Introduction
Classified as a provisional entity in the 4th edition of the World Health Organization (WHO) classification of hematolymphoid tumors, IRF4-Large B cell lymphoma (LBCL) was recently upgraded to a separate entity [Citation1]. This uncommon subtype of LBCL is characterized by strong IRF4/MUM1 expression resulting from the translocation of an enhancer gene, most commonly IGH, to IRF4 regulatory sequences [Citation2]. IRF4-LBCL prevails in children and young adults and is characterized by a favorable clinical outcome [Citation3].
IRF4 is located at the terminal end of the short arm of chromosome 6 (6p25.3) and is frequently involved in cryptic cytogenetic aberrations, necessitating detection using break-apart Fluorescent in Situ Hybridization (FISH) [Citation3].
Due to the rarity of this lymphoma subgroup, the number of IRF4-rearranged LBCL reported to date is still limited [Citation4]. We hereby present the first case of a high-grade IRF4-LBCL with spontaneous remission resulting from dysregulation by IGL translocation. We describe the complex genetic rearrangement that led to IRF4 transposition adjacent to the rare partner gene IGL along with Gene Expression Profiling (GEP) and mutational analysis.
Case presentation
A 34-year-old man, previously healthy, presented for the investigation of an isolated right cervical lymphadenopathy. Peripheral blood count parameters were within normal range. Serum protein electrophoresis did not reveal a dominant electrophoretic band. EBV and CMV serologies were in favor of previous (IgG positive), but not active (IgM negative) infections. Fluorine-18-2-fluoro-2-deoxy-d-glucose Positron Emission with Computed Tomography (FDG-PET/CT) identified one region of increased uptake in the cervical area. The single PET-positive lymph node measured 2.3 cm at its largest and 1.5 cm at its smallest diameter. Microscopic examination of the lymph node biopsy (partial lymphadenectomy; incisional biopsy) showed a mixed diffuse and nodular lymphoid proliferation, composed of sheets of monotonous medium to large-sized cells with open chromatin and prominent nucleoli, morphologically suggestive of LBCL (; panels A–B). Immunohistochemical (IHC) stains revealed positivity for CD10 (; panel C), CD20, BCL2, BCL6, strong nuclear MUM1 (; panel D), and a high proliferation index (Ki-67: 80%). Tumor cells were negative for CD3, CD5, CD23, CD30, c-MYC, cyclin D1, EBER, IgD, PDL1, and SOX11. A LBCL with IRF4 rearrangement was suspected.
Figure 1. panels A–B: Hematoxylin and eosin stain original magnification ×500 [a], ×1000 [B]; panel C: Immunohistochemistry CD10; panel D: Immunohistochemistry nuclear MUM1.
![Figure 1. panels A–B: Hematoxylin and eosin stain original magnification ×500 [a], ×1000 [B]; panel C: Immunohistochemistry CD10; panel D: Immunohistochemistry nuclear MUM1.](/cms/asset/7cb70add-f183-4184-8f24-6d084a9be23a/ionc_a_2238546_f0001_c.jpg)
Cytogenetics
R-banding analysis of chromosomes from nodal tissue obtained by short-term culture without mitogens revealed a complex karyotype, with loss of chromosomes Y and 6, deletion 13q confirmed by FISH (XL DLEU/LAMP Metasystems, Germany) (; panel A), a derivative of chromosome 22 and a marker chromosome possibly containing material originating from chromosome 6 (; panel D). Break-apart probes targeting IRF4 (XL IRF4; Metasystems, Germany) showed the presence of 2 fusions and one small red split signal indicating a break in the region upstream of the 5′ portion of IRF4 and subsequent translocation into a group G chromosome (; panel B). Translocation/Dual-fusion probes (XL t(8;14) MYC/IGH; Metasystems, Germany) showed no rearrangements in MYC and IGH. No BCL2 or BCL6 rearrangements were detected (XL BCL2, XL BCL6; Metasystems, Germany). IGL break-apart probes (XL 22q11 IGL; Metasystems, Germany) showed the presence of a break and translocation of the region distal to the 3′ portion of IGL into a group C chromosome (; panel C). Chromosome painting FISH for chromosomes 6, 13, and 22 (XCP 6 Orange, XCP 13 Green, XCP 22 Green; Metasystems, Germany) showed the absence of material originating from chromosome 13 in the marker chromosome (; panel E). The short arm of the marker chromosome was found to contain material from chromosome 6 in tandem with material from chromosome 22 (; panel F).
Figure 2. panel A: D-FISH 13q14.2(red)/13q34(Green). panel B: Break apart FISH 5’IRF4(red)/3’IRF4(green). panel C: Break apart FISH 5’-IGL(red)/3’IGL(green). panel D: R-banding karyotype. panel E: Whole chromosome painting FISH chromosome 6 (red) and chromosome 13 (Green). panel F: Whole chromosome painting FISH chromosome 6 (red) and chromosome 22 (Green).
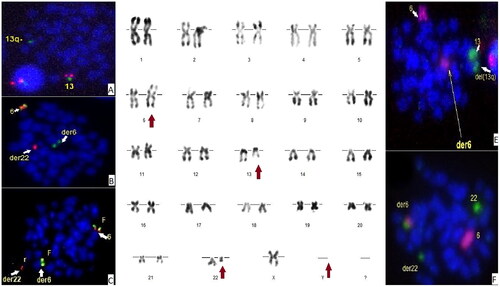
Together, these results indicated the presence of a reciprocal translocation t(6;22)(p25;q11) resulting in the rare IGL::IRF4 fusion. This translocation involves a derivative 22 containing the 5′-IGL fused to part of the 5′ region upstream IRF4, and the reciprocal fusion on a marker chromosome. The ISCN2020 nomenclature of our patient’s karyotype is as follows: 44,X,-Y,del(2)(q2?4q33),-6,del(13)(q13q2?2),der(22)t(6;22)(p25;q11),+mar[8]/46,XY[4].ish del(13)(DLEU-,LAMP1+),der(22)(wcp22+,5’IGL+),+mar(wcp22+,BCR+,wcp6+,3’IGL+,wcp6-,wcp13-,wcp22-)[4].nuc ish(DLEUx1,LAMP1x2)[80/100].
Gene expression profiling
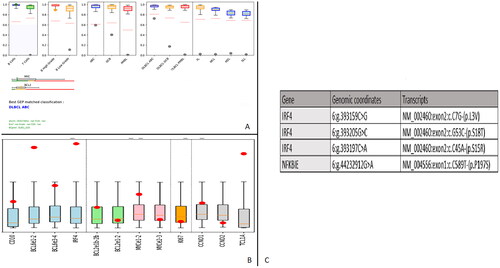
After RNA extraction from Formalin-Fixed Paraffin-Embedded (FFPE) tissue using the Maxwell 16 system (Promega, Manheim), GEP of 137 genetic markers was performed using a combination of ligation-dependent Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR), Next Generation Sequencing (NGS) and machine learning technology. This in-house assay allows the differentiation of the 7 most common subtypes of B-cell Non-Hodgkin Lymphomas (NHL) [Citation5]. Based on expression profiles, our case was classified as an activated B-cell Diffuse LBCL (ABC DLBCL), with a few genetic markers shared by Germinal Center DLBCL (; panel A). The strong expression of CD10 and BCL6 coupled with very high expression of IRF4 detected by GEP confirmed the diagnosis of IRF4-rearranged LBCL (; panel B).
Figure 3. A: Gene expression profiling classifying our case as an activated B-cell diffuse LBCL (ABC DLBCL), with a few genetic markers shared by Germinal Center DLBCL. Green dot: B cell proliferation, Orange dot: high grade B cell proliferation, grey dot: DLBCL ABC profile. B: Gene expression profiling showing strong expression of CD10 and BCL6 coupled with very high expression of IRF4, confirming the diagnosis of IRF4-rearranged LBCL. Red dot: GEP of our lymphoma, boxplot: normal expression from 25th to 75th percentile. C: Mutational profile results.
Mutational profiling
Mutational profiling was performed using a domestic gene panel [Citation6]. The LymphoPanel allows the detection of mutations in 34 genes (69 gene regions), known to play an important role in lymphomagenesis. The NextSeq 550 (Illumina, California, United States) and UMI-VarCall bioinformatics pipeline [Citation7] were used for NGS and mutational analysis. Three mutations were detected in IRF4 with a high Variant Allele Frequency (VAF), indicating their presence in the predominant (main) clone. In addition, 1 mutation was detected in NFKBIE but not in other NFkB pathway genes, including CARD11/CD79B/MYD88 (; panel C).
Treatment and follow-up
The patient was planned to undergo 4 cycles of R-CHOP (Rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine, prednisolone). However, the pretreatment FDG-PET/CT showed a complete absence of metabolically active tissue, indicating spontaneous regression of the disease. No chemotherapy was administered. The patient has been followed up with laboratory and radiologic testing for the past 28 months, with no recorded events.
Discussion
IRF4-LBCL accounts for 0.05% of all lymphoma cases. More common in children and young adults, it is known to present at early stages, typically localized in the head and neck region. IGH is the most frequent partner of IRF4, constituting between 57% and 74% of translocations [Citation3,Citation4]. Our case describes the involvement of IGL in high-grade disease. IGL::IRF4 fusion is rare, with only 4 cases reported to date [Citation3,Citation8,Citation9]. IRF4 dysregulation by light chain immunoglobulin was long thought to be exclusive of low-grade lymphoma, possibly accounting for the superior outcomes [Citation8,Citation10]. The rarity of this occurrence, however, prohibits drawing any conclusions about the effect of IGL involvement on the clinical behavior of IRF4-LBCL.
IRF4 high-grade lymphomas share common phenotypic features. Morphologically, most cases show a diffuse pattern of proliferation, but can also present as a nodular or mixed follicular and diffuse growth [Citation11]. Tumor cells express strong MUM1/IRF4 and BCL6 [Citation11]. CD10 and BCL2 expression is not uniform and was detected in around 60% of reported cases [Citation3,Citation9]. Ki-67 expression is variable but overall high [Citation3]. Based on the Hans Algorithm (HA) [Citation12], LBCL aberrantly coexpressing CD10+/BCL6+/MUM1+ are classified in the Germinal Center (GC) group. However, GEP is suggestive of an Activated B-Cell (ABC) origin in 30% of those lymphomas, including our case [Citation2,Citation9]. This reflects the inability of the IHC classification to accurately determine the cell of origin of all IRF4-LBCL, especially in adults [Citation9]. Around 22% of LBCL with aberrant coexpression were found to harbor IRF4 rearrangements [Citation9]. LBCL with CD10+/BCL6+/MUM1+ should thus be screening for IRF4 breaks using FISH. Because CD10 expression is not uniform in IRF4-LBCL, double expressors (strong MUM1 and BCL6) negative for BCL2 and MYC rearrangement by FISH should be considered for IRF4 screening in the right clinical context such as young age at presentation and the presence of isolated lymphadenopathy or tonsillar enlargement. In this setting, the choice of adequate probes is crucial. Around 15% of LBCL with IRF4 rearrangement are negative by FISH [Citation2,Citation3,Citation9]. IRF4 dysregulations frequently involve breaks encompassing only a small centromeric [Citation3] or telomeric portion of the gene (as in our case) or are the result of the positioning of the immunoglobulin gene regulatory sequence far upstream of IRF4, resulting in the elevated negativity rate. FISH probes should be selected to cover a wide region, extending to sequences upstream and downstream of the 5′ and 3′ portions of IRF4.
IRF4-LBCL patients exhibit a good clinical outcome, with event-free survivals (EFS) of 100% at 2 and 5 years (versus 88.5% and 64.9% respectively in IRF4-negative patients) [Citation3,Citation4]. However, the difference in age was found to account for this observation after adjustment by COX regression analysis [Citation3]. Indeed, among IRF4-positive patients, pediatrics (≤ 18 years) was shown to have a better 5-year EFS (76%) than young adults (19–25 years) (46%) [Citation2]. Adult IRF4-LBCL was found to carry a higher mutational burden, including multiple IRF4 mutations and downstream dysregulation of the NF-κB pathway, more frequently CARD11/CD79B/MYD88. The latter was described in lymph node biopsies with diffuse morphology [Citation2,Citation9]. The low mutational load of our adult lymphoma (3 IRF4 + 1 NFKBIE mutations) could explain the low-risk behavior of our patient’s lymphoma. The association between CARD11/CD79B/MYD88 mutations and diffuse morphology is uncertain, demonstrated by our wild-type lymphoma case exhibiting mixed morphology.
Currently, IRF4-positive lymphoma is treated with the same high-intensity regimens as IRF4-negative high-grade NHL [Citation11]. As previously mentioned, reported cases show low-risk disease [Citation2,Citation4]. Our adult patient underwent spontaneous remission, despite the elevated proliferation index and ABC subtype. Among the 20 IRF4-LBCL reported by Ramis-Zaldivar et al. 4 were treated with surgical excision only [Citation2]. This raises the possibility of therapy de-escalation for IRF4-positive NHL, hence the importance of screening LBCL with strong IRF4/MUM1 and BCL6/CD10.
Conclusion
Chromosome banding analysis is important for the accurate elucidation of the complex genetic aberrations frequently observed in IRF4-LBCL. Screening of BCL6+/MUM1+ LBCL for IRF4 rearrangements using wide coverage FISH probes is essential for potential therapy de-escalation. Immunohistochemistry (HA) is insufficient for the classification of IRF4-LBCL, hence the importance of publishing GEP results of this rare entity. Further research is required to establish the molecular profile of IRF4-LBCL, its transition along the age continuum, and its effect on clinical behavior.
Ethics approval
Institutional board review (IRB) is not required for single case reports as per the IRB of the American University of Beirut.
Patient consent statement
Non opposition of data sharing was signed by the study participant.
Acknowledgements
The authors would like to thank Mrs Vinciane Rainville, Mrs Elodie Bohers and Dr Philippe Ruminy for their valuable contributions in the molecular profiling of our case including testing and data interpretation.
Disclosure statement
The authors do not have any relationships/activities/interests with for-profit or not-for-profit third parties whose interests may be affected by the content of the manuscript.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Additional information
Funding
References
- Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36(7):1720–1748. doi: 10.1038/s41375-022-01620-2.
- Ramis-Zaldivar J, Gonzalez-Farre B, Balague O, et al. Distinct molecular profile of IRF4-rearranged large B-cell lymphoma. Blood. 2020;135(4):274–286. doi: 10.1182/blood.2019002699.
- Salaverria I, Philipp C, Oschlies I, et al. Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood. 2011;118(1):139–147. doi: 10.1182/blood-2011-01-330795.
- Au-Yeung R, Padilla L, Zimmermann M, et al. Experience with provisional WHO-entities large B-cell lymphoma with IRF4-rearrangement and Burkitt-like lymphoma with 11q aberration in paediatric patients of the NHL-BFM group. Br J Haematol. 2020;190(5):753–763. doi: 10.1111/bjh.16578.
- Bobée V, Drieux F, Marchand V, et al. Combining gene expression profiling and machine learning to diagnose B-cell non-Hodgkin lymphoma. Blood Cancer J. 2020;10(5):59. doi: 10.1038/s41408-020-0322-5.
- Dubois S, Viailly P-J, Mareschal S, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin Cancer Res. 2016;22(12):2919–2928. doi: 10.1158/1078-0432.CCR-15-2305.
- Sater V, Viailly P, Lecroq T, et al. UMI-VarCal: a new UMI-based variant caller that efficiently improves low-frequency variant detection in paired-end sequencing NGS libraries. Bioinformatics. 2020;36(9):2718–2724. doi: 10.1093/bioinformatics/btaa053.
- Takeuchi K, Sakata S, Asaka R, et al. A low-grade B-cell lymphoma with prolymphocytic/paraimmunoblastic proliferation and IRF4 rearrangement. Haematologica. 2013;98(3):e32-5–e35. doi: 10.3324/haematol.2012.076851.
- Frauenfeld L, Castrejon-de-Antra N, Ramis Zaldivar J, et al. Diffuse large B-cell lymphomas in adults with aberrant coexpression of CD10, BCL6, and MUM1 are enriched in IRF4 rearrangements. Blood Adv. 2021;6(7):2361–2372. doi: 10.1182/bloodadvances.2021006034.
- Nasim M, Chalif D, Demopoulos A, et al. Primary low-grade B-cell lymphoma of skull with translocation between immunoglobulin and interferon regulatory factor 4 genes. Int J Surg Pathol. 2020;28(3):330–335. doi: 10.1177/1066896919883013.
- Swerdlow S. WHO classification of tumors of hematopoietic and lymphoid tissues. Revised 4th edition ed. Lyon: WHO; 2017.
- Hans C, Weisenburger D, Greiner T, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545.
