Abstract
A real-time polymerase chain reaction (PCR) assay was developed to specifically amplify infectious laryngotracheitis virus (ILTV) DNA from field samples. The 222-base-pair PCR fragment was amplified using primers located in a conserved region of the infected cell protein 4 gene that was demonstrated in this work to encompass a single nucleotide polymorphism. Subsequent restriction fragment length polymorphism (RFLP) analysis of real-time PCR amplified fragments from a range of ILTV isolates using the restriction endonuclease MspI enabled differentiation between older ILTV isolates that were prevalent in the 1960s prior to the availability of vaccine strains and more recent isolates that predominantly are identical to vaccine strains. The assay, using real-time PCR, RFLP and sequence analysis, was used to characterize two recent field cases of infectious laryngotracheitis from Northern Ireland. One of the field cases was demonstrated to be similar to older “wild-type” isolates, while the other field case was identified to have a concurrent ILTV infection of both “wild-type” and vaccinal type origin. The assay described here using real-time PCR and RFLP provides a rapid, specific method that enables detection and characterization of ILTV directly from field cases.
Détection et caractérisation rapides du virus de la laryngotrachéite infectieuse (ILTV) à partir de cas du terrain par PCR en temps réel et par polymorphisme de taille des fragments de restriction (RFLP)
Un test d'amplification en chaîne par polymérase (PCR) en temps réel a été développé pour amplifier spécifiquement l'ADN du virus de la laryngotrachéite infectieuse (ILTV) à partir d'échantillon du terrain. Le fragment PCR de 222 bp a été amplifié en utilisant des amorces localisées dans une région conservée du gène de la protéine 4 des cellules infectées (ICP-4). Il a été démontré dans cette étude que cette région comprenait un seul polymorphisme nucléotidique (SNP). L'analyse ultérieure du polymorphisme de taille des fragments de restriction (RFLP) amplifiés par PCR en temps réel à partir de souches différentes de ILTV en utilisant l'endonucléase de restriction MspI, a permis la différentiation des souches anciennes de ILTV qui étaient prévalentes dans les années 1960 avant la mise à disposition des souches vaccinales, des souches plus récentes qui de façon prédominante sont identiques aux souches vaccinales. La méthode utilisant la PCR en temps réel, la RFLP et l'analyse de la séquence a été employée pour caractériser deux cas récents de ILT en Irlande du Nord. Il a été démontré qu'un des cas du terrain était de type similaire aux souches “sauvages” anciennes, alors que l'autre cas présentait une infection concomitante par deux virus ILT de type sauvage et d'origine vaccinale. L'essai décrit ici, utilisant la PCR en temps réel et la RFLP fournit une méthode spécifique et rapide qui permet la détection et la caractérisation des ILTV directement à partir des cas du terrain.
Schnellnachweis und Charakterisierung des Virus der infektiösen Laryngotracheitis (ILTV) aus Feldinfektionen mittels Real Time-PCR und Restriktionsfragmentlängenpolymorphismus (RFLP)
Es wurde ein Real Time-Polymerasekettenreaktions (PCR)-Test zur spezifischen Amplifizierung von DNS des Virus der infektiösen Laryngotracheitis (ILTV) aus Feldproben entwickelt. Das 222-bp-PCR-Fragment wurde unter Verwendung von Primern amplifiziert, die sich in einer konservierten Region des ICP 4 (infected cell protein 4) befanden, bei dem in dieser Arbeit gezeigt werden konnte, dass es einen einzelnen Nukleotidpolymorphismus umfasste. Die nachfolgende RFLP-Analyse der amplifizierten Real Time-PCR-Fragmente aus einer Reihe von ILTV-Isolaten mittels des Restriktionsendonuklease MspI ermöglichte die Differenzierung zwischen älteren Isolaten, die in den 1960iger Jahren vor der Verfügbarkeit von Vakzinestämmen vorkamen, und neueren Isolaten, die überwiegend mit den Impfstämmen übereinstimmten. Dieses Testverfahren mit Real Time-PCR, RFLP und Sequenzanalyse wurde zur Charakterisierung von zwei neueren ILT-Feldinfektionsfällen in Nordirland angewendet. Bei einem dieser Feldisolate konnte gezeigt werden, dass es einem der älteren ,,Wildtyp“-Isolate ähnlich war, während bei dem anderen Fall zwei konkurrierende ILTV-Infektionen mit einem “Wildtyp”- und einen Vakzinetypisolat auftraten. Mit dem hier beschriebenen Verfahren der Real Time-PCR und der RFLP steht eine schnelle und spezifische Methode zum Nachweis und zur Charakterisierung von ILTV direkt aus Feldinfektionsfällen zur Verfügung.
Rápida detección y caracterización del Virus de Laringotraqueítis Infecciosa (ILTV) procedente de casos de campo mediante PCR a tiempo real y polimorfismo de la longitud de los fragmentos de restricción (RFLP)
Se puso a punto una técnica de reacción en cadena de la polimerasa (PCR) a tiempo real para amplificar específicamente DNA de virus de laringotraqueítis infecciosa (ILTV) de muestras de campo. Se amplificó el fragmento de PCR de 222pb mediante el uso de cebadores localizados en una región conservada del gen de la proteína 4 de la célula infectada (ICP 4) para la cual se demostró en este estudio que incluye polimorfismos de un solo nucleótido (SNP). El análisis del polimorfismo de la longitud de los fragmentos de restricción (RFLP) de los fragmentos amplificados de un rango de aislamientos de ILTV mediante el uso de la endonucleasa de restricción MspI, permitió la diferenciación entre aislamientos antiguos de ILTV que predominaron en los años 60 antes de la disponibilidad de cepas vacunales, y aislamientos más recientes que son en su mayoría idénticos a las cepas vacunales. El estudio, utilizando PCR a tiempo real, RFLP y análisis de las secuencias, sirvió para caracterizar dos casos recientes de campo de ILT de Irlanda del Norte. Se demostró que uno de los casos de campo era similar a antiguos aislamientos de tipo ‘campo’ mientras que el otro caso de campo se identificó como una infección concurrente de ambos tipos de ILTV, el de origen campo y vacunal. La técnica descrita aquí, basada en el uso de PCR a tiempo real y RFLP, proporciona un método rápido y específico que permite la detección y caracterización de ILTV directamente de casos de campo.
Introduction
Avian infectious laryngotracheitis (ILT) is a viral respiratory disease of chickens with worldwide distribution, caused by infectious laryngotracheitis virus (ILTV), a double-stranded DNA member of the family of Herpesviridae, subfamily Alphaherpesvirinae. Clinical characteristics of the disease include decreased egg production, ill-thrift, oral and ocular discharge and conjunctivitis (Guy & Bagust, Citation2003). The disease is of economic importance as virulent strains can cause high levels of mortality and reduced productivity, while recovery from the disease can result in birds with a latent carrier status (Hughes et al., Citation1987 Citation1991b). Although strains may vary considerably in their virulence, there is recent evidence that vaccine-derived strains have become established in the field (Guy et al., Citation1989; Graham et al., Citation2000). This ability of ILT vaccine strains to recirculate may also be responsible for some outbreaks in susceptible birds, as passage in birds has been reported to result in increasing virulence (Guy et al., Citation1991), while stress in latently infected birds has also been demonstrated to be responsible for the re-excretion of ILTV (Hughes et al., Citation1987; Bagust et al., Citation2000).
The use of DNA amplification methods in recent years has proved effective in investigating the epidemiology of field cases of ILTV. These methods have included the detection of field strains using polymerase chain reaction (PCR) coupled with non-radioactive probes (Abbas et al., Citation1996), while differentiation between field and vaccine strains has been demonstrated through the use of PCR coupled with restriction fragment length polymorphism (RFLP) (Chang et al., Citation1997; Clavijo & Nagy, Citation1997; Graham et al., Citation2000; Han & Kim, Citation2001a). Differentiation between field isolates of high and low virulence has also been achieved through the use of PCR, RFLP and sequence analysis (Han & Kim, Citation2001b), while multiplex PCR has been used for ILT diagnosis (Pang et al., Citation2002). The use of PCR for ILTV has also been described for the evaluation of vaccine purity (Vögtlin et al., Citation1999).
The present paper describes the development and application of real-time PCR, targeting a relatively conserved region of the infected cell protein 4 (ICP 4) gene (Johnson et al., Citation1995) that, when combined with RFLP, provides a rapid, specific test for the detection and characterization of field isolates of ILTV.
Materials and Methods
Viruses
Field samples were derived from two cases in Northern Ireland (NI) during 2004 where ILTV was subsequently isolated. The first NI field case submitted (04 14677) was obtained from a commercial, free-range, layer flock that was showing low but increasing mortality rates and an approximate 30% drop in egg production with some birds exhibiting symptoms of upper respiratory tract infection, which included conjunctivitis, sneezing and nasal and ocular discharge. The second field case (04 16713) was submitted from a commercial broiler breeder flock that demonstrated a 25% drop in egg production over 5 days. A high percentage of the birds (20%) subsequently showed symptoms of respiratory distress and depression, and within 72 h a mortality of 116 birds from the flock of 5000 was noted. The latter case was originally submitted as a precautionary investigation for avian influenza virus (AIV). All samples were prepared according to previously described methods (Office International des Epizooties [OIE], Citation2004) and the tissues used with their passage history are itemized in . Other previously characterized ILTV isolates used in this study are also presented in .
Table 1. Details of virus strains and comparison of the MspI RFLP patterns
Experimental infection
An attempt to reproduce disease using the recent NI isolate 04 16713 was carried out by experimental infection of specific pathogen free chickens (Valo, Germany). Each of eight 9-week old birds was inoculated intranasally using 0.2 ml volumes of this isolate after one passage in chicken embryo liver (CEL) prepared using previously described methods (OIE, Citation2004). The birds were checked each day for clinical signs of disease until they were humanely euthanized 6 weeks post inoculation. Before euthanasia, blood samples were removed from the wing vein of each bird and serum samples were evaluated to detect ILTV antibodies using the indirect immunofluorescent antibody test using dilutions of the experimental sera on ILTV infected cell culture as previously described (OIE, Citation2004).
DNA isolation
Total DNA was isolated from 200 µl samples of tissue homogenate, egg-passaged chorioallantoic membrane, cell culture or swab homogenate as appropriate using the QIAamp DNA mini kit (Qiagen) according to manufacturer's instructions. Total DNA was also isolated from uninfected CEL cells to provide a negative control for the PCR assay. To evaluate the specificity of the ILTV primers, DNA was also isolated from strains of other avian DNA viruses. These include egg drop syndrome 127, chicken anaemia virus CUX-1, turkey haemorrhagic enteritis virus, duck hepatitis virus and chick embryo lethal orphan virus PHELPS strain. All of the additional avian viruses were propagated in cell culture using the standard procedures for each of these pathogens as previously described (Swayne et al., Citation1998). All DNA was eluted in 200 µl elution buffer and stored at −20°C until required.
Amplification and RFLP analysis of the thymidine kinase gene
To determine the molecular basis of virulence of the ILTV strains, DNA amplification of a 1.296-kbp fragment located in the ILTV thymidine kinase (TK) gene was carried out using primers and conditions as previously described (Han & Kim, Citation2001a Citationb; OIE, Citation2004). All isolates and field strains presented in were amplified using HotStarTaq DNA polymerase (Qiagen) with amplification carried out in a GeneAmp PCR system 9700 Thermal Cycler (Applied Biosystems). Aliquots of the PCR products were resolved by electrophoresis using a 1.5% horizontal agarose gel using established procedures. Subsequent to electrophoretic analysis, PCR products were analysed for RFLP using HaeIII, Sau96I and NciI endonucleases as described previously (Han & Kim, Citation2001a Citationb). Nucleic acid sequencing of a 649-bp fragment within the 1.296-kbp TK fragment was carried out using primers described by Han & Kim (Citation2001a Citationb) and the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems). Following cycle sequencing, the reactions were purified using the Montage SEQ96 Sequencing reaction Cleanup Kit (Millipore). Sequence reactions were run on an ABI 30100 Genetic Analyser (Applied Biosystems) and analysed using the Vector NTI Advance 9 software (Informax, Invitrogen).
Amplification and RFLP analysis of the ICP4 gene
A 4.9-kbp fragment of the ICP4 gene was amplified from all field cases and isolates presented in using the Elongase Amplification System (Invitrogen) and primers as previously described (Chang et al., Citation1997) in a GeneAmp PCR System 9700 thermal cycler. The PCR products were purified using a QIAQuick PCR purification system (Qiagen), digested using MspI as previously described (Graham et al., Citation2000) and electrophoresed on a 1.5% agarose gel.
Identification of a “wild-type” 724-bp fragment within the ICP4 gene MspI digest of the 4.9-kbp ICP4 PCR product from the ILTV PRC isolate (see ) was electrophoresed on a 3% NuSieve GTG low melting point gel, and a 724-bp band that is unique to the “wild-type” strain (Graham et al., Citation2000) was excised and purified using the QIAQuick gel extraction kit system (Qiagen). The purified DNA fragment was labelled using the ECL direct nucleic acid labelling and detection system (Amersham). Individual restriction endonuclease digests of the ILTV PRC strain, using PstI, HindIII and BamHI, were carried out. Following electrophoresis on a 1.5% agarose gel, the digested DNA fragments were then Southern blotted onto Hybond ECL membrane (Amersham) using established methods and ECL detection, using the “wild-type” 724 bp ECL-labelled probe was carried out according to the manufacturer's instructions.
Real-time PCR amplification of the ILTV ICP4 gene
Using the information derived from the Southern blot analysis together with previously published sequence data (accession number L32139), PCR primers within the 724-bp “wild-type” region of the 4.9-kbp ICP4 PCR fragment were designed using Vector NTI Advance 9 software (Informax). The primers were designed to flank the MspI restriction site in this region, which was missing in the “wild-type” strains. The primer sequences and location were as follows: sense primer, 3239-CTCTTCCTCCTCTTCCTCAT-3258 and antisense primer, 3460-GTTACTGACTGAACCGACCC-3441—with sequence numbering according to a previously published ILTV sequence (Johnson et al., Citation1995; accession number L32139). Amplification of this 222-bp fragment followed by melt curve analysis was carried out in a DNA Engine Opticon 2 (MJ Research) Real Time Thermal Cycler using the Quantitect Sybr Green PCR kit system (Qiagen) under the following conditions: denaturation in 15 min at 95°C for one cycle; amplification in 40 cycles of 94°C for 10 sec, 58°C for 15 sec, 72°C for 10 sec, 78°C for 1 sec (data acquisition step) and final product extension in one cycle at 72°C for 7 min. Melt curve analysis was carried out by increasing PCR product temperature from 55 to 95°C in 1°C increments. Sensitivity of the real-time ILTV assay was quantified as follows. A 4.9-kbp ICP4 PCR fragment of the ILTV UK vaccine was generated using the methods described earlier. This fragment was selected as the standard for absolute quantification as it encompassed the target 222-bp fragment. The fragment was then purified using QIAquick purification system (Qiagen) and quantified spectrophotometrically, enabling the copy number of standard DNA molecules to be calculated. The dilutions used for this standard ranged from 14×106 to 14×101 molecules/µl.
Restriction endonuclease and sequence analysis of 222-bp ICP4 fragments
Real-time PCR amplicons were cleaned-up post-amplification using the QIAquick PCR Purification system (Qiagen), were digested with MspI and electrophoresed on Novex precast 10% TBE polyacrylamide gels (Invitrogen) using established methods. PCR products were also sequenced and analysed as described previously.
Results
Amplification of the TK gene
In order to examine the molecular determinants of virulence of field strains 04 14677 and 04 16713, a 1.296-kbp fragment of the TK gene was amplified from the DNA. Digestion of the PCR amplicons with HaeIII, Sau96 and NciI, gave a RFLP pattern indicative of low virulence, according to RFLP patterns previously described (Han & Kim, Citation2001b; OIE, Citation2004). Sequence analysis of these field isolates in the 649-bp region of the TK gene showed the presence of a threonine at amino acid position number 252, which again was indicative of isolates of low virulence (Han & Kim, Citation2001a Citationb). Identification of NI 04 16713 as a low virulent isolate by RFLP and sequence analysis was confirmed by the data from the experimental infection where infected birds did not show any signs of clinical disease, although detection of ILTV antibodies in all birds indicated evidence of infection.
“Wild-type” 724-bp fragment mapping
Southern blots of restriction endonuclease digested vaccine and “wild-type” strains of ILTV ICP4 fragments, when probed with the 724-bp ECL probe, mapped the location of the “wild-type” specific 724-bp fragment to be between nucleotide position 2026 and 3067 within the ICP4 gene (numbering according to accession number L32139). The MspI site missing in the “wild-type” strains was identified to be at nucleotide position 2335 and PCR primers were designed to encompass this site.
Real-time PCR and restriction enzyme analysis of the 222-bp ICP4 fragment from ILTV strains
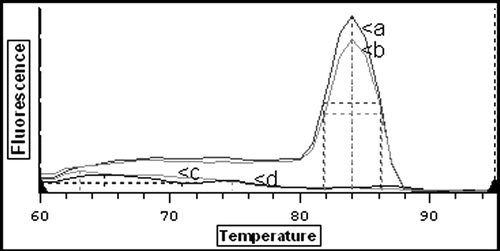
A 222-bp PCR product was amplified using real-time PCR from all isolates and field strains of ILTV. Characterization of the real time products was achievable as melt curve analysis of the amplified product indicated that the melting temperature of the product in all cases was 84°C (). Specificity of the assay was confirmed from the absence of products from reactions with no template controls, uninfected CEL cells or from the previously described unrelated avian DNA viruses. Although no Marek's disease virus DNA was available for direct evaluation of specificity of ILTV primers, the sequence comparison of the primers against the Marek's disease virus type 1 genome (accession number AY510475) did not show homology for any non-specific annealing to occur.
Figure 1. Detection of ILTV DNA using real-time PCR. Melt curve analysis of amplified PCR products. Curve a, UK vaccine strain; curve b, NI 04-16713 field sample from cloacal swab; curve c, negative faeces sample; curve d, uninfected CEL cells.
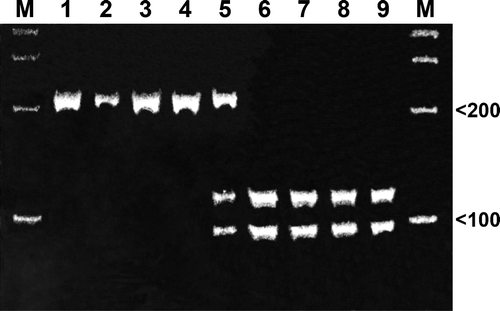
Results of RFLP analysis using MspI are detailed in . The NI field isolate 04 16713 demonstrated the RFLP A pattern, identical to that of older “wild-type” isolates such as A96 and PRC from the 1960s, whereas with the exception of one 1991 field isolate from the Republic of Ireland, all recent isolates showed a RFLP pattern B similar to the vaccinal strains (). The RFLP pattern of the recent NI field isolate 04 14677, however, indicated the presence of two populations of ILTV (patterns A and B) containing a “wild-type” and vaccine-type strain not only in viral DNA derived from all egg-passaged tissue and faeces material but also in the original faeces material. The sensitivity of the assay was quantified and, against the standard previously described, field samples containing dilutions down to 14×101 molecules/µl could be reproducibly detected.
Figure 2. MspI digestion of real-time PCR products. Electrophoretic analysis of restriction endonuclease digested PCR fragments on a 10% polyacrylamide gel. Lane 1, A96 England; lane 2, 1106B ROI; lane 3, Scottish PRC; lane 4, 04-16713 NI field strain; lane 5, NI 04-14677 field strain; lane 6, UK vaccine strain; lane 7, 1031A Midmoor England; lane 8, 1031D Malpas England; lane 9, 97-4842 NI. M, 100-bp DNA ladder.
Sequence analysis of the 222 bp ILTV ICP4 fragment
Details of the nucleic acid and amino acid sequence analysis of the 222-bp fragment of the ICP4 gene are shown in Figures and 4. These results confirm those achieved by real-time PCR and RFLP analysis. The loss of the MspI restriction endonuclease site in the “wild-type” strain is due to a single nucleotide polymorphism at nucleotide 81 (), which also results in a corresponding change at amino acid 27 indicated in from an arginine (R) in the vaccine strains to a tryptophan (W) in the “wild-type” strains.
Figure 3. Nucleic acid sequence of ILTV isolates. The isolates presented in were amplified to give a 222-bp product using PCR primers in the ICP4 gene. The region in bold identifies the MspI restriction endonuclease site. *Y = C/T. **Sequence numbering is according to a previously published ILTV sequence (Johnson et al., Citation1995; accession number L32139).
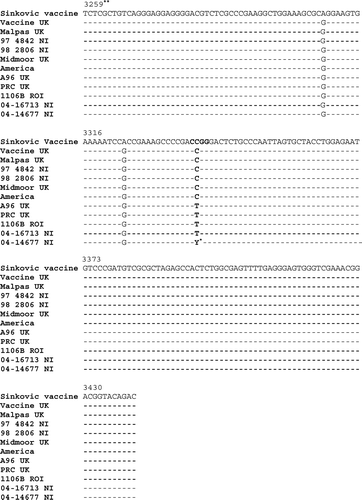
Figure 4. Amino acid translation of the ILTV sequences shown in . The amino acids in bold type are located at the MspI restriction endonuclease site.

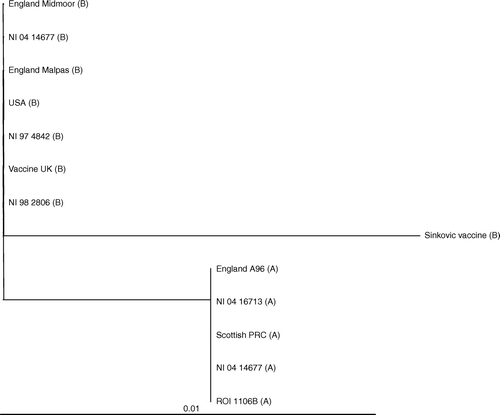
The relationship between strains is demonstrated in the phylogenetic tree shown in where, based upon sequence, the ILT isolates can be segregated into three distinct groups. The cell culture derived Australian Sinkovic vaccine strain is on its own in a group, while the more recent isolates from England, America and the 1997 and 1998 NI isolates are grouped with the UK Fort Dodge vaccine strain of chick embryo origin. The third group consists of “wild-type”, mostly older, isolates that were circulating prior to the introduction of vaccination in the UK and Republic of Ireland. Also included in this group is the recent NI isolate (04 16713) that caused higher levels of mortality. The NI isolate 04 14677 is present in both “wild-type’” and recent vaccine-type branches of the tree, as these represent the two subtypes within this isolate.
Figure 5. Phylogenetic tree of field cases and isolates based on the 222-bp nucleic acid sequence of the PCR product amplified from the ILTV ICP4 gene. The Tree calculation is based on a sequence distance method and utilizes the neighbour-joining (NJ) algorithm of Saitou & Nei (Citation1987). The scale bar indicates the number of nucleotide substitutions per site. The letters following the isolate description identifies the MspI RFLP pattern.
Discussion
The TK gene is associated with ILTV virulence, and previous studies have demonstrated that differentiation between virulent and vaccine-like field strains can be achieved through RFLP and sequence analysis of this region (Han & Kim, Citation2001a Citationb). Determination of virulence in the two recent NI field isolates was considered necessary since up to 20% of birds from the infected flocks from which one of isolates was obtained showed respiratory distress and increasing levels of mortality over a 72-h period. However, RFLP and sequence analysis of the TK gene demonstrated that the two recent NI isolates were not highly virulent according to the criteria previously described (Han & Kim, Citation2001b; OIE, Citation2004). The identification of the isolates from NI cases as low virulent seems appropriate, even though the clinical symptoms and mortality levels associated with the cases (such as the one involving 04 16713) increasing, as mortality rates in highly virulent cases have been reported to reach 80% (Han & Kim, Citation2001b). The low virulence of these isolates was subsequently confirmed through experimental infection, where no clinical signs of the disease were observed. The apparent inconsistency between clinical symptoms and mortality levels in the field and no mortality with experimental infection of chickens could, however, be explained through the interaction of environmental or nutritional factors (e.g. stock density, air quality) in conjunction with infection in the field that can contribute to increased mortality levels. However, although a low virulent phenotype was identified through analysis of the TK gene, no differences were identified in the TK region between field isolates, older “wild-type” ILTV isolates and vaccinal strains, necessitating the development of a PCR assay based on another region of the viral genome. Assays targeting the ICP4 gene, which enables differentiation between vaccinal and non-vaccinal strains, have been previously described (Chang et al., Citation1997; Graham et al., Citation2000; OIE, Citation2004), and therefore this gene was selected as a region likely to be suitable for development for a real-time PCR assay.
The real-time PCR assay developed here amplifies a 222-bp fragment of the ICP4 gene. The sensitivity of the primers designed was able to be confirmed, as amplicons were observed from all of the ILTV isolates, including those of the field cases with quantification of the assay, indicating that down to 14×101 molecules/µl could be reproducibly detected. Sensitivity of the real-time assay was further demonstrated through amplification of ILTV DNA from both original and egg-passaged material from field cases. This ability of the real-time test to amplify ILTV DNA from original material including cloacal swabs and faeces is worthy of note as identification and characterization of the virus could be achieved ante mortem. It is also notable that detection was carried out using original faecal material, as difficulties in amplification of viral nucleic acid have previously been described from faeces due to the potential presence of inhibitors (Creelan et al., Citation2002). The assay described here was confirmed to be specific since no products could be amplified from the other viral and cellular sources of DNA using these ILTV primers.
The characterization of the isolate 04 16713 obtained from a field case and the identification of this as an ILTV similar to “wild-type” is significant as this isolate represents the first reported case of “wild type” ILTV in NI. Since ILT vaccination of poultry has been in use in mainland UK from the 1970s, it has been reported that many vaccinated birds have become latently infected, subsequently excreting the virus periodically, and therefore may be responsible for the widespread presence of vaccine-type strains in the field (Hughes et al., Citation1991a). This spread of vaccine-type ILTV has also been demonstrated from two outbreaks in 1998 in NI (Graham et al., Citation2000), whereas isolates from Southern Ireland, where vaccination is prohibited, have to date been of “wild-type” in nature (Graham et al., Citation2000). The results from the other recent NI field case (04 14677) indicated a co-infection with both vaccine and “wild-type” ILTV strains. Such an occurrence does not appear to have been previously reported. The finding that this RFLP pattern was able to be demonstrated in both the original faeces sample and then subsequent faecal and tissue egg-passaged material strongly indicates that this is not a case of reversion of a SNP, but that both virus strains had been present in the birds. The possibility of the occurrence of cross-contamination from other isolates within the laboratory was able to be eliminated by repeatability of the results of the DNA isolation procedure, PCR and RFLP analysis from the original tissues, in zones that were separate from all other ILTV strains. The NI field isolate 04 16713 of “wild-type” origin was obtained from a location within the same county and a relatively short distance from this farm, and with both cases being diagnosed approximately 5 weeks apart would tend to indicate that the “wild-type” virus was likely to be circulating within the region at that time. The additional fact that in the first case (04 14677) the birds were free range also raises the possibility that wild birds might have facilitated spread of the disease. Although ILTV does not infect wild birds, it has been reported that they may act as mechanical carriers of the disease (Venugopal et al., Citation2002).
In this paper, we report an improved diagnostic method for the specific detection of ILTV DNA using a real-time PCR assay that, when coupled with RFLP, has the ability to distinguish between “wild-type” and vaccinal type strains. The two recent field cases of ILTV studied were able to be identified of “wild-type” origin and a co-infection of “wild type” and vaccine origin, respectively. This real-time PCR assay, including amplification and melt curve analysis, can be carried out in less than 2 h. This provides a very rapid test when compared with virus isolation procedures or even standard block-based PCR methods followed by gel electrophoresis. It has potential to be a valuable tool for the rapid diagnosis of ILTV from infected flocks, with important applications with respect to differential diagnosis as many clinical symptoms of the disease bear close resemblance to other notifiable avian viral pathogens. This is particularly noteworthy as the second NI field case (04 16713) was originally submitted to the laboratory as a precautionary investigation for AIV.
Acknowledgments
ILTV strains used in this work were kindly provided by Mr W. Allen (Veterinary Laboratories Agency, Weybridge) (PRC-UK), Dr W. Baxendale (Intervet UK) (A96), Dr R. Jones (University of Liverpool) (Midmoor, Malpas) and Mrs H. de Geus (Veterinary Research Laboratory, Dublin) (PV114/91). The authors acknowledge virus isolation, experimental infection and serology carried out by DSIB staff, Veterinary Sciences Division, Stormont. The authors also acknowledge Dr Brian Meehan for his valuable comments on the manuscript.
References
- Abbas , F. , Andreasen , J.R. Jr and Jackwood , M.W. 1996 . Development of a polymerase chain reaction and a non-radioactive DNA probe for Infectious laryngotracheitis virus . Avian Diseases , 40 : 56 – 62 .
- Bagust , T.J. , Jones , R.C. and Guy , J.S. 2000 . Avian infectious laryngotracheitis . Revue Scientifique et Technique Office International des Epizooties , 19 : 483 – 492 .
- Chang , P.-C. , Lee , Y.-L. , Shien , J.-H. and Shieh , H.K. 1997 . Rapid differentiation of vaccine strains and field isolates of infectious laryngotracheitis virus by restriction fragment length polymorphism of PCR products . Journal of Virological Methods , 66 : 179 – 186 .
- Clavijo , A. and Nagy , E. 1997 . Differentiation of infectious laryngotracheitis virus strains by polymerase chain reaction . Avian Diseases , 41 : 241 – 246 .
- Creelan , J.L. , Graham , D.A. and McCullough , S.J. 2002 . Detection and differentiation of pathogenicity of avian paramyxovirus serotype 1 from field cases using one-step reverse transcriptase-polymerase chain reaction . Avian Pathology , 31 : 493 – 499 .
- Graham , D.A. , McLaren , I.E. , Calvert , V.M. , Torrens , D. and Meehan , B.M. 2000 . RFLP analysis of recent Northern Ireland isolates of infectious laryngotracheitis virus: comparison with vaccine virus and field isolates from England, Scotland and the Republic of Ireland . Avian Pathology , 29 : 57 – 62 .
- Guy , J.S. and Bagust , T.J. 2003 . “ Laryngotracheitis ” . In Diseases of Poultry , 11th edn , Edited by: Calnek , B.W. , Barnes , H.J. , Beard , C.W. , McDougald , L.R. and Saif , Y.M. 121 – 134 . Ames : Iowa State University Press .
- Guy , J.S. , Barnes , H.J. , Munger , L.L. and Rose , L. 1989 . Restriction endonuclease analysis of infectious laryngotracheitis viruses; comparison of modified live-vaccine viruses and North Carolina field isolates . Avian Diseases , 33 : 316 – 323 .
- Guy , J.S. , Barnes , H.J. and Smith , L. 1991 . Increased virulence of modified-live infectious laryngotracheitis vaccine virus following bird-to-bird passage . Avian Diseases , 35 : 348 – 355 .
- Han , M.G. and Kim , S.J. 2001a . Comparison of virulence and restriction endonuclease cleavage patterns of infectious laryngotracheitis viruses isolated in Korea . Avian Pathology , 30 : 337 – 344 .
- Han , M.G. and Kim , S.J. 2001b . Analysis of Korean strains of infectious laryngotracheitis virus by nucleotide sequences and restriction fragment length polymorphism . Veterinary Microbiology , 83 : 321 – 331 .
- Hughes , C.S. , Jones , R.C. , Gaskell , R.M. , Jordan , F.T.W. and Bradbury , J.M. 1987 . Demonstration in live chickens of the carrier state in infectious laryngotracheitis . Research in Veterinary Science , 42 : 407 – 410 .
- Hughes , C.S. , Gaskell , R.M. , Bradbury , J.M. , Jordan , F.T.W and Jones , R.C. 1991a . Survey of field outbreaks of infectious laryngotracheitis in England and Wales . Veterinary Record , 129 : 258 – 260 .
- Hughes , C.S. , Williams , R.A. , Gaskell , R.M. , Jordan , F.T.W. , Bradbury , J.M. , Bennett , M. and Jones , R.C. 1991b . Latency and reactivation of infectious laryngotracheitis vaccine virus . Archives of Virology , 121 : 213 – 218 .
- Johnson , M.A. , Tyack , S.G. , Prideaux , C. , Kongsuwan , K. and Sheppard , M. 1995 . Nucleotide sequence of infectious laryngotracheitis virus (gallid herpesvirus 1) ICP4 gene . Virus Research , 35 : 193 – 204 .
- Office International des Epizooties . 2004 . “ Avian infectious laryngotracheitis ” . In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Mammals, Birds and Birds) , 5th edn , 889 – 895 . Paris : World Organisation for Animal Health .
- Pang , Y. , Wang , H. , Girshick , T. , Xie , Z. and Khan , M.I. 2002 . Development and application of a multiplex polymerase chain reaction for avian respiratory agents . Avian Diseases , 46 : 691 – 699 .
- Saitou , N. and Nei , M. 1987 . The neighbor-joining method: a new method for reconstructing Guide trees . Molecular Biology Evolution , 4 : 406 – 425 .
- Swayne , D.E. , Glisson , J.R. , Jackwood , M.W. , Pearson , J.E. and Reed , W.M. 1998 . A Laboratory Manual for the Isolation and Identification of Avian Pathogens , 4th edn , Pennsylvania, , USA : American Association of Avian Pathologists .
- Venugopal , K. , Jones , R.C. and Gough , R. 2002 . “ Herpesviridae ” . In Poultry Diseases , 5th edn , Edited by: Jordan , F. , Pattison , M. , Alexander , D. and Faragher , T. 221 – 240 . London : W.B. Saunders .
- Vögtlin , A. , Bruckner , L. and Ottiger , H.-P. 1999 . Use of polymerase chain reaction (PCR) for the detection of vaccine contamination by infectious laryngotracheitis . Vaccine , 17 : 2501 – 2506 .