Abstract
The cellular receptor of subgroup B avian leukosis virus (ALVB) is encoded by a gene at the tumour virus B (TVB) locus. TVB alleles encode specific receptors permitting infection by exogenous ALVB or avian leukosis virus subgroup D (ALVD) as well as endogenous avian leukosis virus subgroup E (ALVE), and thus susceptibility is dominant to resistance. Two single nucleotide polymorphisms at the TVB locus have been reported distinguishing three TVB alleles (TVB*S1, TVB*S3 and TVB*R). We have developed a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay using the two single nucleotide polymorphisms to define three observed allelic haplotypes and to identify the six possible TVB genotypes consisting of the three haplotypes in defined laboratory strains of chickens. One additional potential allelic haplotype and four genotypes were also briefly discussed. Chickens from parents heterozygous for different TVB alleles were challenged with Rous sarcoma viruses of subgroup ALVB and ALVE to induce wing-web tumours. Tumour incidences were evaluated between chickens of the genotypes determined with this newly developed PCR-RFLP assay. Importantly, chickens typed with this assay as TVB*S3/*S3 were resistant to infection by ALVE only, and those TVB*R/*R were resistant to both ALVE and ALVB. Furthermore, a vast majority of chickens with the susceptible TVB*S1/– genotypes developed a tumour. This PCR-RFLP assay enables a relatively rapid assessment of all six anticipated TVB genotypes in experimental strains of chickens undergoing segregation for TVB*S1, TVB*S3, and TVB*R alleles. This non-infectious assay should be further evaluated for the capacity to select and breed commercial chickens for genetic resistance to infections by ALVB, ALVD and ALVE.
Développement et validation d'un test PCR-RFLP pour évaluer les haplotypes TVB codant les récepteurs des sous groupes B et E des virus de la leucose aviaire chez les Leghorn blanche
Le récepteur cellulaire du sous groupe B du virus de la leucose aviaire (ALVB) est encodé par un gène au niveau du locus tumeur virus B (TVB). Les allèles TVB encodent les récepteurs spécifiques permettant l'infection par les virus exogènes appartenant au sous groupe B, ou au sous groupe D (ALVD) ainsi que par les virus endogènes appartenant au sous groupe E (ALVE) et ainsi la sensibilité est dominante à résistante. Deux polymorphismes nucléotidiques simples (SNP) au niveau du locus TVB ont été rapportés distinguant trois allèles TVB (TVB*S1, TVB*S3 et TVB*R). Nous avons développé un test PCR-RFLP utilisant les deux SNPs pour définir trois haplotypes allèliques observés et pour identifier les six génotypes TVB possibles consistant en trois haplotypes définis chez les souches de poulet du laboratoire. Un haplotype allèlique complementaire et quatre génotypes ont également fait l'objet d'une courte discussion. Les poulets, issus de parents hétérozygotes pour différents allèles TVB, ont été éprouvés avec les virus du sarcome de Rous (RSVs) des sous groupes ALVB et ALVE pour induire des tumeurs à l'aile. Les incidences des tumeurs ont été évaluées chez les poulets de génotypes déterminés à l'aide du nouveau test développé PCR-RFPL. De façon importante, les poulets typés avec ce test comme étant TVB*S3/*S3 ont été résistants uniquement à l'infection par l'ALVE, et ceux typés TVB*R/*R ont été résistants aux deux virus ALVE et ALVB. De plus, une forte majorité de poulets avec les génotypes TVB*S1/- ont développé une tumeur. Ce test PCR-RFLP permet une évaluation relativement rapide des 6 génotypes TVB attendus chez les souches expérimentales de poulets en examinant la ségrégation des allèles TVB*S1, TVB*S3, et TVB*R. La capacité de ce test à sélectionner les croisements commerciaux de poulets pour leur résistance aux infections par des ALVB, ALVD, et ALVE doit être davantage évalué.
Entwicklung und Validierung eines PCR-RFLP-Tests zur Untersuchung von TVB-Haplotypen mit kodierenden Rezeptoren für die aviären Leukoseviren der Subgruppen B und E bei weißen Leghorn-Hühnern
Der zelluläre Rezeptor des aviären Leukosevirus Subgruppe B (ALVB) wird von einem Gen auf dem Tumorvirus B (TVB) kodiert. TVB-Allele kodieren spezifische Rezeptoren, die die Infektion mit exogenen ALVB oder ALV Subgruppe D (ALVD) sowie mit endogenenem ALV Subgruppe E (ALVE) zulassen, und deshalb ist der Empfänglichkeitsfaktor dominant über dem Resistenzfaktor. Der beschriebene Polymorphismus bei zwei einzelnen Nukleotiden (SNP) am TVB-Lokus ermöglichte die Unterscheidung von drei TVB-Allelen (TVB*S1, TVB*S3 und TVB*R). Wir haben einen PCR-RFLP-Test unter Verwendung der beiden SNPs entwickelt, um die drei beobachteten allelen Haplotypen zu bestimmen und die sechs möglichen aus diesen drei Haplotypen bestehenden TVB-Genotypen in definierten Hühner-Laborstämmen zu identifizieren. Ein weiterer potentieller alleler Haplotyp und vier Genotypen werden ebenfalls kurz besprochen. Hühnerküken von für verschiedene TVB-Allelen heterozygoten Elterntieren wurden mit Rous Sarkom-Viren (RSVs) der Subgruppen ALVB und ALVE infiziert, um Wing-Web-Tumoren zu induzieren. Bei diesen Hühnern, deren Genotyp mit Hilfe des neu entwickelten PCR-RFLP-Tests bestimmt wurde, wurde das Auftreten von Tumoren ermittelt. Bedeutsamerweise waren die Hühner, die mit dem Test als TVB*S3/*S3 typisiert wurden, nur resistent gegen die ALVE-Infektion und die als TVB*R/R typisierten waren resistent sowohl gegen ALVE Und ALVB. Ferner entwickelten die Mehrzahl der Hühner mit dem empfänglichen TVB*S1/- -Genotyp einen Tumor. Dieser PCR-RFLP-Test ermöglicht eine relativ schnelle Bestimmung aller sechs vorkommenden TVB-Genotypen durch Abgrenzung der TVB*S1-, TVB*S3- und TVB*R-Allele. Dieses nicht infektiöse Testverfahren sollte hinsichtlich seiner Eignung für die Selektion und Zucht kommerzieller Hühner auf genetische Resistenz gegen Infektionen mit ALVB, ALVD und ALVE weitergehend untersucht werden.
Desarrollo y validación de una técnica de PCR-RFLP para evaluar los haplotipos TVB codificantes de receptores para los subgrupos B y E de virus de leucosis aviar en aves White Leghorn
El receptor celular del subgrupo B del virus de leucosis aviar (ALVB) es codificado por un gen en el locus (TVB) del virus tumoral B. Los alelos TVB codifican para los receptores específicos permitiendo la infección por virus exógenos ALVB o ALV subgrupo D (ALVD) así como los ALV endógenos del subgrupo E (ALVE) y, por lo tanto, la susceptibilidad es dominante respecto a la resistencia. Se han descrito dos únicos polimorfismos de nucleótidos (SNP) en el locus TVB y se distinguen tres alelos TVB (TVB*S1, TVB*S3 y TVB*R). En el presente trabajo se ha desarrollado una técnica de PCR-RFLP utilizando los dos SNPs para definir tres haplotipos alélicos observados para identificar lo seis posibles genotipos de TVB que consistían en los tres haplotipos definidos en razas de laboratorio de pollos. Un haplotipo alélico potencial adicional y cuatro genotipos también se discuten brevemente. Pollos de padres heterocigotos de diferentes alelos TVB fueron infectados con virus del sarcoma de Rous (RSVs) de los subgrupos ALVB y ALVE para inducir tumores vía inoculación al ala. Se determinó la incidencia de tumores entre los pollos de los genotipos determinados por este nuevo método de PCR-RFLP. Es remarcable que los pollos tipificados con esta técnica como TVB*S3/*S3 fueron resistentes únicamente a la infección por ALVE, mientras que los TVB*R/*R fueron resistentes tanto a los ALVE como a los ALVB. Además, una vasta mayoría de pollos con los genotipos susceptibles TVB*S1/- desarrollaron un tumor. Este método de PCR-RFLP permite una determinación rápida de los seis genotipos TVB anticipados en cepas experimentales de pollos con segregación para los alelos TVB*S1, TVB*S3, y TVB*R. Esa técnica no infecciosa debe ser evaluada con más detalle para saber si se puede seleccionar y cruzar las diferentes razas comerciales de pollos para mejorar la resistencia genética a las infecciones por ALVB, ALVD y ALVE.
Introduction
After decades of comprehensive studies, avian leukosis-sarcoma virus (ALV) has been classified into six well-defined major subgroups of A to E and J based on receptor usage, host range, and infection interference patterns (Crittenden et al., Citation1967; Vogt, Citation1970; Payne & Pani, Citation1971; Crittenden & Motta, Citation1975; Weiss, Citation1981 Citation1993; Crittenden, Citation1991; Payne et al., Citation1991; Barnard & Young, Citation2003). Five of the subgroups (ALVA to ALVD and ALVJ) are exogenous avian viruses and the other subgroup (ALVE) is an endogenous virus. The exogenous viruses of subgroups ALVA to ALVD and the endogenous virus ALVE initiate the cell entry phase of infection through an interaction between surface units on viral subgroup-specific glycoprotein envelope and subgroup-specific surface receptors on host cells.
A harboured endogenous virus was recognized in chicken cells decades ago when ALV group-specific (GS) antigens were found in uninfected cells from the chicken embryo (Dougherty & DiStefano, Citation1966) and the inheritance could be attributed to one gene (Payne & Chubb, Citation1968). Subsequently, over 22 DNA insertion sites or ALVE genes have been identified in White Leghorns, and individuals may possess one to three or more ALVE genes (Crittenden, Citation1991).
Three autosomal tumour viral (TV) loci have been identified that are responsible for coding subgroup-specific surface receptors on host cells that mediate or block viral entry by subgroups ALVA through ALVE. TVA and TVC encode the cellular receptors for ALVA and ALVC viruses, respectively. Both TVA and TVC have been mapped to chicken Chr28 (Pani, Citation1974; Schmid et al., Citation2000; Elleder et al., Citation2004). TVB is one complex locus and encodes receptors for ALVB, ALVD, and ALVE (Crittenden & Motta, Citation1975; Weiss, Citation1993; Barnard & Young, Citation2003). TVB has been mapped to chicken Chr22 (Smith & Cheng, Citation1998). No receptor locus has been identified for ALVJ.
The TVB locus is complex because alleles at this locus encode several types of receptors to accommodate the viral entry of different subgroups. The TVB locus transcribes a set of three different alleles, TVB*S1, TVB*S3, and TVB*R. TVB*S1 encodes receptors that support viral entry of three subgroups: ALVB, ALVD and ALVE. TVB*S3 encodes a receptor that permits viral entry for both subgroups ALVB and ALVD, but not for ALVE. The TVB*R allele encodes a defective (incomplete) receptor due to a premature stop codon within its DNA sequence. Therefore, the TVB*R-encoded receptor permits no viral entry to any of the ALV subgroups (Crittenden & Motta, Citation1975; Weiss, Citation1993; Barnard & Young, Citation2003).
Different receptors encoded by different TVB alleles have distinct cellular functions (Crittenden & Motta, Citation1975; Brojatsch et al., Citation1996; Adkins et al., Citation2001; Klucking et al., Citation2002) and the cDNA sequences for different alleles were cloned and sequenced (Brojatsch et al., Citation1996; Adkins et al., Citation2001; Klucking et al., Citation2002). Based on GenBank accession numbers AF161713, AF161712, and AF507016.1, two single nucleotide polymorphisms (SNPs) at nucleotide positions (following the numbering system of AF507016.1) 172 (C/T) and 184 (T/A) together differentiate the allelic transcripts for TVB*S1, TVB*S3, and TVB*R. Based on this sequence information, Klucking et al. (Citation2002) generated a Southern blot procedure that distinguished the TVB*R/*R genotype from the TVB*S1/*S1 and TVB*S3/*S3 genotypes but did not differentiate the TVB*S1/*S1 from TVB*S3/*S3 genotypes.
The goal of this study was to develop a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay using both of the SNPs to identify all TVB genotypes in chickens and to validate the PCR-RFLP assay in chickens produced from breeders segregating for the three alleles, TVB*S1, TVB*S3, and TVB*R. This study consisted of two experiments. Chickens in the first experiment were sequentially infected with two subgroups of Rous sarcoma viruses (RSV) to evaluate TVB*S1 and TVB*R determined by the PCR-RFLP assay, and chickens in the second experiment were infected with subgroup ALVE RSV to evaluate TVB*S1 and TVB*S3. Experiment 2 was also designed to examine any impact that endogenous virus ALVE21 may impose on tumour incidences in chickens of various TVB genotypes.
Materials and Methods
PCR-RFLP assay development
Gene-specific primers (forward, 5′-CAG ACC TCC AGA AGC CAG AC; reverse, 5′-GGG TGG TCA GGT AAG CAG AA) were designed based on the published chicken TVB cDNA sequence (GenBank accession number AF507016.1) before the chicken genome sequence was released (Hillier et al., Citation2004). These primers were used to amplify the TVB gene using genomic DNA from a single White Leghorn chicken as template. The PCR product was cloned using the pCR®2.1-TOPO® Cloning kit (Invitrogen, Carlsbad, California, USA) and sequenced on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, California, USA) using T7 and M13-reverse primers. This partial TVB genomic sequence was 797 base pairs (bp) in length and aligns with the draft chicken genome sequence at chr22:1,252,854-1,253,650 (Hillier et al., Citation2004). Two pairs of PCR primers were subsequently designed based on the 797 bp of the TVB DNA sequence. The first pair, designated TVB303 primers (forward, 5′-ACC CCT TCT TGC AGG CAC CTA TGA; reverse, 5′-GGA TGC TGT GCT GCG TGG AGA), flanked a 303 bp segment of the TVB genomic DNA. This PCR product named TVB303 encompasses the SNPs at positions 172 and 184. The TVB303 PCR amplification reaction consisted of 25 ng genomic DNA, 5 pmol each TVB303 primer, 50 µM each dNTP, 1 µl of 10×Taq buffer, 1.5 mM MgCl2, and 0.5 U Taq DNA polymerase (Promega, Madison, Wisconsin, USA). This mixture was immediately subjected to the cycling conditions of 94°C for 3 min for initial denaturation, then 30 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, and a final extension cycle for 5 min at 72°C.
The second primer pair (forward, 5′-GGT AAG GCA GTC ACA AGC ATC ACT C; reverse, 5′-TAC TCG TCT TTC TTA CAT GGG AGG CTC T) was designed to flank the TVB 172 SNP and it generated a 202 bp PCR product designated as TVB202. The underlined base close to the 3′ end of the TVB202 reverse primer indicates a designed single A→T substitution. This substitution was made during primer synthesis. If the reverse primer was synthesized exactly as the TVB gene sequence i.e. TAC TCG TCT TTC TTA CAT GGG AGG CAC T, the third from the last base is an A→T. When this base was synthesized as a T, an XbaI restriction site was formed. This allowed one to distinguish the TVB*R allele from the TVB*S1 and TVB*S3 alleles. PCR amplification of TVB202 was performed in a similar fashion as TVB303 described earlier with the following modifications: each PCR reaction included 5 pmol each TVB202 primer and was subjected to 30 cycles for 1 min at 94°C, 1 min at 56°C, and 45 sec at 72°C. The PCR products, TVB303 and TVB202, were subsequently subjected to separate endonuclease digestion for RFLP analyses.
Experimental animals
Experiment 1 used TVB congenic lines 72 and 100B, which both possess ALVE1 and ALVE2. Line 72 is TVB*R/*R, whereas line 100B is maintained as heterozygous for TVB*S1/R and expresses endogenous virus ALVE2 inherited from parental line 72 according to R2 antibody agglutination and GS antigen tests (Bacon et al., Citation2000). 100B males and hens were mated to produce chickens in six hatches at 13-day to 16-day intervals. Lines 0 and 72 were used as controls. Line 0 chicken lacks ALVE genes and is TVB*S3/*S3 (Bacon et al., Citation2000).
Experiment 2 used chicks from line 0.44-21 breeders that were heterozygous for TVB*S1/*S3 alleles as well as for the endogenous virus ALVE21+/– (Bacon et al., Citation2000). The ALVE21 gene is invariably linked to the gene K determining slow feathering (SF) (Bacon et al. Citation1988) on the Z chromosome, and therefore chicks from this mating were also classified into rapid feathering (RF) and SF groups. RF chicks were expected to be free of ALVE21 but SF chicks could be either homozygous or heterozygous for ALVE21 (Bacon et al., Citation1988). The feathering status (phenotype) was determined at hatch by the ADOL farm staff. In addition, chicks were produced from heterozygous breeders of a new rapid feather endogenous virus susceptible (RFS) line under development. The RFS breeders were comparable with line 0.44-21 in that they were segregating for TVB*S1 and TVB*S3 alleles, but all the RFS chickens were free of all endogenous viral genes (Bacon et al., Citation2000 Citation2004). Both 0.44-21 and RFS chicks were produced in four separate hatches at 14-day to 22-day intervals. Chicks from lines 15B1 (homozygous for ALVE1 and TVB*S1/*S1) and 0 were used as ALVE susceptible and resistant controls. All the breeder chickens for experiments 1 and 2 were free of pathogens including ALV (Bacon et al., Citation2000). All chicks in experiments 1 and 2 were vaccinated against Marek's disease with the FC126 strain of turkey herpes virus (Witter et al., Citation1970) at 1 day of age. The chicks of various genotypes were intermingled in Horsfall–Bauer isolators after hatch until experimental termination.
Viral tumour induction assays
In experiment 1 the TVB haplotypes of chicks were defined by the PCR-RFLP assay and then each chick was evaluated by ALV challenges and observation for tumours. The first injection used 500 focus forming units (ffu) RSVE [RSV (RAV-60)] (Smith & Crittenden, Citation1988) into the right wing-web of chicks at 6 to 15 weeks of ages. Chicks that did not develop a wing-web tumour after 21 days of age were injected a second time with 1500 ffu RSV (RAV-60). Forty-three days after the second challenge, chicks that remained free of a wing-web tumour were injected a third time with 500 ffu subgroup B RSV (RAV-2) into the left wing-web. Chicks were checked for wing-web tumours at 9, 14, and 21 days after each inoculation. Wing-web tumours were measured following an established routine (Bacon et al., Citation1983 Citation2004). In experiment 2 the chicks received two injections of the same dosages of RSV-(RAV-60) used in experiment 1. The chicks were 7 to 14 weeks old when the first injection of 500 ffu was given, and those that did not develop a wing-web tumour by 48 days received a second injection of 1500 ffu of RSV (RAV-60). Chickens were terminated in a timely manner using CO2 following the provisions of ADOL Animal Use Guidelines regarding euthanasia.
R2 and GS tests
Breeder chickens from lines 100B and 0.44-21 were heterozygous for the TVB*S1 allele, and therefore possessed the cellular receptor necessary for ALVE infection. In addition, these breeders express a complete endogenous virus and test R2 positive by erythrocyte agglutination (Bacon et al., Citation1996). A positive R2 test indicates the presence of the TVB*S1 allele-encoded receptor coupled with the endogenous viral envelope glycoprotein (Bacon et al., Citation1996). The R2 test was performed on all chickens of experiment 1 at about 4 weeks of age. In experiment 2, the R2 test was made on all slow-feathering and a few RF 4-week-old to 5-week-old chicks from the 0.44-21 mating. The GS antigen of ALVE was also evaluated for chickens from the line 100B mating (experiment 1) at the same ages that the R2 tests were conducted using an enzyme-linked immunosorbent assay procedure described by Smith et al. (Citation1986).
R2 flow cytometric assay
An R2 flow cytometric (FC) assay was performed to access the presence or absence of an ALVE receptor on erythrocytes for RF chicks both from 0.44-21 and RFS matings following procedures described by Bacon et al. (Citation2004). This test requires incubating erythrocytes from chickens lacking ALVE with plasma containing ALVE followed by mixing the cells with R2 antibody and conducting flow cytometric analysis. Test samples with a specific binding index greater than 1.33 were considered positive in this study.
Endogenous viral gene tests by PCR
To ensure that a chicken has or does not have an endogenous viral gene, all chicks from 100B mating were tested for the expected presence of ALVE2, and chicks from 0.44-21 and RFS matings were tested for ALVE21 using published primers (Benkel, Citation1998) with a modified PCR condition. A final volume of 10 µl PCR reaction was prepared for each of the tested chicks with 25 ng genomic DNA, 1 pmol each primer, 50 µM each dNTP, 1 µl of 10×Taq buffer, 1.5 mM MgCl2, and 0.5 U Taq DNA polymerase (Promega). The PCR mix was subjected to cycling conditions of 94°C for 2 min for initial denaturation, then 30 cycles of 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C and a final extension cycle for 5 min at 72°C. The primers used in the ALVE2 PCR test were LTRE (5′-GTG TTC GCA ATC GTT AGG GAC TC) and EV2.UP (5′-GCA CCA CTG ATG GGA TTC TTG TTC TC). ALVE2-negative chicks gave no PCR product and the positive ones gave a 320 bp band when visualized on 1% of agarose gel. For ALVE21, all chicks were tested with two pairs of primers. ALVE21-negative was tested with EV21.UP (5′-GTG GGA ATG GTA CTA CAG AGA AGG) and EV21.DWN (5′-CAT TTC AAG CAA GGG ACT GGC) primers, which flanked a 510 bp of PCR product. ALVE21-positive was tested with LTRB (5′-ACC TGA ATG AAG CTG AAG GCT TC) and EV21.DWN primers, which produce a 390 bp product if the genome of the tested chick has ALVE21.
Statistical analysis
Data of tumour incidences between genotypes and ages at inoculation were statistically tested for difference of significance with a chi-square test adjusted for non-continuity with degrees of freedom = 1 (Geng & Hills, Citation1989).
Results
TVB PCR-RFLP assay
The TVB PCR-RFLP assay consists of two PCR products (TVB303 and TVB202) followed by two separate endonuclease digestions and electrophoresis. Electrophoresis patterns representing the presence or absence of a specific SNP at nucleotide position 172 (in TVB202) or 184 (in TVB303) constitute allelic haplotypes. Allelic haplotypes, in turn, define the genotypes. The first PCR product is TVB303. Digestion of TVB303 by NlaIII produces three electrophoresis patterns, P1, P2, and P3 (a). The second PCR product is TVB202, which was amplified including a reverse primer integrating an XbaI restriction site through primer mutagenesis. Digestion of TVB202 with XbaI produces another three patterns, P4, P5, and P6 (b). Depicted electrophoresis patterns of endonuclease-restricted TVB303 together with TVB202 are given in in an effort to assist easy readings of the TVB genotypes from electrophoresis patterns. The six electrophoresis patterns formulate four allelic haplotypes representing specific combinations of the TVB SNPs at nucleotide positions 172 and 184. P3 and P4 constitute allelic haplotype S1, representing a SNP combination of C and T at nucleotide positions 172 and 184, respectively. P1 and P4 form haplotype S3, representing a C and A combination at nucleotides 172 and 184, whereas P3 and P6 make haplotype R representing a SNP combination of T and T at the nucleotide 172 and 184 positions. In addition, a fourth haplotype is conceivable (R′). R′ consists of P1 and P6 representing a SNP combination of T and A at positions 172 and 184. Although R′ and R are distinct haplotypes () they both result in a premature stop codon due to the single base substitution at position 172. The R′ haplotype has not been observed. Thus, there are 10 possible TVB genotypes that now can be differentiated with this PCR-RFLP assay, and six have been observed. They are summarized in in relation to the haplotype(s), SNPs, and electrophoresis patterns for each of the 10 genotypes.
Figure 1. a. Electrophoresis patterns (P1, P2, and P3) of the PCR product TVB303 digested with endonuclease NlaIII (CTAG). When the TVB303 is homozygous for base T at nucleotide position 184, TVB303 is digested into two fragments of 212 and 91 bp (pattern P1); when the same base is homozygous for A, TVB303 is digested into three fragments of 158, 91, and 54 bp (P3); when the TVB303 is heterozygous for T and A at the position, the NlaIII digestion yields all the four fragments (P2). Since the 54 bp band under P2 should have about half the DNA mass comparing to P3, it was not visible under P2. b. Electrophoresis patterns (P4, P5 and P6) of the PCR product TVB202 digested with endonuclease XbaI (TCTAGA). When the PCR product TVB202 is homozygous for base C at nucleotide position 172, the TVB202 is not digestible (P4); when the same base is homozygous for T, it is digested into two fragments of 172 and 30 bp (P6) but the 30 bp band is not visible in the P6 pattern; whenTVB202 is heterozygous for C and T at the position, the XbaI digestion of TVB202 gives three fragments of 202, 172 and 30 bp, where the 30 bp band again is not visible (P5).
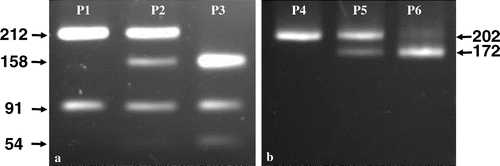
Figure 2. Depicted electrophoresis patterns (P1–P6) of both NlaIII digested TVB303 and XbaI digested TVB202 for each of the TVB genotypes. Note that a 30-bp band was not shown under patterns P5 and P6 since it was not visible on 2.5% Agarose gels.
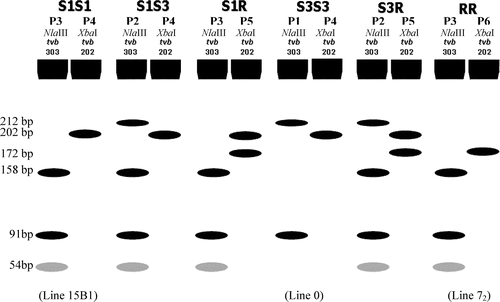
Table 1. TVB haplotype and defined genotypes in relation to the TVB SNPs and electrophoresis patterns
Evaluation of the allelic haplotype S1 and R in experiment 1
The expected genotypes of the chickens from 100B mating were TVB*S1/*S1, TVB*S1/*R, or TVB*R/*R with an expected ratio of 1:2:1. The numbers of chickens that were determined by this PCR-RFLP assay as TVB*S1/*S1, TVB*S1/*R, or TVB*R/*R genotypes are presented in , but did not confirm the expected 1:2:1 ratio (i.e. there was a deficiency of heterozygotes compared with homozygotes, P < 0.05). Of the chickens identified by the PCR-RFLP assay as TVB*S1/*S1 and TVB*S1/*R, 25% and 23% respectively, of them developed a wing-web tumour 21 days after the first inoculation with RSV (RAV-60). An additional 18% of the remaining TVB*S1/*R chickens developed a tumour after the second challenge with a three times dosage of the same virus. Overall, after both challenges with RSV (RAV-60), 36% of TVB*S1/*R chickens developed a wing-web tumour. In the TVB*S1/*S1 and TVB*S1/*R genotypes combined, 31% of the chickens developed a tumour. In contrast, none of TVB*R/*R chickens from the same mating developed a tumour. None of the control chickens from line 0 (tested as TVB*S3/*S3), or line 72 (typed as TVB*R/*R), developed a tumour, in full agreement with expectation.
Table 2. Subgroup E RSV (RAV-60) and subgroup B RSV (RAV-2) induced tumour incidences among chickens from a line 100B mating segregating for haplotypes of TVB*S1 and TVB*R alleles
After two consecutive challenges with RSV (RAV-60), a total of 42 chickens from the 100B mating remained free of any wing-web tumour. Fifteen of the 42 chickens were TVB*S1/*S1, 13 chickens were TVB*S1/*R, and the remaining 14 chickens were TVB*R/*R. All 26 chickens of the control line 0 remained free of any tumour. Following the second challenge with RSV (RAV-60), all of the chickens from 100B mating lacking a tumour were superinfected with exogenous subgroup B RSV (RAV-2). Twenty-one days after the superinfection, 73% of the TVB*S1/*S1 and 85% of the TVB*S1/*R chickens developed a tumour. In contrast, 0% of the TVB*R/*R chickens had a tumour (P<0.01). In the control lines, 67% of line 0 chickens (TVB*S3/*S3) developed a tumour whereas none of the line 72 chickens (TVB*R/*R) had a tumour ().
Chickens typed as TVB*S1/*S1 and TVB*S1/*R from 100B mating were also 100% R2-positive and GS-positive. All chickens typed as TVB*R/*R were R2-negative and GS-negative, as were chickens of the line 0 and 72 (), which was in full agreement with expectation. All of the 100B chickens regardless of their TVB genotypes had EV2 based on a PCR test, which was in full agreement with expectation.
Evaluation of the TVB allelic haplotype S1 and S3 in experiment 2
Experiment 2 was designed to test and evaluate allelic haplotypes S1 and S3 for their influence on resistance to an endogenous virus-induced tumour. In chickens from the 0.44-21 mating, at 21 days after inoculation with RSV (RAV-60) the tumour incidences in chickens typed either as TVB*S1/*S1, TVB*S1/*S3, or TVB*S3/*S3 were not significantly different from each other regardless of their EV21 status (EV21 + /– or EV21–/–). This remained true even after the second challenge with a three times dosage of the same virus. It appeared that more RF chickens lacking EV21 from this 0.44-21 mating tended to develop a tumour than those that had inherited EV21, but the differences were not statistically significant. It is important that none of the TVB*S3/*S3 chickens from this mating developed a tumour ().
Table 3. Genotypes and tumour incidences induced by RSV (RAV-60) in White Leghorns carrying or lacking endogenous virus
In the RFS mating following infection with RSV (RAV-60), tumour incidences of 77% and 45% were observed in chickens typed as TVB*S1/*S1 and TVB*S1/*S3, respectively, and these incidences differed significantly from the 0% tumour incidence in chickens typed TVB*S3/*S3 (P<0.05; ). A second three times dosage challenge increased the tumour incidence in TVB*S1/*S3 chickens but not in TVB*S1/*S1 or TVB*S3/*S3 chickens. Most importantly, none of the 17 TVB*S3/*S3 chickens developed a tumour, in contrast to 77% of TVB*S1/*S1 and 68% of TVB*S1/*S3 chickens (P<0.01). All the chickens of the control line 15B1 were typed as TVB*S1/*S1 and developed a tumour after the two consecutive challenges with RSV (RAV-60). In contrast, no chickens of the control line 0 (TVB*S3/*S3) developed tumours, in full agreement with expectation.
All of the chickens from different matings in each line were produced by multiple hatches and were infected at the same time within each experiment. The effect of age at inoculation was examined using the chickens from RFS mating. Tumour incidences between the groups inoculated at 52 to 102 days within each genotype group were not significantly different from one another (P>0.05; data not shown). Therefore the data of all ages within each genotype group were tabulated together for further statistical analysis.
In chickens from the 0.44-21 mating, 71% of the chickens were identified as ALVE21+/– by the PCR test, close to the 75% expectation (). ALVE21 and feathering status were in good agreement, but three of 70 were not in accordance. Two of the ALVE21+/− chickens were recorded as RF, and one chicken that tested by PCR as ALVE21–/– was recorded as SF. All chickens from the RFS mating were ALVE21–/– by PCR tests and their feather status was recorded as RF ().
Discussion
The aim of this study was to develop a molecular genetic tool to access the TVB genotype in chickens and to provide experimental data to support the validity of this technology. Our data show that a newly developed PCR-RFLP assay can be used to distinguish TVB genotypes with relative ease and high accuracy necessary for research and industrial application. The combined data from both experiments 1 and 2 provided distinctive evidence sustaining the identity of each of the six observed TVB genotypes and their relationship to a viral-induced tumour phenotype. In addition, the total agreement between the expected genotypes and the PCR-RFLP determined genotypes of the control lines (line 0 and line 72 in experiment 1; line 0 and line 15B1 in experiment 2) also validate the developed PCR-RFLP assay. The expected genotype of line 0 chickens is TVB*S3/S3, and the line 0 chickens in experiment 1 and experiment 2 were all typed as TVB*S3/S3 and developed no tumour even after two consecutive challenges with RSV (RAV-60). However, 67% of line 0 chickens (TVB*S3/*S3) developed a tumour after a one-time RSV (RAV-2) challenge. As discussed elsewhere, the RSV (RAV-2) virus does not always result in 100% of chickens with tumours where expected (Bacon et al., Citation2004). Line 72 chickens were typed as TVB*R/*R, and remained tumour free throughout experiment 1. All line 15B1 chickens tested TVB*S1/S1 as expected and all developed a tumour in experiment 2.
This PCR-RFLP assay was developed based on published TVB SNPs (Brojatsch et al., Citation1996; Adkins et al., Citation2001; Klucking et al., Citation2002). Natural restriction sites exist for both of the TVB SNP sites. Although the endonuclease BfaI separates the C/T SNP at nucleotide position 172 of the TVB gene in genomic DNA, it does not hydrolyse the same interior bond between the two nucleotides within any PCR-generated DNA product, such as TVB303. A cumbersome, time-consuming and expensive way to get around this technical obstacle is to purify every PCR product before digestion with BfaI in a PCR-RFLP analysis. To avoid this purification step we have designed a primer through primer mutagenesis that provides a new restriction site so that an alternative endonuclease can separate the SNP. An XbaI restriction site was engineered into the 3′ primer of the TVB202 product to accommodate the necessary analysis of C/T SNP at position 172, and this has been shown to work accurately.
None of 17 chickens typed as TVB*R/*R genotype from 100B mating developed a tumour after two consecutive challenges with RSV (RAV-60). Subsequently, 15 (from both 100B and 72 matings) TVB*R/*R chickens that were superinfected with RSV (RAV-2) remained tumour free. These results are in complete agreement with expectation and support the hypothesis that the newly developed PCR-RFLP assay typed the TVB genotypes of the chickens in experiment 1 correctly. R2 and GS test results also gave 100% agreement with the PCR-RFLP determined genotypes (). As expected, all TVB*S1/*S1 and TVB*S1/*R chickens were positive for R2 and GS. All TVB*R/*R chickens were negative. However, a tumour developed in only 31% of the chickens typed either as TVB*S1/*S1 or TVB*S1/*R, although both of these genotypes should be susceptible to RSV (RAV-60). The factor that most probably explains the reduction of tumour incidence in this group of chickens is that the expression of ALVE2 interfered with the TVB*S1 receptor so RSV (RAV-60) could not attach to the TVB*S1 receptor. Viral interference at the receptor level is well recognized (Vogt & Ishizaki, Citation1966; Vogt, Citation1970). ALVE1 has little or no known influence on tumour incidence, probably because it is a defective provirus and does not code for an infectious ALVE (Crittenden, Citation1991). Our data from line 15B1 chickens (ALVE1+ only) in experiment 2 fully support the failure of ALVE1 to interfere with RSV (RAV-60) infection ().
Line 0.44-21 and the RFS chickens that typed as TVB*S3/*S3 in experiment 2 were all free of tumour after two consecutive infections with RSV (RAV-60). This again fully supports the integrity of the newly developed PCR-RFLP assay, and verified that the TVB genotype was assessed properly. In chickens from the 0.44-21 mating that possessed ALVE21, none of the TVB*S1/*S1 and only 7% of the TVB*S1/*S3 chickens had a tumour. As explained earlier, the presence of infectious ALVE21 may have blocked infection by RSV (RAV-60). In the TVB*S1/*S1 and TVB*S1/*S3 chicks lacking ALVE21 from this same mating, only 38% and 25% developed tumours. These chicks were grown together with their ALVE21-carrying hatchmate siblings and may have been infected with ALVE21 resulting in this low tumour incidence. In contrast, RFS chicks typed as TVB*S1/*S1 and TVB*S1/*S3 had a high level of tumours (77% and 68%, respectively). Tumour incidences of the control lines in experiment 2 were 100% in 15B1 (TVB*S1/*S1) and 0% in line 0 (TVB*S3/*S3), which also supported the integrity of the PCR-RFLP assay (). The 15B1, RFS and line 0 chicks were housed in separate isolators from the ALVE21 positive chicks, and therefore contact transmission by ALVE21 could not interfere with tumour development in these chicks.
Only 4% discrepancy was seen between the PCR tests for ALVE21 and the recorded feather status of chicks at hatch. Two of the 20 chickens assigned the RF status from the 0.44-21 mating were PCR tested as ALVE21+/–, and one of 45 chickens assigned the SF phenotype from the same mating tested as ALVE21–/– (). A clear explanation for this discrepancy can not be provided. However, sex assigned by feather phenotyping is usually only 95% to 98% accurate due to the presence of additional genes that affect feather growth (Bacon et al., Citation1988). We expect that mistaken identity of feather status at hatch explains this discrepancy.
The PCR test results for the endogenous viral gene ALVE21 were highly consistent with the R2 tests in SF chicks from the 0.44-21 matings that possessed ALVE21 (). However, in RF TVB*S1/*S1 and TVB*S1/*S3 chicks lacking ALVE21, about one-half of the chickens were unexpectedly positive using the R2 test. We postulate that the chicks that were susceptible to ALVE may have become infected with ALVE21 from their slow feather cage-mates, and therefore became R2-positive. On the other hand, the two TVB*S3/*S3 chicks were negative by the R2 test. The R2 FC assay was developed and used to detect susceptibility to ALVE (Bacon, Citation2000; Bacon et al., Citation2004). In the RFS chicks there was full agreement between the PCR-RFLP assay and the R2 FC assay for the TVB*S3/*S3 chicken's cells that tested negative in the R2 FC assay. However, only about one-half of the RFS chickens classified as TVB*S1/*S1 or TVB*S1/*S3 were positive in the R2 FC assay (). This shows that the R2 FC assay is highly accurate in identifying ALVE-resistant chickens but is only moderately accurate for identifying chickens susceptible to ALVE. Thus, while this R2 FC assay will accurately identify TVB*S3/*S3 chickens, it may lead one to falsely conclude that some TVB*S1/*S1 and TVB*S1/*S3 chickens are ALVE resistant. Therefore, this fairly sophisticated R2 FC assay requiring expensive machinery and unique antisera is not fully accurate for identification of genetic resistance to ALVE.
It is critical to define chickens resistant to ALVE. The expression of ALVE decreases productivity of eggs and meat (Smith & Nelsen, Citation1993), depresses immunity following infection with ALVA, or ALVJ, and may be detrimental in fertile eggs used for vaccine production (see Bacon et al., Citation2004). This study presents the first molecular genetics tool that is fully accurate in determining the precise TVB genotypes coding for receptors mediating viral entry of endogenous ALVE and exogenous ALVB. This new tool detects chickens genetically resistant or susceptible to ALVE irrespective to the presence of ALVE, and should be evaluated as a practical way to determine the TVB genotypes in chickens of commercial strains. It is noted, however, that in an earlier report a single SNP of the TVA gene distinguished susceptibility to ALVA between two experimental inbred lines of chickens, but a subsequent PCR-sequencing analysis showed that the SNP of the TVA gene did not exclusively predict susceptibility to ALVA in a commercial broiler line (Bates et al., Citation1998). In a preliminary analysis using the PCR-RFLP assay described here we have identified the TVB*S1, TVB*S3, and TVB*R alleles in commercia l layer-type chickens and, in contrast to other TVB genotypes, the chickens typed as TVB*R/*R were all free of tumours 21 days after RSV (RAV-2) infection (Zhang et al., unpublished data).
Compiling all data from the experimental chickens in this study (), following subgroup E RSV (RAV-60) infection a tumour incidence of 41% was observed among 155 chickens that were typed by this PCR-RFLP assay as either TVB*S1/*S1, TVB*S1/*S3, or TVB*S1/*R, in comparison with 0% tumour incidence among 67 chickens typed as TVB*S3/*S3 or TVB*R/*R. Following subgroup B RSV (RAV-2) infection, a tumour incidence of 77% was observed among 34 chickens typed as TVB*S1/*S1, TVB*S1/*S3, TVB*S1/*R, or TVB*S3/*S3. In contrast, no tumour was observed in 15 chickens typed as TVB*R/*R. This is the first PCR-based molecular genetics assay that empowers simultaneous assessment of the TVB genotypes in chickens validated by viral-induced tumour experiments.
Table 4. Subgroup E and subgroup B RSV-induced tumour incidences in chickens differ according to TVB genotypes determined by PCR-RFLP defined haplotypes
Translations of the abstract in French, German and Spanish are available on the Avian Patholgy website.
Acknowledgments
The authors thank Evelyn Young for her excellent technical assistance in chicken virus inoculation, recording of data and performance of laboratory tests. They thank Tom Goodwill and Jonathan Kenyon for assistance with chicken handling and sample collections. They are grateful to Lyman Crittenden for careful manuscript review and helpful suggestions.
References
- Adkins , H.B. , Blacklow , S.C. and Young , J.A.T. 2001 . Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses . Journal of Virology , 75 : 3520 – 3526 .
- Bacon , L.D. 2000 . Detection of endogenous avian leukosis virus envelope in chicken plasma using R2 antiserum . Avian Pathology , 29 : 153 – 164 .
- Bacon , L.D. , Crittenden , L.B. , Witter , R.L. , Fadly , A. and Motta , J. 1983 . B5 and B15 associated with progressive Marek's disease, Rous sarcoma, and avian leukosis virus-induced tumours in inbred 15I4 chickens . Poultry Science , 62 : 573 – 578 .
- Bacon , L.D. , Smith , E. , Crittenden , L.B. and Havenstein , G.B. 1988 . Association of the slow feathering (K) and an endogenous viral (ev21) gene on the Z chromosome of chickens . Poultry Science , 67 : 191 – 197 .
- Bacon , L.D. , Smith , E.J. , Fadly , A.M. and Crittenden , L.B. 1996 . Development of an antiserum (R2) that detects susceptibility of chickens to subgroup E endogenous avian leukosis virus . Avian Pathology , 25 : 551 – 568 .
- Bacon , L.D. , Hunt , H.D. and Cheng , H.H. 2000 . A review of the development of chicken lines to resolve genes determining resistance to diseases . Poultry Science , 79 : 1082 – 1093 .
- Bacon , L.D. , Fulton , J.E. and Kulkarni , G.B. 2004 . Methods for evaluating and developing commercial chicken strains free of endogenous subgroup E avian leukosis virus . Avian Pathology , 33 : 233 – 243 .
- Barnard , R.J. and Young , J.A. 2003 . Alpharetrovirus envelope-receptor interactions . Current Topics in Microbiology and Immunology , 281 : 107 – 136 .
- Bates , P. , Rong , L. , Varmus , H.E. , Yong , J.A. and Crittenden , L.B. 1998 . Genetic mapping of the cloned subgroup A avian sarcoma and leukosis virus receptor gene to the TVA locus . Journal of Virology , 72 : 2505 – 2508 .
- Benkel , B.F. 1998 . Locus-specific diagnostic test for endogenous avian leukosis-type viral loci in chickens . Poultry Science , 77 : 1027 – 1035 .
- Brojatsch , J. , Naughton , J. , Rolls , M.M. , Zingler , K. and Young , J.A. 1996 . CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis . Cell , 87 : 845 – 855 .
- Crittenden , L.B. 1991 . Retroviral elements in the genome of the chicken: implications for poultry genetics and breeding . Critical Review in Poultry Biology , 3 : 73 – 109 .
- Crittenden , L.B. and Motta , J.V. 1975 . The role of the tvb locus in genetic resistance to RSV(RAV-O) . Virology , 67 : 327 – 334 .
- Crittenden , L.B. , Stone , H.A. , Reamer , R.H. and Okazaki , W. 1967 . Two loci controlling genetic cellular resistance to avian leukosis-sarcoma viruses . Journal of Virology , 1 : 898 – 904 .
- Dougherty , R.M. and DiStefano , H.S. 1966 . Lack of relationship between infection with avian leukosisi virus and the presence of COFAL antigen in chick embryos . Virology , 29 : 586 – 595 .
- Elleder , D. , Plachy , J. , Hejnar , J. , Geryk , J. and Svoboda , J. 2004 . Close linkage of genes encoding receptors for subgroups A and C of avian sarcoma/leukosis virus on chicken chromosome 28 . Animal Genetics , 35 : 176 – 181 .
- Geng , S. and Hills , F.J. 1989 . Biometrics in Agriculture Science , Dubuque, Iowa, IA : Kendall/Hunt Publishing Company .
- Hillier , L.W. , Miller , W. , Birney , E Warren , W. 2004 . Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution . Nature , 432 : 695 – 716 .
- Klucking , S. , Adkins , H.B. and Young , J.A.T. 2002 . Resistance to infection by subgroups B, D, and E avian sarcoma and leukosis viruses is explained by a premature stop codon within a resistance allele of the tvb receptor gene . Journal of Virology , 76 : 7918 – 7921 .
- Pani , P.K. 1974 . Close linkage between genetic loci controlling response of fowl to subgroup A and subgroup C sarcoma viruses . Journal of General Virology , 22 : 187 – 195 .
- Payne , L.N. and Chubb , R.C. 1968 . Studies on the nature and genetic control of an antigen in normal chick embryos which reacts in the COFAL test . Journal of General Virology , 3 : 379 – 391 .
- Payne , L.N. and Pani , P.K. 1971 . Evidence for linkage between genetic loci controlling response of fowl to subgroup A and subgroup C sarcoma viruses . Journal of General Virology , 13 : 253 – 259 .
- Payne , L.N. , Brown , S.R. , Bumstead , N. , Howes , K. , Frazier , J.A. and Thouless , M.E. 1991 . A novel subgroup of exogenous avian leukosis virus in chickens . Journal of General Virology , 72 : 801 – 807 .
- Schmid , M. , Nanda , I. , Guttenbach , M. , Steinlein , C. , Hoehn , M. , Schartl , M. , Haaf , T. , Weigend , S. , Fries , R. , Buerstedde , J.M. , Wimmers , K. , Burt , D.W. , Smith , J. , A'Hara , S. , Law , A. , Griffin , D.K. , Bumstead , N. , Kaufman , J. , Thomson , P.A. , Burke , T. , Groenen , M.A. , Crooijmans , R.P. , Vignal , A. , Fillon , V. , Morisson , M. , Pitel , F. , Tixier-Boichard , M. , Ladjali-Mohammedi , K. , Hillel , J. , Maki-Tanila , A. , Cheng , H.H. , Delany , M.E. , Burnside , J. and Mizuno , S. 2000 . First report on chicken genes and chromosomes 20000 . Cytogenetics and Cell Genetics , 90 : 169 – 218 .
- Smith , E.J. and Cheng , H.H. 1998 . Mapping chicken genes using preferential amplification of specific alleles . Microbial & Comparative Genomics , 3 : 13 – 20 .
- Smith , E.J. and Crittenden , L.B. 1988 . Endogenous viral genes in a slow-feathering line of White Leghorn chickens . Avian Pathology , 15 : 395 – 406 .
- Smith , E.J. and Nelsen , T.C. 1993 . Effect of tumour virus susceptibility alleles on the late-feathering gene K on early growth of White Leghorns . Poultry Science , 72 : 1400 – 1404 .
- Smith , E.J. , Fadly , A.M. and Crittenden , L.B. 1986 . Observations on an enzyme-linked immunosorbent assay for the detection of antibodies against avian leukosis-sarcoma viruses . Avian Disease , 30 : 488 – 493 .
- Vogt , P.K. 1970 . Envelope classification of avian RNA tumour viruses . Bibliotheca Haematologica , 36 : 153 – 167 .
- Vogt , P.K. and Ishizaki , R. 1966 . Patterns of viral interference in the avian leukosis and sarcoma complex . Virology , 30 : 368 – 374 .
- Weiss , R.A. 1981 . “ Retrovirus receptors and their genetics ” . In Receptors and Recognition: Virus Receptors Part 2 , Edited by: Lonberg-Holm , K. and Philipson , L. 186 – 202 . London : Chapman and Hall .
- Weiss , R.A. 1993 . “ Cellular receptors and viral glycoproteins involved in retrovirus entry ” . In The Retroviridae , Edited by: Levy , J.A. Vol. 2 , 1 – 108 . New York : Plenum Press .
- Witter , R.L. , Nazerian , K. , Purchase , H.G. and Burgoyne , G.H. 1970 . Isolation from turkeys of a cell-associated herpesvirus antigenically related to Marek's disease virus . American Journal of Veterinary Research , 31 : 325 – 538 .