcription. Hence, we utilized Spearman correlation analysis to measure the relationship between APOE promoter methylation and APOE transcription. A strong association with negative coefficient (r=–0.52, p = 4.55*10−26) was identified between APOE promoter methylation and APOE transcription in HCC (Supplementary Figure S2(B)). We investigated the association of the 12 CpG sites with common clinical features, such as race, BMI and survival outcomes, which was listed in Supplementary Figure S2(C).
Correlation between APOE methylation and immune activation
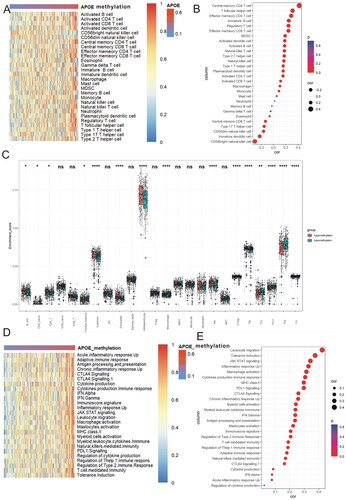
As the survival outcomes of HCC are closely linked to the infiltration of immune cells [Citation17], we measured the association between APOE methylation and infiltration of immune cells in HCC. As listed in the heat map (), the immune cells tend to be hot with the rise of APOE methylation. Methylation of APOE was significantly correlated with central memory CD4 T cell (r = 0.42, p < 0.0001), T follicular helper cell (r = 0.32, p < 0.0001) and effector memory CD4 T cell (r = 0.29, p < 0.0001) (). Due to the close relationship between APOE methylation and infiltration of T cells, we utilized a novel immune algorithm via ImmuCellAI to quantitatively measured the abundances of 24 kinds of tumor infiltrating immune cells in HCC. The result demonstrated that the majority of tumor infiltrating immune cells were remarkably different between APOE hypermethylation and hypomethylation subgroups (). Then we also investigated the relationship between APOE methylation and immune phenotypes. The heat map revealed that the immune phenotypes tend to be hot along with the increase of APOE methylation (). Correlation analysis () highlighted the close association between APOE methylation with leukocyte migration (r = 0.47, p < 0.0001), tolerance induction (r = 0.38, p < 0.0001), JAK STAT signaling (r = 0.35, p < 0.0001), inflammatory response up (r = 0.33, p < 0.0001) and macrophages activation (r = 0.31, p < 0.0001).
Figure 3. APOE methylation is associated with immune infiltration and immune activation in HCC. (A) Heatmap displayed APOE methylation associated relative abundance of 28 immune cells in HCC. (B) The relationship between the methylation of APOE and 28 immune cells in HCC. (C) The comparison of TILCs in APOE hypermethylation and hypomethylation subgroups. (D) Heatmap showing a relative association between APOE methylation and 26 immunity-related gene sets. (E)The relationship between 26 immunity-related gene sets and APOE methylation in HCC
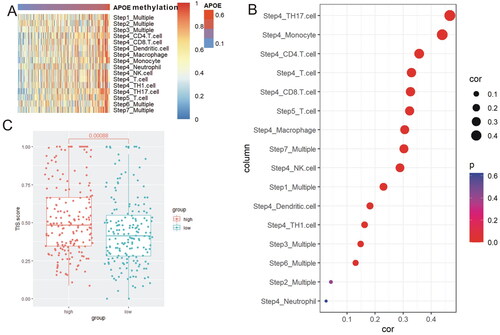
The cancer immunity cycle could represent the anti-cancer response, and consists of seven important steps. In the APOE hypermethylation subgroup, most steps in the cancer immunity cycle were up-regulated (), including step 4 (Th17 cell, Monocyte, CD4 T cell), step 5 (T cell) and step 7 (multiple) (). Considering most types of T cells were up-regulated in the APOE hypermethylation group, and anti-cancer immune response was up-regulated in APOE hypermethylation group, we wondered whether levels of PD-1, PD-L1 and CTLA4 were also over-expressed in APOE hypermethylation group. Levels of PD-1 (p = 0.0054, Supplementary Figure S3(A)), PD-L1 (p < 0.0001, Supplementary Figure S3(B)) and CTLA4 (p = 0.048, Supplementary Figure S3(C)) were significantly higher in APOE hypermethylation group than that in APOE hypomethylation group. The methylation level of APOE was positively correlated with PD-1 (r = 0245, p < 0.001, Supplementary Figure S3(G)), PD-L1 (r = 0.325, p < 0.0001, Supplementary Figure S3(H)) and CTLA4 (r = 0.1695, p = 0.0005, Supplementary Figure S3(I)) in HCC. Subsequently, we adopted T cell inflammatory signature (TIS) scores to evaluate the immune significance in APOE hypermethylation and hypomethylation subgroups. HCC sufferers with APOE hypermethylation obtained higher TIS scores than those with APOE hypomethylation (Supplementary Figure 4(C)) (p = 0.00088).
Association between APOE expression and immune activation
We also measured the relationship between APOE expression and infiltration of immune cells to determine which is closer to immune microenvironment in HCC. As exhibited in the heat map (Supplementary Figure S4(A)), the immune cells tend to be hot with the decrease of APOE expression. APOE expression was weakly correlated with CD56 bright NK cell (r = 0.25, p < 0.0001), CD56 dim NK cell (r = 0.24, p < 0.0001), and inversely correlated with effector memory CD4 T cell (r=–0.28, p < 0.0001) and central memory CD4 T cell (r=–0.27, p < 0.0001) (Supplementary Figure S4(B)). Then, we utilized ImmuCellAI to quantitatively measure the abundances of 24 kinds of tumor infiltrating immune cells in HCC. The result demonstrated that several tumor infiltrating immune cells were remarkably different between APOE low and high expression subgroups (Supplementary S4(C)). Subsequently, we investigated the relationship between APOE expression and immune phenotypes. The heat map revealed that immune phenotypes tended to be hot along with the decrease of APOE transcription (Supplementary Figure S4(D)). Correlation analysis highlighted the weak association between APOE expression NK cell mediated immunity (r=–0.14, p < 0.01, Supplementary Figure S4(E)).
In the APOE over-expression subgroup (Supplementary Figure S5(A)), fewer steps in the cancer immunity cycle were up-regulated, including step 1 (multiple), and step 5 (T cell) (Supplementary Figure S5(B)). When we divided these HCC individuals into low APOE and high APOE expression groups, we found that only levels of PD-L1 (p < 0.0001) were significantly higher in APOE low expression group compared to that in APOE high expression group (Supplementary S3(D–F)). Correlation analysis (Supplementary Figure S3(J–L)) proved that APOE expression was negatively correlated with PD-L1 (r=–0.32, p < 0.0001), while there was little correlation with PD-1 (r=–0.07, p = 0.156) and CTLA4 (r=–0.085, p = 0.083) in HCC. Finally, we adopted TIS scores to assess the immune significance in low and high APOE expression subgroups. HCC sufferers with over-expression of APOE mRNA obtained similar TIS scores compared to those with low expression of APOE mRNA (p = 0.5, Supplementary Figure S5(C)).
Association between APOE protein and clinical features
We used the 92 HCC tissues from Shanghai Outdo Biotech Company to validate the protein expression of APOE, and to verify the association between APOE protein and PD-L1 and CTLA4 in HCC. APOE protein was mainly expressed in the hepatoma cell cytoplasm other than nucleus (), and we also observed that expression of APOE protein is stronger in HCC tissues than the expression of APOE protein in adjacent tissues (). In our experiments, we also detected the expression of PD-L1 and CTLA4 protein in HCC (Supplementary Figure S6). Spearman correlation analysis proved that expression of PD-L1 protein is correlated with expression of CTLA4 (r = 0.292, p = 0.005). However, there was no association of APOE protein with PD-L1(r = 0.085, p = 0.428) or CTL4A (r = 0.087, p = 0.4154) in HCC tissues.
Figure 5. Expression and survival significance of APOE protein in HCC patients. Representative figures of APOE staining in HCC tissues (A-B) and adjacent liver tissues (C-D). APOE protein is highly expressed in HCC tissues (E). HCC patients with APOE over-expression displayed favorable overall survival (F) and disease-free survival (G) than those with low APOE expression.
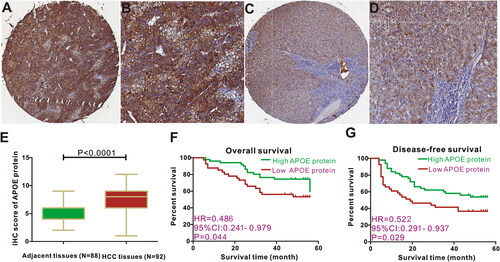
Then, we applied Chi-square test to determine the distribution of clinical metrics between the low APOE protein group and the high APOE protein group based on the threshold value derived from X-tile. Survival analysis showed that HCC individuals with high expression of APOE protein possessed longer OS time compared to those with low APOE protein expression (HR = 0.486, 95%CI: 0.241–0.979, p = 0.044, ). Similarly, Kaplan–Meier curve also exhibited that HCC individuals with high expression of APOE protein experienced relatively favorable DFS compared to those with low APOE protein expression (HR = 0.522, 95%CI: 0.291–0.937, p = 0.029, ). Collectively, we concluded that high expression of APOE protein is a protective factor for favorable survival outcomes in HCC sufferers, and was in line with the transcription level of APOE in HCC.
Discussion
Abnormal lipid metabolism is implicated in the occurrence and progression of HCC [Citation18,Citation19]. Dysfunction of lipid metabolism in hepatocytes induces aberrant expression of a series of metabolic genes [Citation20–22]. Our team previously reported the clinical and prognostic significance of APOA-1 in HCC, and we deemed that low levels of APOA-1 mRNA might be a reliable molecular of predicting survival in HCC sufferers [Citation23]. APOE protein is a major apoprotein of the chylomicron, and is essential for the normal catabolism of triglyceride-rich lipoprotein constituents. Our study systematically explored the role of APOE mRNA and DNA methylation in HCC, and we found a negative correlation between APOE mRNA expression and DNA methylation. Moreover, we also investigated the correlation between immune activation status and APOE mRNA as well as APOE methylation. Surprisingly, APOE methylation, rather than APOE mRNA was significantly correlated with immune activation status in HCC. IHC assay was conducted to determine the clinical and prognostic values of APOE protein and to verify the associations between APOE expression and PDL1 as well as CTLA4 in HCC. Hence, this is the first research to systematically elucidate the role of APOE both from transcription and protein levels in HCC patients.
The pathogenesis of HCC is a complex process involving the progressive accumulation of key gene alterations and epigenetic modification [Citation24]. DNA methylation is the most common epigenetic phenomenon in HCC, and aberrant DNA methylation will result in abnormal expression of oncogenes, exerting drastic effects on the biological behaviors of HCC [Citation25]. In our analysis, we found that the methylation level of APOE in HCC tissues was much lower than that in correspondingly normal tissues. Moreover, we also observed a negative correlation between APOE mRNA expression and DNA methylation in HCC. The differential methylation of APOE between HCC tissues and normal tissues and the negative correlation could well explain why APOE mRNA expression was remarkably higher in HCC specimens than that in normal tissues. Our study provides an area for subsequent research in understanding the clinical significance of APOE DNA methylation in the progression of HCC.
The secreted APOE possesses pleiotropic effects on human immunity [Citation26]. Wolters et al. [Citation27] collected data from 38,537 participants from six population-based cohort studies, and found that APOE-ε2 is correlated with prolonged survival time, but APOE-ε4 is associated with a high risk of mortality. Recent research [Citation28] pointed out that common germline variants of the APOE gene can significantly affect the progression and survival outcomes in patients with melanoma. Chen et al. [Citation29] demonstrated that expression of APOE in nuclei was significantly correlated with a higher 5-year survival rate in ovarian cancer patients with peritoneal effusion. Moreover, Su and coworkers [Citation30] clarified that high expression of APOE was a strong risk factor for more unfavorable survival in lung cancer patients presented with malignant pleural effusions. Although all these related studies indicate that the APOE gene is crucial in the progression of multiple cancers, the actual significance for survival in HCC has not been documented to date. Our study is the first analysis to report that high expression of APOE mRNA was associated with better overall survival and disease-free survival. Moreover, we also utilized IHC assay to explore the prognostic value of the APOE protein in HCC, and found that HCC patients with over-expression of APOE exhibited superior overall survival. In a word, we threw light on the prognostic significance of APOE mRNA and protein in HCC for the first time.
It is known that the survival outcomes of HCC are closely correlated with the infiltration status of immune cells [Citation31,Citation32]. APOE is highly expressed in tumor-associated macrophages, which play a critical role in the process of immunosuppression. A previous study [Citation33] put forward that APOE can suppress the proliferation of T cells, regulate the functions of macrophages, and facilitate the presentation of lipid antigens to natural killer T cells. Kemp et al. [Citation11] found that silence of the APOE gene in mice leads to the up-regulation of CD8+ T cells in tumor cells. Moreover, activation of APOE induced the over-expression of immunosuppressive factors, such as CXCL1 and CXCL5. In HCC, we found that expression of APOE was positively correlated with the infiltration of natural killer cells, and negatively correlated with the infiltration of effect memory CD4 T cells. As for immunological pathways, activation of APOE is positively correlated with cytokine production, and negatively correlated with natural killer cells' medicated immunity. However, these correlations are relatively weaker than correlations between APOE methylation and immune cells. APOE hypermethylation in HCC is positively correlated with the infiltration of central effect memory CD4 T cells, and leukocyte immigration. Finally, we found that APOE hypermethylation rather than APOE over-expression is closer to immune infiltration in HCC. Taken together, APOE methylation plays a critical role in the regulation of the immune component of TIME of HCC, indicating that APOE methylation might play an important role in the immune microenvironment in HCC.
Several limitations should not be neglected in our analysis. First, our bioinformatic analysis was mainly derived from the TCGA data set, and we lacked our own clinical cohort to validate the role of APOE methylation in HCC patients. Then, due to the available data, we could not verify the predictive significance of APOE methylation for predicting the immunotherapy response. Moreover, immunotherapy is effective for HCC patients with advanced stage, while we included HCC individuals with early-stage from Outdo cohort. It might be the reason why APOE protein is not correlated with PD-L1 or CTLA4. Therefore, clinical cohorts from different medical centers are warranted in our future research. Subsequently, cellular experiments and multi-center clinical trials are still needed to investigate the regulatory mechanism of APOE in HCC.
Conclusion
Our analysis revealed that APOE expression is correlated with survival outcomes, and negatively regulated by APOE DNA methylation in HCC. HCC patients with APOE hypermethylation are more likely to be associated with the activation of the immune response. These findings shed novel insights into the regulation of APOE DNA methylation associated with the immune response in HCC.
Ethics statement
Our study plan was reviewed and approved by the Ethics Committee of Renmin Hospital of Wuhan University (No. WDRY-2019-K104).
Author contributions
Peng PL and Liu Q designed this study; Li J and Tian S completed the bioinformatic analysis; Li J and Tian S wrote the manuscript; Peng PL revised this manuscript.
Supplemental Material
Download MS Word (82.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analyzed during this study are included in this article and its supplementary information files.
Additional information
Funding
References
- Scheiner B, Pomej K, Kirstein MM, et al. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy–development and validation of the CRAFITY score. J Hepatol. 2022;76(2):353–363. doi: 10.1016/j.jhep.2021.09.035.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492.
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2.
- Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382(20):1894–1905. doi: 10.1056/NEJMoa1915745.
- Getz GS, Reardon CA. Apoprotein e and reverse cholesterol transport. Int J Mol Sci. 2018;19(11):3479.
- Kulminski AM, Culminskaya I, Arbeev KG, et al. Trade-off in the effect of the APOE gene on the ages at onset of cardiocascular disease and cancer across ages, gender, and human generations. Rejuvenation Res. 2013;16(1):28–34. doi: 10.1089/rej.2012.1362.
- Zhao N, Ren Y, Yamazaki Y, et al. Alzheimer’s risk factors age, APOE genotype, and sex drive distinct molecular pathways. Neuron. 2020;106(5):727–742.e6. doi: 10.1016/j.neuron.2020.02.034.
- Butterbrod E, Sitskoorn M, Bakker M, et al. The APOE epsilon4 allele in relation to pre- and postsurgical cognitive functioning of patients with primary brain tumors. Eur J Neurol. 2021;28(5):1665–1676. doi: 10.1111/ene.14693.
- Carroll JE, Small BJ, Tometich DB, et al. Sleep disturbance and neurocognitive outcomes in older patients with breast cancer: interaction with genotype. Cancer-Am Cancer Soc. 2019;125(24):4516–4524. doi: 10.1002/cncr.32489.
- Sienski G, Narayan P, Bonner JM, et al. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci Transl Med. 2021;13(583):eaaz4564. doi: 10.1126/scitranslmed.aaz4564.
- Kemp SB, Carpenter ES, Steele NG, et al. Apolipoprotein e promotes immune suppression in pancreatic cancer through NF-kappa B-Mediated production of CXCL1. Cancer Res. 2021;81(16):4305–4318. doi: 10.1158/0008-5472.CAN-20-3929.
- Zheng P, Luo Q, Wang W, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional apolipoprotein E. Cell Death Dis. 2018;9(4):434. doi: 10.1038/s41419-018-0465-5.
- Tavazoie MF, Pollack I, Tanqueco R, et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell. 2018;172(4):825–840.e18. doi: 10.1016/j.cell.2017.12.026.
- Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T-Cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci. 2020;7(7):1902880. doi: 10.1002/advs.201902880.
- Xu L, Deng C, Pang B, et al. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res. 2018;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689.
- Tian S, Peng P, Li J, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/beta-catenin signaling pathway. Aging. 2020;12(4):3574–3593. doi: 10.18632/aging.102831.
- Zhao Y, Kong LX, Feng FS, et al. A simple CD4+ T cells to FIB-4 ratio for evaluating prognosis of BCLC-B hepatocellular carcinoma: a retrospective cohort study. BMC Cancer. 2022;22(1):311. doi: 10.1186/s12885-022-09433-3.
- Zhao M, Bu Y, Feng J, et al. SPIN1 triggers abnormal lipid metabolism and enhances tumor growth in liver cancer. Cancer Lett. 2020;470:54–63. doi: 10.1016/j.canlet.2019.11.032.
- Li XJ, Li QL, Ju LG, et al. Deficiency of histone methyltransferase SET domain-containing 2 in liver leads to abnormal lipid metabolism and HCC. Hepatology. 2021;73(5):1797–1815. doi: 10.1002/hep.31594.
- Seo J, Jeong DW, Park JW, et al. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun Biol. 2020;3(1):638. doi: 10.1038/s42003-020-01367-5.
- Gu L, Zhu Y, Lin X, et al. Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and hepatocarcinogenesis. Oncogene. 2020;39(11):2437–2449. doi: 10.1038/s41388-020-1156-0.
- Jiang X, Qian H, Ding WX. New glance at the role of TM6SF2 in lipid metabolism and liver cancer. Hepatology. 2021;74(3):1141–1144. doi: 10.1002/hep.31851.
- Guo Y, Huang B, Li R, et al. Low APOA-1 expression in hepatocellular carcinoma patients is associated with DNA methylation and poor overall survival. Front Genet. 2021;12:760744. doi: 10.3389/fgene.2021.760744.
- Ho DW, Lo RC, Chan LK, et al. Molecular pathogenesis of hepatocellular carcinoma. Liver Cancer. 2016;5(4):290–302. doi: 10.1159/000449340.
- De Zhu J. The altered DNA methylation pattern and its implications in liver cancer. Cell Res. 2005;15(4):272–280. doi: 10.1038/sj.cr.7290296.
- Shi Y, Holtzman DM. Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol. 2018;18(12):759–772. doi: 10.1038/s41577-018-0051-1.
- Wolters FJ, Yang Q, Biggs ML, et al. The impact of APOE genotype on survival: results of 38,537 participants from six population-based cohorts (E2-CHARGE). PLoS One. 2019;14(7):e0219668. doi: 10.1371/journal.pone.0219668.
- Ostendorf BN, Bilanovic J, Adaku N, et al. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat Med. 2020;26(7):1048–1053. doi: 10.1038/s41591-020-0879-3.
- Chen YC, Pohl G, Wang TL, et al. Apolipoprotein E is required for cell proliferation and survival in ovarian cancer. Cancer Res. 2005;65(1):331–337. doi: 10.1158/0008-5472.331.65.1.
- Su WP, Chen YT, Lai WW, et al. Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer. 2011;71(1):28–33. doi: 10.1016/j.lungcan.2010.04.009.
- Dai Y, Qiang W, Lin K, et al. An immune-related gene signature for predicting survival and immunotherapy efficacy in hepatocellular carcinoma. Cancer Immunol Immunother. 2021;70(4):967–979. doi: 10.1007/s00262-020-02743-0.
- Xue R, Zhang Q, Cao Q, et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature. 2022;612(7938):141–147. doi: 10.1038/s41586-022-05400-x.
- Zhang H, Wu LM, Wu J. Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm. 2011;2011:949072. doi: 10.1155/2011/949072.