Abstract
As part of an epidemiological surveillance of infectious bronchitis virus (IBV) in Spain, four Spanish field isolates showed high S1 spike sequence similarities with an IBV sequence from the GenBank database named Italy 02. Given that little was known about this new emergent IBV strain we have characterized the four isolates by sequencing the entire S1 part of the spike protein gene and have compared them with many reference IBV serotypes. In addition, cross-virus neutralization assays were conducted with the main IBV serotypes present in Europe. The four Spanish field strains and the Italy 02 S1 sequence from the NCBI database were established as a new genotype that showed maximum amino acid identities with the 4/91 serotype (81.7% to 83.7%), the D274 group that included D207, D274 and D3896 strains (79.8% to 81.7%), and the B1648 serotype (79.3% to 80%). Furthermore, on the basis of these results, it was demonstrated that the Italy 02 genotype had been circulating in Spain since as early as 1997. Based on the average ratio of synonymous:non-synonymous (dS/dN) amino acid substitutions within Italy 02 sequences, no positive selection pressures were related with changes observed in the S1 gene. Moreover, phylogenetic analysis of the S1 gene suggested that the Italy 02 genotype has undergone a recombination event. Virus neutralization assays demonstrated that little antigenic relatedness (less than 35%) exists between Italy 02 and some of the reference IBV serotypes, and indicated that Italy 02 is likely to be a new serotype.
Caractérisation antigénique et moléculaire des souches de virus de la bronchite infectieuse de génotype Italy 02
Lors d'une étude épidémiologique de virus de la bronchite infectieuse en Espagne, quatre souches espagnoles isolées sur le terrain ont montré de très fortes similitudes au niveau de la séquence de la spicule S1 avec la séquence d'un IBV nommé Italy 02, obtenue dans la base de données de GenBank. Etant donné que l'on connaît peu de chose sur cette nouvelle souche émergente d'IBV, nous avons caractérisé les quatre souches par séquençage de la totalité de la partie S1 du gène de la protéine de spicule et nous les avons comparées à de nombreux sérotypes d'IBV de référence. De plus, des essais de séroneutralisations croisées (VN) ont été réalisés avec les principaux sérotypes d'IBV présents en Europe. Il a été établi que les quatre souches espagnoles du terrain et de la souche Italy 02 dont la séquence de S1 a été obtenue auprès de la base de données du NCBI, représentaient un nouveau génotype qui avait un maximum d'identité en acides aminés avec le sérotype 4/91 (81,7–83,7%), le groupe D274 incluant les souches D207, D274 et D3896 (79,8–81,7%), et le sérotype B1648 (79,3%-80%). De plus, sur la base de ces résultats, il a été démontré que le génotype Italy 02 a circulé en Espagne dès 1997. En se basant sur le rapport moyen des substitutions en acides aminés synonymes : non-synonymes (dS/dN) au sein des séquences des souches de type Italy 02, aucune pression de sélection positive n'était associée aux changements observés au niveau du gène S1. De plus, l'analyse phylogénétique du gène S1 suggère que le génotype Italy 02 a subi un évènement de recombinaison. Les essais de neutralisation ont montré qu'une relation antigénique faible (moins de 35%) existe entre les sérotypes Italy 02 et quelques souches d'IBV de référence, et indiquaient que la souche Italy 02 représente probablement un nouveau sérotype.
Antigenetische und molekulare Charakterisierung von Isolaten des Italien 02-Genotyps des Virus der infektiösen Bronchitis
Im Rahmen einer Überwachungsstudie zum Vorkommen des Virus der infektiösen Bronchitis (IBV) in Spanien zeigten vier spanische Feldisolate große Übereinstimmung der S1-Spikesequenz mit einer IBV-Sequenz aus der Gendatenbank namens Italien 02. Da wenig über diesen neuen IBV-Stamm bekannt war, haben wir die vier Isolate mittels Sequenzierung des gesamten S1-Teils des Spikeprotein-Gens charakterisiert und sie mit vielen IBV-Referenzserotypen verglichen. Außerdem wurden mit den Haupt-IBV-Serotypen in Europa Kreuzvirusneutralisationstests (VN) durchgeführt. Die vier spanischen Feldisolate und die S1-Sequenz des Italien 02-Stamms aus der NCBI-Datenbank wurden als zu einem neuen Genotyp gehörend eingeordnet. Dieser wies mit dem 4/91-Serotyp eine maximale Aminosäurenübereinstimmung von 81,7–83,7 %, mit der D274-Gruppe, zu der die Stämme D207, D274 und D3896 gehören, von 79,8–81,7 % und mit dem B1648-Serotyp von 79,3–80 % auf. Weiterhin wurde auf der Basis dieser Ergebnisse festgestellt, dass der Italien 02-Genotyp seit 1997 in Spanien zirkuliert. Basierend auf dem mittleren Verhältnis von übereinstimmenden zu nicht übereinstimmeneden (dS/dN) Aminosäurensubstitutionen innerhalb der Italien 02-Sequenz konnte kein Zusammenhang zwischen einem verstärkten Selektionsdruck und den beobachteten Veränderungen im S1-Gen hergestellt werden. Überdies legte die phylogenetische Analyse des S1-Gens nahe, dass der Italien 02-Genotyp eine Rekombination erfahren hat. Die VN-Tests ließen eine nur geringe antigenetische Verwandtschaft (<35 %) zwischen dem Italien 02-Stamm und einigen der IBV-Referenz-Serotypen erkennen, was darauf hinweist, dass es sich um einen neuen Serotyp handelt.
Caracterización antigénica y molecular de aislamientos del genotipo Italy 02 del virus de la bronquitis infecciosa aviar.
Dentro de un estudio epidemiológico del virus de la bronquitis infecciosa llevado a cabo en España, cuatro aislamientos de campo Españoles mostraron una elevada similitud de las secuencias de la espícula S1 con una secuencia de IBV de la base de datos del GenBank conocida como Italy 02. Dado que poco se conoce respecto a esta nueva cepa emergente de IBV, hemos caracterizado los cuatro aislamientos mediante la secuenciación completa de la parte S1 del gen de la proteína de la espícula y las hemos comparado con un gran número de serotipos de IBV de referencia. Además, se llevaron a cabo estudios de neutralización viral cruzada (VN) con los principales serotipos de IBV presentes en Europa. Las cuatro cepas de campo Españolas y la secuencia del S1 de Italy 02 de la base de datos del NCBI establecieron un nuevo genotipo que mostraba máxima similitud aminoacídica con el serotipo 4/91 (81.7–83.7%), el grupo D274 formado por las cepas D207, D274 y D3896 (79.8–81.7%), y el serotipo B1648 (79.3%-80%). Es más, se demostró, basándonos en estos resultados, que el genotipo Italy 02 había estado circulando en España desde 1997. Teniendo en cuenta la ratio media entre las secuencias Italy 02 de las sustituciones aminoacídicas sinónimas: no-sinónimas (dS/dN) , no se estableció ninguna relación entre presiones selectivas positivas y los cambios observados en el gen S1. Además, los estudios filogenéticos del gen S1 sugirieron que el genotipo Italy 02 ha sufrido un proceso de recombinación. Los estudios de neutralización viral demostraron que existe una baja relación antigénica (menos de 35%) entre Italy 02 y algunos de los serotipos de referencia de IBV, e indicaron que Italy 02 es probablemente un nuevo serotipo.
Introduction
Infectious bronchitis is an acute, highly contagious, viral disease of poultry with worldwide distribution (Cavanagh & Naqi, Citation2003; Cavanagh, Citation2005). The most characteristic clinical signs are those derived from the respiratory disease. However, infectious bronchitis can also show renal (Liu & Kong, Citation2004), enteric and reproductive clinical signs (Raj & Jones, Citation1997). This disease causes major economic losses not only because of poor performance or decreased egg production and quality, but also because of secondary infections (Cavanagh & Naqi, Citation2003). Infectious bronchitis virus (IBV), the aetiological agent of infectious bronchitis, belongs to Group 3 of the genus Coronavirus (Guy, Citation2000; Gonzalez et al., Citation2003; Cavanagh, Citation2005). Its genome consists of a 27.6 kb single-stranded positive-sense RNA molecule, and encodes four structural proteins: a nucleocapsid protein (N), a surface spike glycoprotein (S), a small integral membrane glycoprotein (M), and the envelope protein (E). The S protein, formed by a globular S1 subunit that is anchored in the membrane by the S2 subunit, has been demonstrated to be a determinant for cell tropism for some other coronaviruses. Moreover, three hypervariable regions comprising amino acid residues 38 to 51, 99 to 115 and 274 to 387, respectively, have been located within the S1 subunit and have been associated with haemagglutination-inhibiting and virus-neutralizing epitopes (Stern & Sefton, Citation1982; Cavanagh, Citation1983; Cavanagh et al., Citation1986 Citation1988 Citation1992b; Casais et al., Citation2003).
Many serotypes have been described for IBV, probably due to the frequent point mutations that occur in RNA viruses and also to recombination events demonstrated for IBV (Cavanagh et al., Citation1992a; Kottier et al., Citation1995; Estevez et al., Citation2003; Gelb et al., Citation2005). In most cases, these new isolates have a restricted distribution and remain associated with particular geographical areas (Gelb et al., Citation2001; Smati et al., Citation2002; Zanella et al., Citation2003). Notwithstanding, in some cases these new strains become widely distributed and become dominant genotypes, as occurred with the 4/91 serotype (Adzhar et al., Citation1997; Cavanagh et al., Citation1998a). This serotype was described for the first time in the United Kingdom in the early 1990s, associated with outbreaks of respiratory disease, and rapidly spread, displacing the D274 serotype that had been dominant in the 1980s (Adzhar et al., Citation1997; Cavanagh et al., Citation1998a). Currently 4/91, also named 793/B and CR88, is one of the most common IBV serotypes in Europe.
In 2002, the S1 subunit sequence of a new and unknown IBV strain named Italy 02 was submitted to the GenBank nucleotide database. Surprisingly, Italy 02 has recently been described to be the third most frequently detected IBV strain and probably the dominant wild type in Western Europe where it has been reported in Spain, France, the United Kingdom, Italy and Germany (Worthington et al., Citation2004; Jones et al., Citation2005). As part of an ongoing molecular epidemiological survey based on the study of the IBV S1 sequence carried out in Spain, four isolates showed high similarity with the Italy 02 sequence. In view of the lack of scientific information available and the increasing epidemiological importance of this strain, further characterization of these isolates was considered necessary in order to establish genetic and antigenic relationships with other IBV strains, and to ascertain whether Italy 02 and closely related Spanish strains represented a new IBV serotype.
Materials and Methods
Viruses
The viruses used in this study are presented in . The four IBV field strains were obtained from chickens and passaged in embryonated chicken eggs. Allantoic fluid samples were kindly provided by CESAC (Centre de Sanitat Avícola de Catalunya i Aragó). The reference IBV serotypes M41, D274, D1466 and 4/91 used in the virus neutralization (VN) assay were purchased from the Animal Health Centre, Deventer, The Netherlands.
Table 1. Epidemiological information of field IBV isolates of the Italy 02 genotype included in the study
Virus growth and titration
Ten-day-old to 11-day-old specific pathogen free (SPF) chicken embryos were used for seed stock production and titration of IBV strains. Allantoic fluid was harvested, frozen at −80°C and titrated. Briefly, 10-fold dilutions were inoculated into the allantoic cavity of five SPF chicken embryos. Inoculated chickens were candled daily and examined 1 week later for characteristic IBV lesions. Endpoint dilutions and titres were determined by the Reed and Muench method (Villegas, Citation1998).
Reverse transcriptase-polymerase chain reaction and nucleotide sequencing
RNA was extracted from 150 µl infectious allantoic fluid with the Nucleospin RNA Virus Kit (Macherey-Nagel) according to the manufacturer's instructions. The reverse transcriptase-polymerase chain reaction (RT-PCR) used to amplify the complete S1 complete with oligonucleotides S1Uni2+ and IBP1− was carried out as previously described (Adzhar et al., Citation1996). Besides the flanking primers used in the RT-PCR, a combination of eight internal primers to various regions of the S1 gene were designed to completely sequence both strands of the S1 gene of the field strains. The sequencing primers and their location are indicated in . The 1800-base-pair RT-PCR products were purified by the QIAquick PCR Purification Kit and Minelute PCR purification Kit (Qiagen Inc.) following the manufacturer's instructions. Purified RT-PCR products were sequenced by the dideoxy-mediated chain-termination method using ABI PRISM BigDye® Terminator v3.1 Cycle Sequencing Kit (PE Biosystems) as described by the manufacturer. Sequences were analysed with an automated nucleic acid analyser (ABI PRISM 3100 Avant; PE Biosystems).
Table 2. Sequence and genome localization of primers used in S1 gene sequencing
Nucleotide and amino acid deduced sequence analyses
Assembly and analysis of sequence data were conducted using BioEdit 5.0 package. Nucleotide and amino acid deduced sequences were aligned using ClustalX software. Phylogenetic analysis was performed by the neighbour-joining method with 1000 bootstrap replicates with the software MEGA version 3.0 (Kumar et al., Citation2004). The average ratio dS/dN of synonymous to non-synonymous nucleotide changes for the S1 gene was conducted by the Nei and Gojobori method (Jukes Cantor distance) also included in the software MEGA version 3.0.
GenBank accession numbers
Complete S1 gene nucleotide sequences, including the cleavage site, of the four field viruses were deposited in GenBank with the following accession numbers: Spain/97/314 (DQ064806), Spain/98/313 (DQ064808), Spain/00/337 (DQ064813) and Spain/00/338 (DQ064814).
Production of monospecific antisera for the VN test
Monospecific antisera to the field isolate Spain/00/337 was raised following a specific immunization protocol previously described (Gelb & Jackwood, Citation1998). Briefly, 4-week-old SPF chickens, obtained from eggs acquired from Charles River Spafas, were intratracheally immunized with about 105 median embryo infective doses (EID50) per bird. Two and 5 weeks later chickens were reinoculated with the same dosage by the intravenous route. Blood samples were collected from chickens 2 weeks after the last inoculation. Sera were harvested and frozen before being used in the VN procedure. Antisera to reference serotypes M41, D274, D1466 and 4/91 were purchased from the Animal Health Centre, Deventer, The Netherlands. Lyophilized antisera were resuspended in 1 ml Hank's solution with penicillin G sodium salt (1000 IU/ml) and streptomycin (1000 µg/ml).
Cross-VN analysis
Reciprocal VN tests were performed on the field isolate Spain/00/337 and reference strains M41, 4/91, D274 and D1466 by the diluted-serum constant-virus method (beta procedure) as previously described (Thayer & Beard, Citation1998). Briefly, four-fold serial dilutions of each antiserum previously incubated for 1 h at 37°C were reacted in equal volumes with viruses suspensions containing 100 EID50/0.1 ml. Serum–virus mixtures were incubated at room temperature for 1 h. Five SPF embryos were inoculated by CAM route with 0.1 ml per embryo for each mixture. For antiserum controls, each antiserum dilution was mixed with Hank's solution. Virus controls were carried out by mixing them with Hank's solution. Moreover, viruses were back-titrated in each neutralization test to confirm that 100 EID50 virus had been used. Chicken embryos were evaluated 24 h after inoculation for non-specific mortality, and 1 week after inoculation for death, dwarfing and curling, indicating that virus had not been neutralized by sera. Endpoint titres were calculated by the Reed and Müench method. The VN titres against homologous and heterologous virus were used to calculate the antigenic relatedness (r values) by the formula of Archetti & Horsfall (Citation1950).
Results
Italy 02 and the Spanish field isolates share a common new genotype based on S1 sequence analyses
The nucleotide and deduced amino acid sequences of the S1 subunit of the four field strains and of Italy 02 isolate obtained from GenBank were compared (). The nucleotide and amino acid similarities ranged from 96.6% (between Spain/98/313 and all the other isolates) to 99.8% (between Spain/00/337 and Spain/00/338) and from 93.8% (between Spain/98/313 and Italy 02) to 99.4% (between Spain/337/00 and Spain/338/00), respectively. Thirty-three was the maximum number of amino acid substitutions observed in the S1 subunit among these strains as a group, whereas three amino acid substitutions was the minimum number observed between two strains isolated in the same year. The average dS/dN ratio for the five sequences, including the Italy 02 and the four field Spanish strains, was estimated to be 1.6. Phylogenetic analyses based on both nucleotide and deduced amino acid sequences of the whole S1 gene demonstrated that the four field strains and Italy 02 isolate formed a separate cluster ().
Figure 1. Neighbour-joining phylogenetic tree with 1000 bootstrap replicates showing the relationships among the deduced S1 amino acid sequences of Italy 02 strains and the IBV strains of the main reference serotypes. Horizontal branches indicate the sequence distance (number of amino acid substitutions per site) and are drawn to scale. Sequences in bold letters are Italy 02 genotype isolates. Accession numbers of the reference strains: Ark-DPI (AF006624); Ark-99 (L10384); B1648 (X87238); Beaudette (X02342); DE072 (U77298); D1466 (M21971); D207 (M21969); D274 (X15832); D3896 (X52084); FR-85131-85 (AJ618985); FR-94047-94 (AJ618987); Gray (L14069); Holte (L18988); H120 (M21970); H52 (AF352315); Italy 02 (AJ457137); JMK (L14070); M41 (X04722); UK/7/93 (Z83979); UK-1233-95 (AJ618984); 4/91 (AF093794).
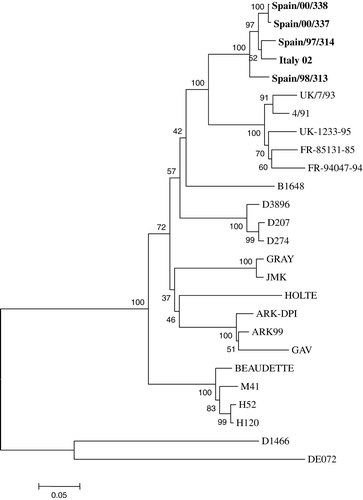
Table 3. Nucleotide and amino acid identities of IBV S1 gene sequences: comparison of the four field strains and the main reference IBV strains of different serotypes obtained from GenBank
Italy 02 S1 sequence is a mosaic
Nucleotide and amino acid sequences identities are presented in . Italy 02 isolates showed maximum nucleotide identities with strains of the 4/91 serotype (83.6% to 85.2%), and secondly with the B1648 strain (81.1% to 81.3%) and D274 strains (79.5% to 80.8%), which included D274, D207 and D3896 isolates. Similarly, when amino acid sequences were compared, Italy 02 strains showed maximum amino acid identities with 4/91 isolates (81.3% to 83.7%), D274 strains (79.8% to 81.7%) and B1640 isolate (79.3% to 80%). S1 nucleotide and amino acid similarities of Italy 02 isolates with the remainder of the IBV strains included in the comparison were lower than 80%.
Amino acid sequence alignment revealed that Italy 02 strains shared some mutated residues in common with 4/91 and D274 strains and the B1648 isolate, but also had unique amino acid mutations (). Particularly, most of the mutations observed in the 3′ region shared residues with the D274 strains and the B1648 isolate, whereas the majority of mutated residues in the 5′ region corresponded with 4/91 isolates and the B1648 isolate. In order to further study these preliminary observations, the S1 analysis and phylogenetic studies were also conducted in both S1 regions separately, the 5′ region comprising the initial 230 amino acids and the 3′ region including the final 310 amino acid residues. Comparison at the 5′ 210 amino acid region showed that Italy 02 sequences shared maximum mutated residues with D274 strains (48 out of 86) (). However, when the 3′ region was compared, Italy 02 shared maximum mutated positions with 4/91 group (47 out of 76, which represents 62% of mutated residues). Mutated residues shared with B1648 or not shared with any of the other three strains group were maintained at a constant rate between both regions.
Figure 2. Alignment of deduced amino acid sequences of the complete S1 gene. Four Spanish field isolates, Italy 02 and main reference IBV strains published in GenBank are compared. Dots indicate identical residues.
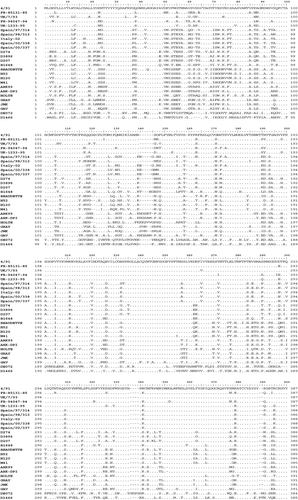
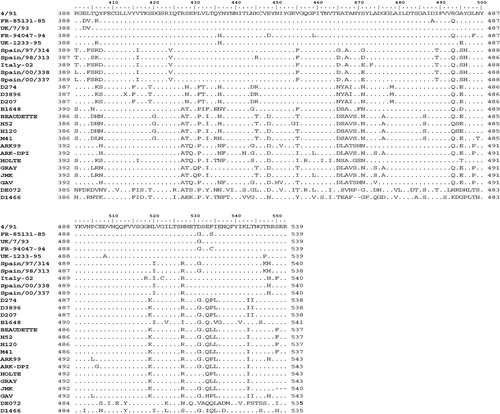
Table 4. Comparison of the S1 amino acid sequence of strain Italy 02 with that of strains 4/91, D274 and B1648
The maximum amino acid identity of Italy 02 isolates when the 5′ S1 region was compared was 79.2% to 82.7% with D274 strains, whereas the 4/91 strains and B1648 showed amino acid identities ranging from 71% to 74.8% and from 72.6% to 74%, respectively. When the 3′ S1 region was compared, however, Italy 02 isolates had maximum amino acid identities with 4/91strains (87.3% to 89.8%), while amino acid identities with D274 strains and the B1648 isolate were 79.3% to 81.5% and 83.4% to 84.7%, respectively.
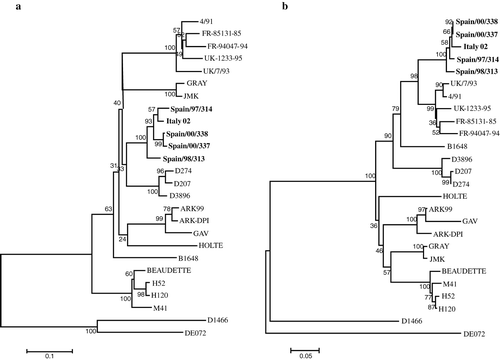
When phylogenetic analysis was performed based on the 5′ S1 region, Italy 02 strains were grouped together and closely related with D274 strains, while phylogenetic analysis based on the 3′ end of the S1 gene revealed that Italy 02 was closely related with 4/91 strains ().
Figure 3. Neighbour-joining phylogenetic trees with 1000 bootstrap replicates showing the relationships among amino acid deduced sequences when phylogenetic analyses are conducted in both S1 regions separately. The length of the bar denotes number of amino acid substitutions per site. 3a: Tree of the 5′ 230-amino-acid region: Italy 02 isolates are clustered with D274 strains group. 3b: Tree of the 3′ 310-amino-acid region: Italy 02 sequences are clustered with 4/91 strains group. Sequences in bold letters are Italy 02 genotype isolates.
All Italy 02 strains had two amino acid insertions, with regard to 4/91 and D274 isolates, located at residues 141 and 144, close to the epitope associated with HVR2. IBV strains share a common sequence pattern for the S1–S2 cleavage site that consists of X1-Arg-X2-Arg-Arg residues. Italy 02 strains had sequences at this site distinct from those in 4/91 and D274 strains. The four isolates Spain/98/313, Spain/97/314, Spain/00/337 and Spain/00/338 had the sequence His-Arg-Ser-Arg-Arg like Ark-99 and GAV strains, whereas Italy 02 isolate had the sequence His-Arg-Phe-Arg-Arg.
VN test shows little antigenic relatedness between the Italy 02 isolate and the reference IBV serotypes
Based on the S1 gene molecular analysis, we selected one isolate from the Italy 02 group to perform the VN test. The percentage of antigenic relatedness values (r) was determined by VN for the field isolate Spain/00/337 and reference strains M41, D274, D1466 and 4/91. Isolate Spain/00/337 shared very low antigenic relatedness with reference strains: 19.3% with M41, 23.3% with D1466, 26.7% with 4-91 and 34.3% with D274.
Discussion
Recent studies have shown that Italy 02 has become one of the most widely distributed IBV strains in Western Europe (Worthington et al., Citation2004). In the present study, it has been demonstrated on the basis of nucleotide and predicted amino acid sequence analyses of the whole S1 gene that Italy 02 isolates form a separate phylogenetic group, showing the closest nucleotide and amino acid S1 identities with 4/91 strains, D274 strains (D207, D274 and D3896) and the B1648 isolate. These results confirm that Italy 02 is a new genotype.
Although the origin of this novel genotype remains currently unidentified, it is well known that there are two major forces that drive coronavirus evolution: mutation due to the high error rates of the viral RNA polymerase, and recombination. According to our results, Italy 02 genotype was most closely related with 4/91 and D274 strains, and the B1648 isolate. Therefore, one possibility is that single point mutations accumulated during time in one of these three genotypes would have induced the emergence of a new genotype that differs from the parental one. However, S1 analysis has shown that the percentage of amino acid substitutions shared with other serotypes varies substantially depending on which S1 region is analysed, whereas the percentage of unique mutations for Italy 02 is maintained constant over the entire S1 gene. Moreover, nucleotide and amino acid identities and phylogenetic analysis performed in both S1 regions independently reinforce the finding that molecular relationships of Italy 02 strains with other isolates are significantly different depending on the S1 region analysed. Thus, taking into account these findings it can be hypothesized that not only point mutations, but also a recombination event might have occurred giving rise to the emergence of a new genotype, both mechanisms having contributed to evolution of the new genotype.
A challenging question arises regarding the way in which the Italy 02 genotype has spread widely throughout Western Europe and has become the predominant genotype in some countries. Although this genotype has been reported to be around in Europe since 2000 (Worthington et al., Citation2004), the isolate that has been established as the prototype, Italy 02, was isolated in Italy in 1999 (Dr Ana Moreno, Instituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna, personal communication). Our results indicate that the Italy 02 genotype has been circulating in Spain since as early as 1997. This observation together with the widespread distribution of the Italy 02 genotype in Europe may suggest that current vaccination strategies have not satisfactorily been able to protect chickens against infection.
However, when proper vaccination programmes are implemented for one genotype, better control of the disease is achieved but also provides a selective advantage for the emergence of new variant genotypes (Cavanagh et al., Citation1998a). This situation where one dominant genotype is substituted by a new genotype has been described previously for the 4/91 serotype in the United Kingdom and for the Delaware genotype in the USA (Cavanagh et al., Citation1998b; Lee & Jackwood, Citation2001). Therefore, to address the question of whether positive selection pressures might have been driving Italy 02 evolution, the dS/dN ratio was calculated. This ratio provides an index to estimate whether a nucleic acid segment is subject to positive selection pressures. Thus, dS/dN values lower than 1 suggest that the segment is under positive selection pressures, while values higher than 1 are indicative of a lack of selective pressures. The average ratio of dS/dN amino acid substitutions for the S1 sequence within the Italy 02 genotype was 1.6 and revealed that amino acid changes observed in the S1 gene were not under positive selective pressures. However, the ds/dN ratio was closer to 1 when compared with dS/dN ratios reported for when one serotype is compared with a different serotype (ranging from 3.4 to 5.5) (Cavanagh et al., Citation2005). This shows a higher rate of non-synonymous change within the Italy 02 group than in other genotypes, probably because Italy 02 is a very novel genotype that is still evolving. Therefore, although no positive selective pressures seems to be related with Italy 02 amino acid changes, it is probable that widespread vaccination against the previous known dominant genotypes associated with high recombination and mutation abilities of IBV have lead to the emergence and broad dissemination of this novel genotype.
Because of high sequence differences between Italy 02 and the reference IBV strains, this new genotype was also suspected to be a new serotype. Partial or total sequencing of the S1 gene has been shown to be useful in order to genotype and identify new serotypes (Keeler et al., Citation1998; Wang & Huang, Citation2000). However it has been demonstrated that in some cases a small number of amino acid differences in the S1 proteins can be sufficient to avoid cross-protection between two viruses and indeed can change serotype (Cavanagh, Citation2003). In spite of being time-consuming and expensive, assays investigating antigenic relatedness of new genotypes by serological tests are necessary to finally classify IBV isolates as new serotypes. We therefore decided to assess antigenic relationships between Italy 02 and reference IBV strains M41, D274, 4/91 and D1466 by cross-VN analysis. Our results show that no cross-neutralization exists between Italy 02 and the reference IBV serotypes tested, suggesting that Italy 02 might be established as a new serotype. However, Worthington et al. (Citation2004) and Jones et al. (Citation2005) reported that infectious bronchitis vaccine combinations with Mass, Ark and D274 serotypes gave excellent protection against challenge with Italy 02. The discrepancy between these findings can be explained because VN assays are more discriminating than cross-challenge, and VN relatedness values tend to be lower than cross-challenge relatedness (Gelb et al., Citation1997). Moreover, it should be taken into account that VN assays only assess humoral immunity, but protection is achieved by both humoral and cellular responses (Raj & Jones, Citation1997).
In summary, the present study has demonstrated that Italy 02 genotype has been circulating in the field since at least as early as 1997. S1 gene analysis has suggested that a potential recombination event might have been involved in the emergence of this new genotype. However, the dS/dN ratio within Italy 02 genotype indicated that mutations in the S1 gene were not related to selective pressures. Moreover, VN assays have indicated that Italy 02 is likely to be a new serotype. Given the epidemiological importance that this serotype has acquired, further in vivo studies are necessary to determine its main pathogenic characteristics.
Acknowledgments
The authors wish to thank Dr Albert Bensaid for helping in primer designing. They also thank Mar Biarnès and Fèlix Ponsa for technical assistance. This work was supported by research grants from Centre de Recerca en Sanitat Animal.
References
- Adzhar , A. , Shaw , K. , Britton , P. and Cavanagh , D. 1996 . Universal oligonucleotides for the detection of infectious bronchitis virus by the polymerase chain reaction . Avian Pathology , 25 : 817 – 836 .
- Adzhar , A. , Gough , R.E. , Haydon , D. , Shaw , K. , Britton , P. and Cavanagh , D. 1997 . Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain . Avian Pathology , 26 : 625 – 640 .
- Archetti , I. and Horsfall , F. L. 1950 . Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum . Journal of Experimental Medicine , 92 : 441 – 462 .
- Boursnell , M.E. , Brown , T.D. , Foulds , I.J. , Green , P.F. , Tomley , F.M. and Binns , M.M. 1987 . Completion of the sequence of the genome of the coronavirus avian infections bronchitis virus . Journal of General Virology , 68 : 57 – 77 .
- Casais , R. , Dove , B. , Cavanagh , D. and Britton , P. 2003 . Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism . Journal of Virology , 77 : 9084 – 9089 .
- Cavanagh , D. 1983 . Coronavirus IBV: structural characterization of the spike protein . Journal of General Virology , 64 ( Pt 12 ) : 2577 – 2583 .
- Cavanagh , D. 2001 . A nomenclature for avian coronavirus isolates and the question of species status . Avian Pathology , 30 : 109 – 115 .
- Cavanagh , D. 2003 . Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus . Avian Pathology , 32 : 567 – 582 .
- Cavanagh , D. 2005 . Coronaviruses in poultry and other birds . Avian Pathology , 34 : 439 – 448 .
- Cavanagh , D. and Naqi , S. 2003 . “ Infectious Bronchitis ” . In Diseases of Poultry , 11th edn , Edited by: Saif , Y.M. , Barnes , H.J. , Glisson , J.R. , Fadly , A.M. , McDougald , L.R. and Swayne , D.E. 101 – 120 . Ames : Iowa State University Press .
- Cavanagh , D. , Davis , P.J. , Darbyshire , J.H. and Peters , R.W. 1986 . Coronavirus IBV: virus retaining spike glycopolypeptide S2 but not S1 is unable to induce virus-neutralizing or haemagglutination-inhibiting antibody, or induce chicken tracheal protection . Journal of General Virology , 67 : 1435 – 1442 .
- Cavanagh , D. , Davis , P.J. and Mockett , A.P. 1988 . Amino acids within hypervariable region 1 of avian coronavirus IBV (Massachusetts serotype) spike glycoprotein are associated with neutralization epitopes . Virus Research , 11 : 141 – 150 .
- Cavanagh , D. , Davis , P.J. and Cook , J.K.A. 1992a . Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype . Avian Pathology , 21 : 401 – 408 .
- Cavanagh , D. , Davis , P.J. , Cook , J.K.A. , Li , D. , Kant , A. and Koch , G. 1992b . Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus . Avian Pathology , 21 : 33 – 43 .
- Cavanagh , D. , Mawditt , K. , Britton , P. and Naylor , C. 1998a . “ Longitudinal studies of infectious bronchitis virus in broilers using genotype-specific quantitative PCRs ” . In Proceedings of the International Symposium on Infectious Bronchitis and Pneumovirus Infections in Poultry , Edited by: Heffels-Redmann , U. and Kaleta , E.F. 215 – 219 . Rauischholzhausen, Germany .
- Cavanagh , D. , Mawditt , K. , Gough , R. , Picault , J.F. and Britton , P. 1998b . “ Sequence analysis of strains of the 793/B genotype (CR88, 4/91) of IBV isolated between 1985 and 1997 ” . In Proceedings of the International Symposium on Infectious Bronchitis and Pneumovirus Infections in Poultry , Edited by: Kaleta , E.F. and Heffels-Redmann , U. 252 – 256 . Rauischholzhausen, Germany .
- Cavanagh , D. , Picault , J. P. , Gough , R. , Hess , M. , Mawditt , K. and Britton , P. 2005 . Variation in the spike protein of the 793/B type of infectious bronchitis virus, in the field and during alternate passage in chickens and embryonated eggs . Avian Pathology , 34 : 20 – 25 .
- Estevez , C. , Villegas , P. and El-Attrache , J. 2003 . A recombination event, induced in ovo, between a low passage infectious bronchitis virus field isolate and a highly embryo adaptedvaccine strain . Avian Diseases , 47 : 1282 – 1290 .
- Gelb , J. Jr and Jackwood , M.W. 1998 . “ Infectious Bronchitis ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 169 – 174 . Pennsylvania : The American Association of Avian Pathologists .
- Gelb , J. Jr , Keeler , C.L. Jr , Nix , W.A. , Rosenberger , J.K. and Cloud , S.S. 1997 . Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus . Avian Diseases , 41 : 661 – 669 .
- Gelb , J. Jr , Ladman , B.S. , Tamayo , M. , Gonzalez , M. and Sivanandan , V. 2001 . Novel infectious bronchitis virus S1 genotypes in Mexico 1998–1999 . Avian Diseases , 45 : 1060 – 1063 .
- Gelb , J. Jr , Weisman , Y. , Ladman , B.S. and Meir , R. 2005 . S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) . Avian Pathology , 34 : 194 – 203 .
- Gonzalez , J.M. , Gomez-Puertas , P. , Cavanagh , D. , Gorbalenya , A.E. and Enjuanes , L. 2003 . A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae . Archives of Virology , 148 : 2207 – 2235 .
- Guy , J.S. 2000 . Turkey coronavirus is more closely related to avian infectious bronchitis virus than to mammalian coronaviruses: a review . Avian Pathology , 29 : 207 – 212 .
- Jones , R.C. , Worthington , K.J. and Gough , R.E. 2005 . Detection of the Italy 02 strain of infectious bronchitis virus in the UK . Veterinary Record , 156 : 260
- Keeler , C.L. Jr , Reed , K.L. , Nix , W.A. and Gelb , J. Jr . 1998 . Serotype identification of avian infectious bronchitis virus by RT-PCR of the peplomer (S-1) gene . Avian Diseases , 42 : 275 – 284 .
- Kottier , S.A. , Cavanagh , D. and Britton , P. 1995 . Experimental evidence of recombination in coronavirus infectious bronchitis virus . Virology , 213 : 569 – 580 .
- Kumar , S. , Tamura , K. and Nei , M. 2004 . Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment . Briefings in Bioinformatics , 5 : 2
- Lee , C.W. and Jackwood , M.W. 2001 . Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus . Virus Research , 80 : 33 – 39 .
- Liu , S.-W. and Kong , X.-G. 2004 . A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China . Avian Pathology , 33 : 321 – 327 .
- Raj , G.D. and Jones , R.C. 1997 . Infectious bronchitis virus: immunopathogenesis of infection in the chicken . Avian Pathology , 26 : 677 – 706 .
- Smati , R. , Silim , A. , Guertin , C. , Henrichon , M. , Marandi , M. , Arella , M. and Merzouki , A. 2002 . Molecular characterization of three new avian infectious bronchitis virus (IBV) strains isolated in Quebec . Virus Genes , 25 : 85 – 93 .
- Stern , D.F. and Sefton , B.M. 1982 . Coronavirus proteins: biogenesis of avian infectious bronchitis virus virion proteins . Journal of Virology , 44 : 794 – 803 .
- Thayer , S.G. and Beard , C.W. 1998 . “ Serologic procedures ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 255 – 266 . Pennsylvania : The American Association of Avian Pathologists .
- Villegas , P. 1998 . “ Titration of biological suspensions ” . In A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Edited by: Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 248 – 254 . Pennsylvania : The American Association of Avian Pathologists .
- Wang , C.H. and Huang , Y.C. 2000 . Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus . Archives of Virology , 145 : 291 – 300 .
- Worthington , K.J. , Savage , C. , Naylor , C.J. , Wijmenga , W. and Jones , R.C. 2004 . “ An RT-PCR survey of infectious bronchitis virus genotypes in the UK and selected European countries between 2002 and 2004 and the results from a vaccine trial ” . In Proceedings of the IV International Symposium on Avian Corona- and Pneumovirus Infections , Edited by: Heffels-Redmann , U. and Kaleta , E.F. 125 – 133 . Rauischholzhausen, Germany .
- Zanella , A. , Lavazza , A. , Marchi , R. , Moreno Martin , A. and Paganelli , F. 2003 . Avian infectious bronchitis: characterization of new isolates from Italy . Avian Diseases , 47 : 180 – 185 .