Abstract
The complete genome of a novel Circovirus isolated from an Australian raven (Corvus coronoides) with feather lesions similar to those that occur in psittacine beak and feather disease is reported. Degenerate polymerase chain reaction primers were designed to amplify and sequence novel Circovirus DNA from affected feathers. Sequence analysis indicated that the tentatively named raven circovirus (RaCV) was 1898 nucleotides in size with two major open reading frames synonymous with other avian circoviruses, ORF C1 and ORF V1, likely to encode a putative capsid protein (Cap) and replicase-associated protein (Rep), respectively. In common with other circoviruses was the conservation of several nucleotide structures and amino acid motifs implicated in virus replication. Comparison with other members of the Circoviridae demonstrated that RaCV shares the greatest sequence homology with canary circovirus (CaCV) and pigeon circovirus (PiCV) and was more distantly related to the beak and feather disease virus, goose circovirus, duck circovirus and the two porcine circoviruses, PCV1 and PCV2. Phylogenetic analysis of the genome and the putative Cap and Rep proteins provided further evidence of the close relationship of RaCV with CaCV and PiCV.
Identification d'un nouveau circovirus chez les corbeaux d'Australie (Corvus coronoides) avec une maladie des plumes
Le génome complet d'un nouveau circovirus isolé d'un corbeau australien (Corvus coronoides) qui présentent des lésions des plumes similaires à celles observées dans la maladie du bec et des plumes des psittacidés est présenté. Les amorces dégénérées de la PCR ont été sélectionnées pour amplifier et séquencer l'ADN du nouveau circovirus à partir des plumes affectées. L'analyse de la séquence a montré que le circovirus, nommé provisoirement circovirus du corbeau (RaCV), avait 1898 nucléotides avec deux principaux cadres ouverts de lecture synonymes des autres circovirus aviaires, ORF C1 et ORF V1 qui respectivement étaient supposés coder une protéine de capside putative (Cap) et une protéine associée à la réplicase (Rep). La conservation de plusieurs structures nucléotidiques et de motifs d'acides aminés impliqués dans la réplication virale sont des caractéristiques communes avec les autres circovirus. La comparaison aux autres membres des Circoviridae a montré que le RaCV présentait la plus grande homologie de séquence avec le circovirus du canari (CaCV) et le circovirus du pigeon (PiCV) et était plus éloigné du virus de la maladie du bec et des plumes (BFDV), du circovirus de l'oie, du circovirus du canard et des deux circovirus porcins PCV1 et PCV2. L'analyse phylogénétique du génome, des protéines Rep et Cap putative ont fourni des preuves complémentaires de la relation étroite du RaV avec le CaCV et le PiCV.
Identifizierung eines neuen Circovirus in australischen Krähen (Corvus coronoides) mit Befiederungsstörungen
Es wird über das komplette Genom eines neuen Circovirus, das aus einer australischen Krähe (Corvus coronoides) mit Federveränderungen ähnlich denen bei der psittazinen Schnabel-und Federkrankheit (PBFD) isoliert worden ist, berichtet. Zur Amplifizierung und Sequenzierung der neuen Circovirus-DNS aus den betroffenen Federn wurden degenerierte PCR-Primer entwickelt. Die Sequenzanalyse zeigte, dass das vorläufig als Krähencircovirus (RaCV) bezeichnete Virus eine Größe von 1898 Nukleotiden hatte. Es wies wie bei den anderen aviären Circoviren zwei bedeutende offene Leserahmen auf, ORF C1 und ORF V1, die wahrscheinlich für das Kapsidprotein (Cap) bzw. für das Replikase assoziierte Protein kodieren. In Übereinstimmung mit anderen Circoviren war auch die Konservierung von mehreren Nukleotidstrukturen und Aminosäurenmustern, die mit der Virusvermehrung im Zusammenhang stehen. Der Vergleich mit anderen Mitgliedern der Familie der Circoviridae ließ erkennen, das das RaCV die größte Sequenzhomologie mit dem Kanariencircovirus (CaCV) und dem Taubencircovirus (PiCV) teilte und nur entferntere Beziehung zum BFDV, dem Gänse-, Enten- und zwei Schweinecircoviren (PCV1 unf PCV2) aufwies. Die phylogenetische Analyse des Genoms sowie der mutmaßlichen Cap- und Rep-Proteine erbrachten weitere Hinweise auf die enge Verwandtschaft des RaCV mit dem CaCV und PiCV.
Identificación de un nuevo circovirus en cuervos Australianos (Corvus coronoides) con la enfermedad de la pluma
Se describe el genoma completo de un nuevo Circovirus aislado a partir de cuervos Australianos (Corvus coronoides) con lesiones en las plumas similares a aquellas encontradas en la enfermedad del pico y las plumas de las psitácidas. Se diseñaron cebadores de PCR degenerados para amplificar y secuenciar el DNA del nuevo Circovirus a partir de las plumas afectadas. Los análisis de las secuencias indicaron que el circovirus llamado provisionalmente circovirus del cuervo (RaCV) tenía un tamaño de 1898 nucleótidos con dos marcos principales de lectura abierta al igual que otros circovirus aviares, ORF C1 y ORF V1, que probablemente codifiquen una supuesta proteína de la cápside (Cap) y una proteína asociada a la replicasa (Rep), respectivamente. La conservación de diversas estructuras nucleotídicas y motivos aminoacídicos implicados en la replicación vírica fueron comunes a otros circovirus. La comparación con otros miembros de Circoviridae demostró que RaCV compartía la mayor homología de secuencia con el circovirus del canario (CaCv) y el circovirus de la paloma (PiCV) y estaba relacionado de manera más distante con el virus de la enfermedad del pico y las plumas (BFDV), el circovirus del ganso, el circovirus del pato y los dos circovirus porcinos, PCV1 y PCV2. El análisis filogenético del genoma y de las supuestas proteínas Cap and rep proporcionaron más evidencias de la estrecha relación de RaCV con CaCv y PiCV.
Introduction
Circoviruses are small, non-enveloped, spherical viruses that contain a circular, single strand DNA genome (Todd et al., Citation2001a). The family Circoviridae presently contains two genera; Gyrovirus and Circovirus. The only member of the Gyrovirus genus based on its unique genome organization is chicken anaemia virus (Todd et al., Citation2004). There are six official members of the Circovirus genus including the two porcine circoviruses, PCV1 (Meehan et al., Citation1997) and PCV2 (Hamel et al., Citation1998; Meehan et al., Citation1998), beak and feather disease virus (BFDV) (Bassami et al., Citation1998; Niagro et al., Citation1998) and the recently assigned canary circovirus (CaCV) (Phenix et al., Citation2001), pigeon circovirus (PiCV) (Mankertz et al., Citation2000; Todd et al., Citation2001b) and goose circovirus (GoCV) (Todd et al., Citation2001b). There has been an increase in the number of tentative circoviruses identified in avian species with duck circovirus (DuCV) (Hattermann et al., Citation2003) recently identified and a number of avian circovirus-like infections based on histopathology, and electron microscopy in doves (Raidal & Riddoch, Citation1997), finches (Mysore et al., Citation1995), gulls (Twentyman et al., Citation1999) and ostrich (Eisenberg et al., Citation2003).
Avian circovirus infections cause different clinical manifestations ranging from severe chronic feather, beak and claw abnormalities to non-specific ill-thrift, lethargy and poor performance, but a common feature shared is lymphoid depletion and associated immunosuppression that exposes the infected birds to secondary opportunistic infections (Todd et al., Citation2001a; Todd, Citation2004). Histologically, circovirus infections are commonly associated with the presence of globular or botryoid, basophilic intracytoplasmic inclusions within macrophages (Pass & Perry, Citation1984). In BFDV-infected psittacine species the lesions are detected in the developing feather shaft and pulp with lymphoid necrosis and atrophy occurring in the thymus and bursa of Fabricius (Pass & Perry, Citation1984), while histological changes in a non-psittacine species are mainly associated with primary and secondary lymphoid tissue.
Viruses from the genus Circovirus possess an ambisense genome with similar genomic organization and conserved nucleotide and amino acid motifs. A stem-loop structure, which contains a nonamer sequence, has been demonstrated to be involved in the initiation of rolling circle replication of the virus (Steinfeldt et al., Citation2001). There are two conserved open reading frames: ORF V1, which is postulated to encode a replicase-associated protein (Rep); and ORF C1, which encodes the structural putative capsid protein (Cap). In this paper, we report the use of polymerase chain reaction (PCR) amplification using degenerate primers to identify a novel circovirus in Australian ravens (Corvus coronoides) exhibiting clinical signs similar to those that occur in psittacine beak and feather disease (PBFD).
Materials and Methods
Source of material
A wild juvenile Australian raven (ID number 04–1131) with a history of progressively developing feather defects was presented for clinical examination by a wildlife rehabilitator. Blood collected from the bird onto filter paper as described by Riddoch et al. (Citation1996) and fragments of broken feathers and developing feathers plucked from follicles were collected for serology and avian circovirus detection by PCR. The bird was anaesthetized with isofluorane in oxygen and several feather follicle biopsies were surgically obtained. These were fixed in formalin and processed for routine paraffin embedding and histological examination.
Feather and/or blood samples were collected from an additional three ravens exhibiting the bilateral pattern of feather depigmentation and dysplasia as the case already described. Similar samples were also collected from five nestling or fledgling ravens that were in contact with each other at a wildlife rehabilitation centre in Perth. Two of these young birds had occasional white-tipped contour and covert feathers scattered throughout their plumage but no structural lesions.
Haemagglutination and haemagglutination inhibition assays
Haemagglutination assay were performed on feather samples as described by Raidal et al. (Citation1993) using galah (Eolophus roseicapillus) erythrocytes sensitive to haemagglutination by BFDV. The haemagglutination inhibition assay was performed on a blood sample collected onto filter paper as described by Riddoch et al. (Citation1996).
Preparation and purification of DNA from feather samples
DNA was extracted from feather tissues using modified Taberlet & Bouvet (Citation1991) and Morin et al. (Citation1994) methods as previously described by Ypelaar et al. (Citation1999).
Amplification of a novel circovirus using degenerate primers
Raven circovirus (RaCV)-specific DNA was amplified using two sets of degenerate primers based on the sequence of BFDV (AF080560 and AF071878), CaCV (NC003410 and AJ301633), DuCV (AY364721 and AY228555), GoCV (NC003054), PCV (U49186 and AF055392) and PiCV (AJ298229 and AF252610) that were designed to amplify two overlapping region in ORF V1 gene. The primer set Deg.prm1.f (TAYTGYTCBAATGARGG; PROLIGO, Australia) and Deg.prm1.r (AGCCABCCRTARAARTCRTC; PROLIGO) reaction consisted of 1×PE buffer II (Perkin Elmer, USA), 2 mM MgCl2 (Perkin Elmer), 0.2 mM each dNTP (Perkin Elmer), 200 ng each primer (PROLIGO), 1 U Taq DNA polymerase, 250 to 500 ng extracted DNA in a final volume of 50 µl Ultra pure water (Fischer Biotec, Australia). The amplification protocol consisted of a denaturation step of 95°C for 5 min, 35 cycles of 95°C for 20 sec, 55°C for 20 sec, 50°C for 20 sec and 72°C for 1 min, and an elongation step at 72°C for 10 min.
The primer set CircoDeg.N (CACSCTKAAYAAYCCTWCC; PROLIGO) and CircoDeg.C (TTGMCCATSATANCCATCC; PROLIGO) reaction consisted of 1×PE buffer II (Perkin Elmer), 2 mM MgCl2 (Perkin Elmer), 0.3 mM each dNTP (Perkin Elmer), 200 ng each primer (PROLIGO), 1 U Taq DNA polymerase, 250 to 500 ng extracted DNA in a final volume of 50 µl Ultra pure water (Fischer Biotec). The amplification protocol consisted of a denaturation step of 95°C for 5 min, 35 cycles of 95°C for 20 sec, 56°C for 20 sec and 72°C for 1 min, and an elongation step at 72°C for 10 min.
Amplification and analysis of raven circovirus genome
The entire RaCV genome was amplified using two different sets of primers designed to produce overlapping fragments. The primer set RaCV.1N (CGCATTCTTGTCTGTA; PROLIGO) and RaCV.1C (CAATGGGCACGGCTAAG; PROLIGO) amplification reaction consisted of 1×PE buffer II (Perkin Elmer), 2 mM MgCl2 (PerkinElmer), 0.2 mM each dNTP (Perkin Elmer), 200 ng each primer (PROLIGO), 1 U Taq DNA polymerase, 250 to 500 ng extracted DNA in a final volume of 50 µl Ultra pure water (Fischer Biotec). The amplification protocol consisted of a denaturation step of 95°C for 5 min, 35 cycles of 95°C for 20 sec, 55°C for 20 sec, 50°C for 20 sec and 72°C for 1 min, and an elongation step at 72°C for 10 min.
The primer set RaCV.2N (CCAGCCGTGCCCATTG; PROLIGO) and RaCV.2C (TACAGACAAGAATGCG; PROLIGO) amplification reaction consisted of 1×PE buffer II (Perkin Elmer), 2 mM MgCl2 (Perkin Elmer), 0.3 mM each dNTP (Perkin Elmer), 200 ng each primer (PROLIGO), 1 U Taq DNA polymerase, 250 to 500 ng extracted DNA in a final volume of 50 µl Ultra pure water (Fischer Biotec). The amplification protocol consisted of a denaturation step of 95°C for 5 min, 35 cycles of 95°C for 20 sec, 56°C for 20 sec and 72°C for 1 min, and an elongation step at 72°C for 10 min.
All PCR products generated by the degenerate and RaCV-specific primer sets were visualized by agar gel electrophoresis (Sambrook & Russell, Citation2001). PCR amp icons generated from raven DNA (04–1131) were purified from the agars using the QIAquick Gel Extraction Kit (QIAGEN) and either were ligated into pCR2.1 vector (Invitrogen) according to manufacturer's protocols or were directly sequenced. The ABI Prism™ Dye Terminator Cycle Sequencing Kit (Applied Biosystem) was used according to manufacturer's protocols except the reaction volumes were halved to 10 µl and the annealing temperature was raised to 58°C. Sequence data were generated from at least two PCR amplicons and three cloned products from distinct reactions in both orientations twice. Sequence information was determined using the Applied Biosystem 3730 DNA Analyzer.
Computer analysis of sequence data
The nucleotide sequence of RaCV was edited and assembled using SeqEd version1.0.3 (Applied Biosystems) and analysed using a range of programs provided by the Australian National Genomic Information Service, the National Center for Biotechnology Information (NCBI) and the European Bioinformatics Institute. The edited sequence was analysed using the BLASTN and BLASTP programmes (Altschul et al., Citation1997) using the non-redundant nucleic acids and protein databases at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). Putative open reading frames were determined using SeqEd version 1.0.3, GenScan, FlipORF and relationship of the predicted gene to other circovirus ORF, and was further analysed using BLASTP. The circovirus sequences were aligned using the ClustalW program (http://www.ebi.ac.uk/clustalw/) and analysed phylogenetically using the TreeCon program with 1000 bootstrap cycles. The phylogenetic trees were generated using the neighbour-joining methods of Tajima & Nei (Citation1984) and Galtier & Gouy (Citation1995).
Accession number
The nucleotide sequence reported in this paper has been deposited into GenBank and assigned the accession number DQ146997.
Results
Histopathology
Histopathological examination of abnormal feathers demonstrated dysplasia of developing follicles and a mild mixed inflammatory cellular infiltration in the pulp in some areas. This revealed a bilaterally symmetrical pattern of patchy feather depigmentation and feather dysplasia characterized by marked thickening of developing feather sheaths, haemorrhages within pulp and distortion and fracture of the developing calamus of affected feathers (a,b). Widespread apoptosis of basilar keratinocytes was present in the epidermis of developing barbs and at the epidermal collar. Scattered globular intracytoplasmic inclusions were present basilar and suprabasilar to midzonal keratinocytes within developing feather barbs and other structures (b).
Figure 1. 1A: Wing feathers from a juvenile Australian raven (Corvus coronoides) demonstrating depigmentation (asterisk) and thickened and retained feather sheaths (arrowhead). 1B: Fractured distorted calami. 1C: Histological examination of a developing feather demonstrated globular intracytoplasmic inclusions within the basilar and suprabasilar keratinocytes (arrowheads).

Haemagglutination and haemagglutination inhibition results
Haemagglutination activity was not detected in feather samples and anti-BFDV haemagglutination inhibition antibody was not detected in blood from the ravens tested.
Isolation and cloning of DNA-specific to RaCV
The two sets of degenerate primers Deg.prm1.f–Deg.prm1.r and CircoDeg.N–CircoDeg.C successfully amplified two circovirus-specific DNA fragments of 951 and 598 base pairs from raven feather material. The nucleotide sequences generated from these 951 and 598 base pair fragments indicated that the RaCV sequence was more closely related to the non-psittacine circoviruses CaCV and PiCV in comparison with BFDV and was distantly related to the porcine circovirus, PCV1 and PCV2; and it was used to design the primer set RaCV.1N and RaCV.1C for further PCR amplification and sequencing.
The four sets of primers were used by PCR to detect RaCV DNA in a further three raven feather and/or blood samples displaying feather depigmentation and dysplasia and in clinically normal juvenile ravens in close contact with affected birds. These PCR amplicons were not sequenced.
Genomic organization of the RaCV genome
The full-length RaCV genome was successfully amplified using overlapping RaCV-specific primer sets. Sequence analysis revealed that the RaCV genome was 1898 nucleotides and circular (). The nucleotide numbering and open reading frame nomenclature adopted to describe the RaCV was similar to that used for the previously described circovirus (Todd et al., Citation2001b). Nucleotide position 1 is the residue A at position 8 of the nonamer sequence (GAGTATT/AC), which is located at the apex of the putative stem-loop structure (). The stem-loop structure is conserved within the circoviruses and is postulated to be involved in the initiation of rolling-cycle replication. The RaCV nonamer sequence differed from other circovirus nonamer sequences with G as the first residue rather than T (b).
Figure 2. Nucleotide sequence of the RaCV genome. The A residue immediately downstream of the putative nick site in the nonanucleotide motif is given as nucleotide position 1. The stem-loop structure is underlined and the nonamer sequence is in italics. The open reading frames are in bold and the direction is indicated by an arrow, and the putative poly A signals are underlined twice.

Figure 3. 3A: The stem-loop sequence present in the RaCV intergenic region demonstrating a nonamer element at the apex of the structure (bold), tandem repeat sequences (underlined) and nick site (arrow). 3B: Relationship between the RaCV nonamer sequence in the stem loop with those described for other circoviruses. Conserved nucleotides in the sequence are indicated by asterisks and the accession number for each different circovirus is indicated. 3C: Schematic representation of the circular RaCV genome indicating the position and length of the two major open reading frames: ORF V1 and ORF C1. The position of the stem-loop structure is indicated with the intergenic region. The positions of the RaCV-specific primer pairs RaCV.1N/RaCV.1C and RaCV.2C/RaCV.2N are shown as closed and open arrowheads, respectively. The positions of degenerate avian circovirus primer pairs are also shown (arrows).
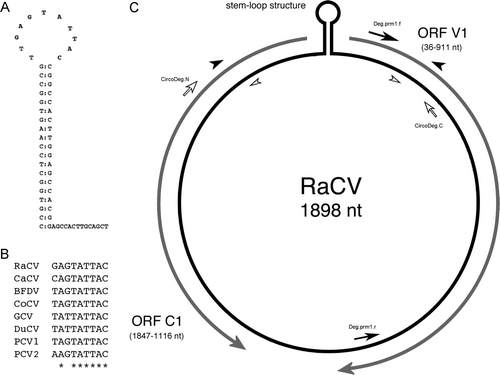
The stem-loop structure was located in a 75-nucleotide intergenic region located between the start codons of the two major open reading frames ORF V1 and ORF C1 (c). Consistent with other circovirus, a series of direct and inverted repeat sequences 5′-CGGCCACTTGGAGCCACGGA-3′were identified at nucleotides 3 to 21 and 1871 to 1889, which form the stem of the set-loop structure (a). In addition, two tandem repeat sequences 5′-GGAGCCAC-3′ were identified at nucleotides 12 to 19 and 20 to 27 and were part of and adjacent to the stem-loop structure. The sequence of the tandem repeats was the same as those published for CaCV (Phenix et al., Citation2001) and consistent with observations in other circoviruses (Todd et al., Citation2001b).
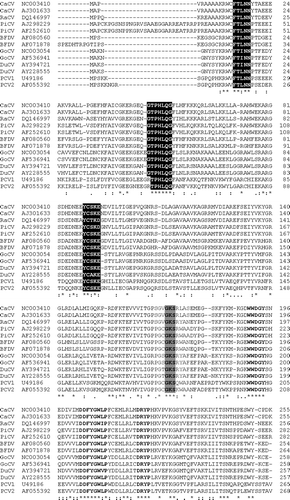
Analysis demonstrated that RaCV displayed an ambisense genomic organization with one large open reading frame located on the viral strand and one major open reading frame on the complementary strand ( and c). The ORF V1 is located on the viral strand, started at nucleotide 36 with an ATG start codon, stopped at nucleotide 911 with a TAA codon and putative poly-A tailed located at 1077 (). The ORF V1 encodes a putative Rep protein of 291 amino acids. BLASTP analysis of the predicted RaCV Rep protein sequence identified significant alignments with the RNA helicase protein family and FtsE protein involved in cell division (Marchler-Bauer & Bryant, Citation2004). Muliple alignment of the putative Rep protein of RaCV with those encoded from other circoviruses, including PCV1, PCV2, BFDV, PiCV, GoCV, DuCV and CaCV, identified the conserved amino acid motifs involved in rolling circle replication (FTLNN, G-HLQG and CSK) and the dNTP-binding domain (G-GSK). In conjunction, three additional motifs—WWDGY, DDFYGWLP and DRYP—reported in the Rep sequence of CaCV (Phenix et al., Citation2001), PiCV and GoCV (Todd et al., Citation2001b) were identified in the RaCV ().
Figure 4. Sequence alignment of the deduced amino acid sequences of the Rep proteins (ORF V1) of RaCV, CaCV PiCV, BFDV, GoCV, DuCV, PCV1 and PCV2. Accession numbers of the sequences are listed alongside the isolates. Alignments were performed and displayed using the ClustalW and Pretty programs. Black shading highlights the three conserved motifs associated with rolling circle replication, and lighter shading highlights the dNTP-binding domain. Three additional conserved motifs mentioned in the text are shown as bold text. Identical amino acids are indicated by asterisks.
As occurs in other avian circoviruses, the complementary strand of RaCV contained one open reading frame, ORF C1, which probably encodes for a putative Cap protein of 243 amino acids. Like in BFDV (Bassami et al., Citation1998), GoCV (Todd et al., Citation2001b) and CaCV (Phenix et al., Citation2001), the start codon for the putative Cap protein is most probably a TCT start codon at nucleotide 1847 or an alternative GTG start codon nearby at nucleotide 1856 (). An ATG sequence near to these is unlikely to be the start codon due to the presence of a stop codon almost immediately downstream at nucleotide position 1116 (). The sequencing of the translation start region was repeated to ensure that the data were correct. Translation studies would be required to clarify which start codon is used by the virus, and this was beyond the scope of our study. A putative poly-A tail signal was identified at nucleotide 1120 (). The N-terminal region of the Cap was highly basic and arginine-rich, similar to that found in the other circoviruses.
Relationship to other circoviruses
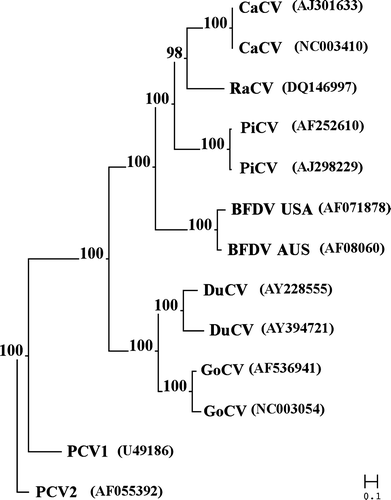
Comparison of the RaCV genome and the nucleotide and amino acid sequences of the two putative open reading frames with the other members of the Circovirus genus demonstrated that the virus was more closely related to the canary and columbid circovirus. Pair-wise alignment of the entire genome of RaCV indicated that the greatest homology was shared with the CaCV (69%) and PiCV (64 to 65%) and the homology decreased with BFDV (56 to 49% ), DuCV (27 to 25% ), GoCV (28 to 25% ), PCV1 (18%) and PCV2 (4%). The highest level of amino acid identity was observed in the putative Rep proteins of the circoviruses. Pair-wise alignment demonstrated that the level of amino acid identity between the RaCV and CaCV (76%) and PiCV (72%) was higher than that observed with RaCV and BFDV (60 to 57% ), DuCV (47%), GoCV (45%), PCV1 (40%) and PCV2 (38%). Comparison of the Cap protein demonstrated that the amino acid identity, overall, was less than observed for the putative Rep protein. The greatest amino acid identity of the RaCV ORF C1 was observed with the pair-wise alignment to the ORF C1 of CaCV (75%). The level of identity between the RaCV ORF C1 and PiCV (41%) BFDV (40%), PCV1 (24%), PCV2 (24%), DuCV (23 to 21% ) and GoCV (22 to 20% ), demonstrated a considerable reduction. The phylogenetic analysis comparing the entire genome and/or the amino acid and nucleotide sequences of the ORFV1 and ORFC1 demonstrated an evolutionary relationship between the RaCV and the other seven members of the Circovirus ().
Discussion
The gross and histopathological lesions presented in the raven we describe in this paper are similar to those that occur in circovirus infections in other avian species (Pass & Perry, Citation1984; Raidal & Riddoch, Citation1997). PCR amplification of a novel circovirus genome from the lesions provides further evidence of the existence of circovirus infection in Australian ravens, which we have tentatively named raven circovirus (RaCV). The data generated indicate that RaCV shares the greatest sequence homology with CaCV, then PiCV, and is more distantly related to BFDV, GoCV, DuCV, PCV1 and PCV2. It may not be surprising that RaCV was most similar to CaCV given that canaries and ravens are both passerine birds, belonging to the suborder Passeri (Sibley & Ahlquist, Citation1985), and are thus potentially more closely related evolutionarily than with columbids, psittacids or anatids, which are the hosts of PiCV, BFDV, GoCV and DuCV.
Of the 115 or so species that are included in the Corvidae family about one-third belong in the genus Corvus. These are among the largest of the passerine birds and are readily identifiable worldwide by mostly being completely black and their distinctive cawing voice. Australia has five endemic Corvus species, and across its range on mainland Australia the Australian raven is the largest and most common. Crows and ravens are relative evolutionary youngsters that appear to be most closely related to the birds of paradise that have a tropical Australasian distribution (Helm-Bychowski & Cracraft, Citation1993; Kriukov & Odati, Citation2000). If avian circoviruses have coevolved with their hosts then we predict that RaCV will be most similar to as yet undiscovered circoviruses in birds of paradise (family Paradisaeidae) or bowerbirds (family Ptilonorhynchidae), which are considered part of the Gondwanan corvid radiation (Ericson et al., Citation2003).
The biological significance of RaCV as a pathogen for the Australian raven and perhaps other Corvus species is debatable. Ravens are a common bird species in Australia and are readily identifiable by members of the public. Unlike most Australian avifauna, many Corvus species are not protected in all jurisdictions across Australia. As in other countries, corvids have a bad reputation among Australian farmers due to their scavenging of lamb carcasses, which can be mistakenly confused with predation. There has been little scientific research into the diseases of Australian corvids but one would consider that plumage defects that result in white depigmentation of feathers would be readily noticed by even untrained wildlife carers and/or the growing number of veterinarians that are taking an interest in the diseases of wildlife. However, not all avian circovirus infections are associated with the development of feather lesions (Todd, Citation2004) and the prevalence of RaCV infection in wild ravens without the occurrence of feather lesions may be much higher. It is also possible that the case we have described represents an aberrant infection by a circovirus adapted to some other avian host. Further epidemiological or pathogenesis studies are warranted to rule out this hypothesis but knowledge of host specificity among other avian circoviruses suggests that this is unlikely (Raidal & Riddoch, Citation1997; Todd, Citation2004).
Feather depigmentation and alterations from normal colouration of feathers are commonly seen in psittacine birds with PBFD but similar changes in pigmentation can sometimes be attributable to malnutrition during the time frame the feathers were being formed, although this would be unlikely in a wild raven. Whereas most Cacatuidae with similar severe feather dystrophy associated with BFDV infection progressively succumb to the disease, the juvenile raven affected in this study made an almost clinical recovery in captivity over 6 to 12 months following its initial presentation. However, it is likely that a spectrum of clinical disease ranging from acute to chronic and clinical remission variations similar to those encountered for a range of psittacine species with PBFD will now be noticed in ravens. Since this first case several other ravens with chronic lesions more severe to those that we have described have been examined by the authors.
References
- Altschul , S.F. , Madden , T.L. , Schaffer , A.A. , Zhang , J. , Zhang , Z. , Miller , W. and Lipman , D.J. 1997 . Gapped BLAST and PSI-BLAST: a new generation of protein database search programs . Nucleic Acids Research , 25 : 3389 – 3402 .
- Bassami , M.R. , Berryman , D. , Wilcox , G.E. and Raidal , S. R. 1998 . Psittacine beak and feather disease virus nucleotide sequence analysis and its relationship to porcine circovirus, plant circoviruses, and chicken anaemia virus . Virology , 249 : 453 – 459 .
- Eisenberg , S.W. , van Asten , A.J. , van Ederen , A.M. and Dorrestein , G.M. 2003 . Detection of circovirus with a polymerase chain reaction in the ostrich (Struthio camelus) on a farm in The Netherlands . Veterinary Microbiology , 95 : 27 – 38 .
- Ericson , P.B.P. , Irestedt , M. and Johansson , U.S. 2003 . Evolution, biogeography and patterns of diversification in passerine birds . Journal of Avian Biology , 34 : 3 – 15 .
- Galtier , N. and Gouy , M. 1995 . Inferring phylogenies from DNA sequences of unequal base compositions . Proceedings of the National Academy of Science USA , 92 : 11317 – 11321 .
- Hamel , A.L. , Lin , L.L. and Nayar , G.P. 1998 . Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs . Journal of Virology , 72 : 5262 – 5267 .
- Hattermann , K. , Schmitt , C. , Soike , D. and Mankertz , A. 2003 . Cloning and sequencing of Duck circovirus (DuCV) . Archives of Virology , 148 : 2471 – 2480 .
- Helm-Bychowski , K. and Cracraft , J. 1993 . Recovering phylogenetic signal from DNA sequences: relationships within the corvine assemblage (class aves) as inferred from complete sequences of the mitochondrial DNA cytochrome-b gene . Molecular Biology and Evolution , 10 : 1196 – 1214 .
- Kriukov , A.P. and Odati , S. 2000 . On the phylogenetic relationship of Corvinae birds (Aves, Corvidae) from data of partial sequencing of cytochrome b gene mitochondrial DNA . Genetika , 36 : 1262 – 1268 .
- Mankertz , A. , Hattermann , K. , Ehlers , B. and Soike , D. 2000 . Cloning and sequencing of columbid circovirus (CoCV), a new circovirus from pigeons . Archives of Virology , 145 : 2469 – 2479 .
- Marchler-Bauer , A. & Bryant , S.H. ( 2004 ). CD-Search: protein domain annotations on the fly . Nucleic Acids Research , 32 ( Web Server issue ), W327 – W331 .
- Meehan , B.M. , Creelan , J.L. , McNulty , M.S. and Todd , D. 1997 . Sequence of porcine circovirus DNA: affinities with plant circoviruses . Journal of General Virology , 78 : 221 – 227 .
- Meehan , B.M. , McNeilly , F. , Todd , D. , Kennedy , S. , Jewhurst , V.A. , Ellis , J.A. , Hassard , L.E. , Clark , E.G. , Haines , D.M. and Allan , G.M. 1998 . Characterization of novel circovirus DNAs associated with wasting syndromes in pigs . Journal of General Virology , 79 : 2171 – 2179 .
- Morin , P.A. , Mesier , J. and Woodruff , D.S. 1994 . DNA extraction, amplification, and direct sequencing from hornbill feathers . Journal of the Science Society of Thailand , 20 : 31 – 41 .
- Mysore , J. , Read , D. and Daft , B. 1995 . “ Circovirus-like particles in finches [Abstract] ” . In Proceedings of the Annual Meeting of the American Association of Veterinary Laboratory Diagnosticians. Histopathology Section , Reno, USA .
- Niagro , F.D. , Forsthoefel , A.N. , Lawther , R.P. , Kamalanathan , L. , Ritchie , B.W. , Latimer , K.S. and Lukert , P.D. 1998 . Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses . Archives of Virology , 143 : 1723 – 1744 .
- Pass , D.A. and Perry , R.A. 1984 . The pathology of psittacine beak and feather disease . Australian Veterinary Journal , 61 : 69 – 74 .
- Phenix , K.V. , Weston , J.H. , Ypelaar , I. , Lavazza , A. , Smyth , J.A. , Todd , D. , Wilcox , G.E. and Raidal , S.R. 2001 . Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus Circovirus of the family Circoviridae . Journal of General Virology , 82 : 2805 – 2809 .
- Raidal , S.R. and Riddoch , P.A. 1997 . A feather disease in Senegal doves (Streptopelia senegalensis) morphology similiar to psittacine beak and feather disease . Avian Pathology , 26 : 829 – 836 .
- Raidal , S.R. , Sabine , M. and Cross , G.M. 1993 . Laboratory diagnosis of psittacine beak and feather disease by haemagglutination and haemagglutination inhibition . Australian Veterinary Journal , 70 : 133 – 137 .
- Riddoch , P.A. , Raidal , S.R. and Cross , G.M. 1996 . Psittacine circovirus antibody detection and an update on the methods for diagnosis of psittacine beak and feather disease . Australian Veterinary Practitioner , 26 : 134
- Sambrook , J. & Russell , D.W. ( Eds. ). ( 2001 ). Molecular Cloning. A Laboratory Manual 3rd edn . New York : Cold Spring Harbor Laboratory Press .
- Sibley , C.G. and Ahlquist , J.E. 1985 . The phylogeny and classification of the Australo-Papuan passerine birds . Emu , 85 : 1 – 14 .
- Steinfeldt , T. , Finsterbusch , T. and Mankertz , A. 2001 . Rep and Rep′ protein of porcine circovirus type 1 bind to the origin of replication in vitro . Virology , 291 : 152 – 160 .
- Taberlet , P. and Bouvet , J. 1991 . A single plucked feather as a source of DNA for bird genetic studies . Auk , 108 : 959 – 960 .
- Tajima , F. and Nei , M. 1984 . Estimation of evolutionary distance between nucleotide sequences . Molecular Biology & Evolution , 1 : 269 – 285 .
- Todd , D. 2004 . Avian circovirus diseases: lessons for the study of PMWS . Veterinary Microbiology , 98 : 169 – 174 .
- Todd , D. , Bendinelli , M. , Biagini , P. , Hino , S. , Mankertz , A. , Mishiro , S. , Niel , C. , Okamoto , H. , Raidal , S. , Ritchie , B.W. and Teo , G.C. 2004 . “ Circoviridae ” . In Virus Taxonomy, VIIIth Report of the International Committee for the Taxonomy of Viruses , Edited by: Fauquet , C.M. , Mayo , M.A. , Maniloff , J. , Desselberger , U. and Ball , L.A. 327 – 334 . London : Elsevier .
- Todd , D. , McNulty , M.S. , Adair , B.M. and Allan , G.M. 2001a . Animal circoviruses . Advances in Virus Research , 57 : 1 – 70 .
- Todd , D. , Weston , J.H. , Soike , D. and Smyth , J.A. 2001b . Genome sequence determinations and analyses of novel circoviruses from goose and pigeon . Virology , 286 : 354 – 362 .
- Twentyman , C.M. , Alley , M.R. , Meers , J. , Cooke , M.M. and Duignan , P.J. 1999 . Circovirus-like infection in a southern black-backed gull (Larus dominicanus) . Avian Pathology , 28 : 513 – 516 .
- Ypelaar , I. , Bassami , M.R. , Wilcox , G.E. and Raidal , S.R. 1999 . A universal polymerase chain reaction for the detection of psittacine beak and feather disease virus . Veterinary Microbiology , 68 : 141 – 148 .