Abstract
Newcastle disease is an important viral infectious disease caused by Newcastle disease virus (NDV), which leads to severe economic losses in the poultry industry worldwide. The molecular mechanisms involved in the pathogenesis of NDV and the host-directed antiviral responses remain poorly understood. In this study, we screened and identified the differentially expressed transcripts from chicken spleen 36 h post NDV infection using suppression subtractive hybridization (SSH). From the SSH library, we obtained 140 significant differentially expressed sequence tags (ESTs), which could be divided into three categories: high homology genes (58), high homology ESTs (62) and novel ESTs (20). The 58 high homology genes could be grouped into nine clusters based on the best known function of their protein products, which involved signalling transduction (HSPC166, PDE7B, GRIA4, GARNL1), transcriptional regulation (ANP32A, LOC423724, SATB1, QKI, ETV6), cellular molecular dynamics (MYLK, MYO7A, DCTN6), cytoskeleton (LAMA4, LAMC1, COL4A1), stress response (DNAJC15, CIRBP), immune response (TIA1, TOX, CMIP), metabolism (RPS15A, RPL32, GLUT8, CYPR21, DPYD, LOC417295), oxidation–reduction (TXN, MSRB3, GCLC), and others. In addition, we found that the 20 novel ESTs provide a clue for the discovery of some new genes associated with infection. In summary, our findings demonstrate previously unrecognized changes in gene transcription that are associated with NDV infection in vivo, and many differentially expressed genes identified in the study clearly merit further investigation. Our data provide new insights into better understanding the molecular mechanism of host–NDV interaction.
Introduction
Newcastle disease is a highly contagious respiratory, neurological or enteric disease caused by Newcastle disease virus (NDV), which leads to severe economic losses in the poultry industry worldwide (Alexander, Citation2000). NDV initiates infection with attachment of the viral haemagglutinin-neuraminidase protein to sialic acid-containing receptors, followed by fusion of viral and cell membranes directed by the fusion F protein, which requires a thiol/disulfide exchange to produce three thiols in the F protein (Jain et al., Citation2007, Citation2008). It has been demonstrated that the V protein of NDV is associated with viral pathogenesis and functions as an IFN-alpha/beta antagonist (Huang et al., Citation2003). In addition, NDV can induce apoptosis in infected host cells including chicken embryo fibroblast cells (CEFs) (Ravindra et al., Citation2008), peripheral blood lymphocytes (Lam & Vasconcelos, Citation1994), heterophils (Lam et al., Citation1996) and macrophages (Lam, Citation1996). A recent study showed that NDV-induced cytopathic effect in infected cells is also caused by apoptosis (Ravindra et al., Citation2009). However, very little is known about the molecular basis of disease pathogenesis and the host's response to infection. Recently, the differentially expressed transcripts of NDV-infected CEFs were analysed by cDNA microarrays, which provided an overview of the gene expression profiles of infected cells and reveal a dynamic host response. This profile includes the regulation of genes with roles in a vast array of cellular functions as well as those that had not been described previously (Munir et al., Citation2005). However, the cell model cannot fully reflect the complexities of the host–virus relationship. Therefore, the spleen was chosen for the subtractive object to screen and identify the differentially expressed genes initiated by in vivo infection with NDV. The object of the present study was to provide useful information for future investigations on the molecular events during avian–NDV interaction.
Materials and Methods
Host systems and virus infection
Six 15-day-old specific pathogen-free White Leghorn chickens were obtained from the China Animal Health and Epidemiology Center. The chickens were divided into two groups: one group was infected intranasally with 100 median embryo lethal dose of NDV/Chicken/China/SC-03/2006NDV/Chicken/China/SC-03/2006a velogenic strain (of VII genotype) . The other group was treated with the same dose of isotonic sodium chloride and served as uninfected controls. Spleen tissue was collected from three chickens per group at 36 h post infection, and was preserved in Sample Protector (Takara, China) at –80°C until used for RNA extraction. The livers and brains of three infected birds were used to detect NDV by specific reverse transcriptase (RT)-polymerase chain reaction (PCR), according to our previous report (Yue et al., Citation2007), in order to investigate whether or not the infections were successful.
RNA extraction and cDNA synthesis
Total RNA of three chickens per group was extracted using TRIZOL reagent (Takara) according to the manufacturer's instructions. The RNA pool was prepared by mixing together equal quantities of three RNA samples per group. Single-stranded and double-stranded cDNA were synthesized from the infected and control RNA pools with the SMART PCR cDNA Synthesis Kit (Clontech, USA) following the manufacturer's protocol. Briefly, 1 µg total RNA was annealed with an oligo dT-T7 RT primer and template switch oligonucleotide at 70°C for 2 min in a total volume of 5 µl. The reaction was followed by the addition of 200 units Prime Script RNase H– reverse transcriptase (Takara) and incubated at 42°C for 1 h. Then representative double-stranded cDNAs were generated by exponential PCR amplification. The optimal number of cycles for each sample was determined by analysing the PCR products of a series of PCR amplifications using different numbers of cycles. Finally, 2 µl from the 50 µl single-stranded cDNA stocks were amplified in 100 µl reactions using the SMART PCR primer and the predetermined exponential number of cycles. Amplified SMART cDNAs were purified using the TIANquick Maxi Purification kit (TIANGEN, China)
Construction of normalized subtracted cDNA libraries by suppression subtractive hybridization
The subtracted cDNA libraries were constructed by suppression subtractive hybridization (SSH) using the PCR-Select cDNA Subtraction Kit (Clontech). As described by the manufacturer's instructions, cDNA from the infected group was used as the tester, and cDNA from the control group as the driver. Both tester and driver cDNAs were digested with RsaI to produce shorter blunt-ended fragments. After digestion with RsaI, the tester cDNA was divided into two portions, each of which was ligated with a different adapter at 16°C for 8 h. After ligation, each tester cDNA was separately hybridized at 68°C for 8 h with an excess of driver cDNA after denaturation at 98°C for 90 sec. Then the two hybridized samples were mixed together and hybridized at 68°C for 12 h with excess of denatured driver cDNA. The resulting mixture was added with 200 µl dilution buffer and amplified by two rounds of suppression PCR. Before the primary PCR, the reaction mixture was incubated at 75°C for 5 min to extend the adaptors. Primary PCR was performed at 94°C for 30 sec, 66°C for 30 sec, and 72°C for 90 sec for 30 cycles in a reaction volume of 25 µl. The PCR product was then diluted 10-fold, and 1 µl diluted product was used as template in the next subsequently nested PCR. The second nested PCR was performed for 15 cycles of 94°C for 30 sec, 68°C for 30 sec, and 72°C for 90 sec. The PCR primers were all in the kit. The subtracted secondary PCR fragments were cloned into the pMD18-T vector (Takara) and transformed into Escherichia coli Top10 to establish subtracted cDNA libraries. Each step of SSH was tested following the manufacturer's instructions.
Screening and authentication of the SSH libraries using the reverse northern dot-blotting method
A total of 1600 individual recombinant clones was chosen randomly and cultured on 100 µl LB broth containing ampicillin in 96-well microtitre plates at 37°C for 6 to 8 h. Cloned inserts in the pMD18-T vector were then amplified by PCR using the nested primers Nested 1 and Nested 2R, which were part of Adaptor 1 and Adaptor 2R, followed by analysis on 1.2% w/v agarose gel. Amplified specific inserts from each clone were denatured by mixing an equal volume of freshly prepared NaOH (0.6 N), and 1 µl volumes were transferred onto a nylon membrane (Millipore) and then fixed by baking in an oven at 80°C for 2 h. RsaI-digested tester and driver cDNA, as mentioned above as the specific probes, were hybridized with two identical blots respectively. The labelling and staining procedures were carried out using the DIG High Prime Labeling and Detection Starter Kit I (Roche Diagnostics, Germany) according to the manufacturer's instructions. The clones that were hybridized to the tester probe alone or had five-fold greater signal intensity over the control were considered to be positive clones that included tester-specific sequences.
DNA sequencing and bioinformatics
The positive clones selected by reverse northern dot-blotting were sequenced by Invitrogen Biotechnology China Corporation. The sequences obtained were edited using Vector NTI software (Invitrogen) to remove the vector sequence contaminations. The sequences of the adaptors were removed manually. Sequence analysis and homology comparisons were carried out using the BLASTn program of NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Classification of sequences homologous to known genes was performed based on the best known function of their protein products in conjunction with the gene function classification program online (http://panther.appliedbiosystems.com). The putative open reading frames (ORFs) of novel expressed sequence tags (ESTs) were predicted using the ORF finder program of the NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
Results
Detection of NDV in infected birds by RT-PCR
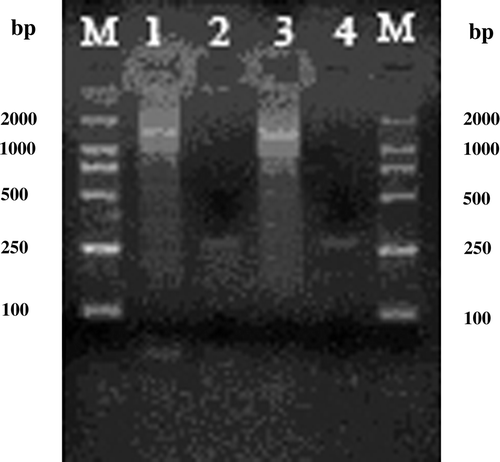
Positive results in the specific RT-PCR showed that the three birds were infected successfully with the NDV SC-03 strain ().
Quality detection of the subtracted cDNA libraries: ligation efficiency analysis of the adaptor
Ligation efficiency of the adaptor is an important factor that determines the success or failure of the SSH. According to the testing method in the manufacturer's instructions, the results of agarose gel electrophoresis showed that the PCR product using one adaptor sequence primer and a gene-specific primer (G3PDH 3′ primer) had two-fold greater signal intensity over the PCR product amplified using two gene-specific primers (G3PDH 3′ and 5′ primers), which indicated that more than 60% of the cDNA had been ligated with the adaptor and there was a high ligation efficiency ().
Analysis of hybridization efficiency
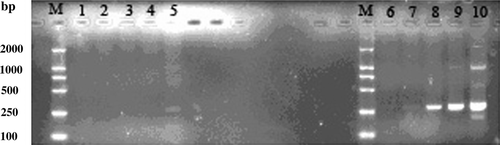
The hybridization efficiency was confirmed by detecting the changes of relative abundance of G3PDH in subtracted and unsubtracted secondary PCR products as described by the manufacturer's instructions. As shown in , using the same cycle parameters for PCR, the product of the unsubtracted cDNA is visible after 21 cycles but the subtracted cDNA product is not visible until after 30 cycles. This result illustrated that the hybridization efficiency was high.
Figure 2. Analysis of hybridization efficiency. PCR was performed on subtracted (lanes 1 to 5) or unsubtracted (lanes 6 to 10) secondary PCR product with the G3PDH 5′ and 3′ primers. Lane M, DNA maker II; lanes 1 and 6, 18 cycles; lanes 2 and 7, 21 cycles; lanes 3 and 8, 24 cycles; lanes 4 and 9, 27 cycles; lanes 5 and 10, 30 cycles.
Screening of the subtracted cDNA libraries
A subtracted cDNA library with 1600 individual recombinant clones was established. The positive recombinant clones of the cDNA library was detected using bacterial PCR; results showed that the positive rate was 95% and the size of inserted cDNA fragments varied from 180 bp to 1 kb. These clones were further screened by reverse northern dot-blotting, with highly restrictive determination of five-fold greater signal intensity or signal only in the tester—313 positive clones were identified.
Bioinformatics analyses of sequence fragments
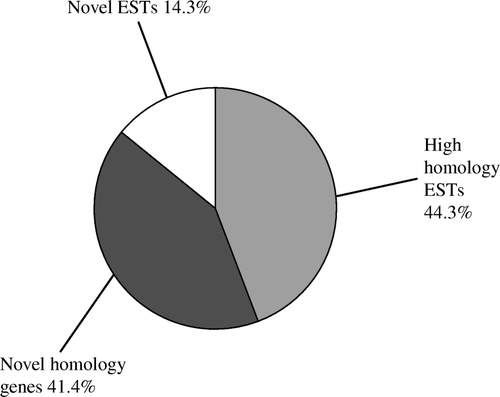
The 313 positive clones were sequenced, resulting in 140 sequences after low-quality and repeat sequences were removed. A search for sequence homology in the GenBank (nr), the EST database (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome) and the Gallus genome database (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=9031) by BLASTn revealed that the sequences obtained in the study could be divided into three categories: high homology genes (58), high homology ESTs (62) and novel ESTs (20) () .
High homology genes
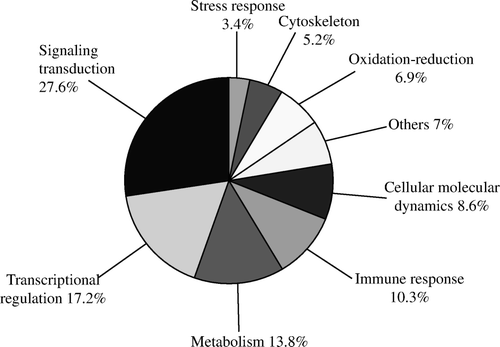
We obtained 58 high homology genes that have high homology with known genes by BLASTn, and they could be grouped into nine clusters based on the best known function of their protein products, which involved signalling transduction, transcriptional regulation, cellular molecular dynamics, cytoskeleton, stress response, immune response, metabolism, oxidation–reduction and others ( and ).
Figure 5. Detection of NDV by PT-PCR. Lane M, 500 bp DNA marker; lanes 1 to 3, PCR product of liver of 1#, 2 and 3# infected birds; lane 4, positive control; lane 5, negative control.
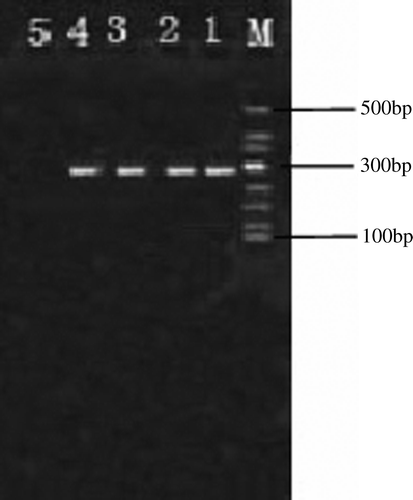
Table 1. List of 58 high homology genes.
High homology ESTs
We obtained 62 high homology ESTs (their sequences have been submitted to NCBI—dbEST_Id: 66693092 to 66693153; GenBank accession numbers: GR884515 to GR884576) that have high homology with known ESTs in the EST database by BLASTn. However, they have no or low homology with known genes, and their functions are also unknown.
Novel ESTs
Twenty novel ESTs (their sequences have been submitted to NCBI—dbEST_Id: 66224039 to 66224058; GenBank accession numbers: GR480711 to GR480730) that have no homology with known genes or known ESTs in the public database, and are reported here for the first time, were obtained in the present study. Through further analysis of chromosome positioning, we found that seven of these novel ESTs (dbEST_Id: 66224039, 66224043, 66224046, 66224047, 66224050, 66224057, 66224058) are located on the chromosome site between two genes. Among these seven ESTs, one (dbEST_Id: 66224046) was predicted to have an ORF using the ORF finder program online. The ORF of this EST starts from the 465th base and ends at the 669th base, encoding 70 amino acids. The third position upstream (–3) of the start codon is A and the fourth position downstream (+4) is G, which correspond with the Kozak principle (A/GNNATGG) (Kozak, Citation1987) and there is a termination codon before the start codon. These results suggest that this EST may be a sequence coding gene.
Discussion
Increasingly, evidence emphasizes that an efficient means of characterizing the molecular basis of the host–pathogen relationship is profiling of genes whose expression is altered during the course of infection (Munir & Kapur, Citation2003). In the present study, we screened and identified the differentially expressed genes initiated by in vivo infection with NDV at the genomics level for the first time, and through analysis of these differentially expressed genes we found that host–NDV interaction is a complex molecular mechanism. This may be reflected as below.
Transcriptional repression of cell genes
During infection by many RNA viruses (such as influenza viruses, micro-RNA virus, rhabdovirus, etc.), an important characteristic is the inhibition of host cell gene expression (Kaariainen & Ranki, Citation1984; Lyles, Citation2000), but this phenomenon has not been reported in NDV infections. In this study, five transcription inhibitor genes including ANP32A (acidic nuclear phosphoprotein 32 family, member A), LOC423724 (PIAS-like protein hZimp10), SATB1 (special AT-rich sequence binding homeobox 1), QKI (quaking homologue, KH domain RNA binding) and ETV6 (ETS variant gene 6) were screened. ANP32A is a nucleocytoplasmic shuttling protein that can repress the expression of cell genes by inhibiting histone acetyltransferases (Kutney et al., Citation2004). In addition, ANP (acidic nuclear phosphoprotein) also plays a role in E4F1-mediated transcriptional repression (Cvetanovic et al., Citation2007). LOC423724 can enhance the transcriptional activity of p53, playing a role in p53-mediated transcriptional repression (Lee et al., Citation2007). SATB1 is a protein found predominantly in the nuclear matrix attachment regions of the thymocytes, it was shown to repress many genes expressed by remodelling chromatin as a “landing platform” for several chromatin remodelling enzymes (Seo et al., Citation2005). QKI is a RNA-binding protein that binds to the 5′-NACUAAY-N(1,20)-UAAY-3′ RNA core sequence. It acts by regulating pre-mRNA splicing, mRNA export and mRNA stability (Galarneau & Richard, Citation2005). Research using Caenorhabditis elegans demonstrated that QKI-6 functions in the same manner as GLD-1 and can specifically bind to tra-2 and GCI elements (TGEs) to repress translation of many genes (Saccomanno et al., Citation1999). ETV6, a member of the ETS (E-Twenty-Six) family, acts as a transcription inhibitor through its ETS domain binding at the promoter regions of the gene (Arai et al., Citation2002). In addition, it can form a co-repressor with SMRT, mSin3A or N-CoR to repress the expression of cell genes (Chakrabarti & Nucifora, Citation1999; Mavrothalassitis & Ghysdael, Citation2000). It has been reported that ETV6 can interact with the acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) and functions as a transcriptional co-repressor to repress the expression of host genes in HIV-1 infection (Nordentoft & Jorgensen, Citation2003) . Based on the results mentioned above, it is suggested that the inhibition of host cell gene expression by NDV infection may provide a facility for NDV synthesis by reducing the output of cell mRNA.
Over-expression of ZEB1 and SATB 1 may lead to inhibition of the interleukin-2 signalling pathway
In this study, two genes including ZEB1 (zinc finger E-box binding homeobox 1) and SATB 1 (special AT-rich sequence binding homeobox 1) that are involved in the interleukin (IL-2) signalling pathway were screened. ZEB1, also named transcription factor 8 (represses IL-2 expression), encodes a zinc finger-containing protein that suppresses IL-2 gene expression by interacting with the negative regulation region of the IL-2 gene in T cells (Seo et al., Citation2005). SATB 1 recruits the histone deacetylase contained in the NURD chromatin remodelling complex to a SATB 1-bound site in the IL-2Rα (interleukin-2 receptor alpha) locus, and mediates the specific deacetylation of histones in a large domain within the locus repressing the expression of the IL-2Rα gene (Yasui et al., Citation2002). Taken together, the transcription factor 8 that acts with the upstream gene of the IL-2 signalling pathway and SATB 1 that acts with the downstream receptor of the IL-2 signalling pathway co-mediate the inhibition of the IL-2 signalling pathway. Interestingly, this phenomenon also exists during HIV-1 infections (Galarneau & Richard, Citation2005).
Novel immune-related genes induced by NDV infection
Three novel immune-related genes, including CMIP (c-Maf-inducing protein), TOX and TIA1 (cytotoxic granule-associated RNA binding protein), which were discovered in recent years, were screened in this research. There are only a few descriptions about them in the immune-related function in mammals (Kawakami et al., Citation1992; Wilkinson et al., Citation2002; Grimbert et al., Citation2004; Nanda et al., Citation2008), but they are still not reported in avian virus infection. CMIP, a new adapter protein involved in the T-helper 2 (Th2) signalling pathway, can trigger a series of Th2-type immune response mediated by IL-4, through activation of c-Maf factors (Grimbert et al., Citation2004). Tox, a factor involved in the regulation of Tcell development, can activate the maturation and differentiation of CD8+-type T cells in immune tissue, possibly enhancing cellular immunity (Wilkinson et al., Citation2002). TIA1, a new class of cytolytic effector molecule, is a cytotoxic granule-associated protein expressed in natural killer (NK) cells and cytotoxic T lymphocytes, and possesses nucleolytic activity against cytotoxic lymphocyte target cells (Kawakami et al., Citation1992). These findings suggest that they participated in the innate immunity and adaptive immunity of chickens that resist NDV infection. Further study of the functions of these genes in host–virus interactions will provide useful information to better understand immunity of avian species.
Stress protein genes induced by NDV infection
Two stress protein genes including DNAJC15 (DnaJ/Hsp40 homolog, subfamily C, member 15) and CIRBP (cold-inducible RNA-binding protein) were screened in this research. Until now, there is no report that the two stress protein genes may be involved in avian–pathogen interaction. DNAJC15 is a member of the DnaJ/Hsp40 family. The function of DnaJ/Hsp40 in mammals has been proven to be an essential cofactor of HSP70, through interaction with HSP70 (Qiu et al., Citation2006). In many viral infections, DnaJ/Hsp40 can interact with viral replication protein or nucleocapsid protein to help the virus complete the protein folding and assembly, and is even involved in the transport of viral proteins. It has been reported that the expression of DnaJ/Hsp40 increased and seems to facilitate viral gene expression in brome mosaic virus, Simian virus 40 and porcine circovirus infections (Sullivan & Pipas, Citation2002; Tomita et al., Citation2003; Finsterbusch et al., Citation2009) but interestingly, it has also been reported that DnaJ/Hsp40 can repress hepatitis B virus replication through destabilization of viral core and X proteins in hepatitis B virus infection (Sohn et al., Citation2006). Previous studies demonstrated that NDV infection can induce an up-regulation in HSP70 (Collins & Hightower, Citation1982), but the DnaJ/Hsp40 gene has not been reported. Whether or not the high expression of such a gene will contribute to virus assembly or prevent virus replication needs further study.
CIRBP is a nuclear 18 kDa protein consisting of an amino-terminal RNA recognition motif and a carboxyl-terminal domain containing several RGG motifs (De Leeuw et al., Citation2007). Previous research demonstrated that the induction of CIRBP can protect against TNF-α-induced apoptosis in BALB/3T3 cells (Sakurai et al., Citation2006). In this study, we showed for the first time that the CIRP could be induced by NDV infection. However, the relationship between cold-inducible RNA-binding protein and NDV infection needs further study.
Genes related with cell molecular motors and cytoskeletal proteins induced by NDV infection
Expression of some cell molecular motors and cytoskeletal proteins increased after NDV infection (). The genes related to cell molecular motors screened in this study included MYLK (myosin light chain kinase), MYO7A (myosin VIIa) and DCTN6 (dynactin 6). The three classes of molecular motors, including kinesins, dyneins and myosins, are involved in a multiplicity of biological movements. But they are often hijacked by intracellular pathogens to reach their site of replication, to leave their host or to control the dynamics of membrane exchanges with their replication compartment (Henry et al., Citation2006). MYLK has been proved to play an important role in HIV-1 release from host cells (Sasaki et al., Citation1995). In addition, it has been reported that the expression of MYLK was higher in Rous sarcoma virus-transformed CEFs than in normal CEFs (Van Eldik et al., Citation1984). Myosins are a diverse category of molecular motors that possess a motor domain and a tail domain involved in cargo binding. MYO7A and its ligand vezatin were demonstrated to be crucial for Listeria entry into epithelial cells (Sousa et al., Citation2004). DCTN6 is a subtype of dynactin that is a multi-subunit cofactor of dynein activating the activity of ATPase of dynein. Dynactin and dynein have been proven to play an important role in many intracellular viral transports. It has been reported that dynein, together with its cofactor dynactin, transports incoming herpes simplex virus type 1 capsids along microtubules to the MT-organizing centre and also transports mature virus from nuclear to host membranes (Dohner et al., Citation2002). In adenovirus infections, dynactin and dynein have been demonstrated to mediate adenovirus binding to microtubules for viral reverse transport (Kelkar et al., Citation2004). Genes related to molecular motors in NDV infection have not been reported; here we confirmed that infection of NDV induces up-regulation in such genes. In accordance with their function in other viral infection, we speculate that high expression of molecular motors will facilitate the transport or membrane budding of NDV.
The genes related to the cytoskeleton include LAMA4 (laminin, alpha 4), LAMC1 (laminin, gamma 1), and COL4A1 (collagen, type IV, alpha 1). Laminin and collagen can be used by a variety of viruses to facilitate their invasion, replication and transportation. Vaccinia virus intracellular mature virus can facilitate entry into the host cell through viral envelope protein A26L by binding to cell surface extracellular matrix protein laminin (Chiu et al., Citation2007). It has been reported that keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells (Culp et al., Citation2006). Rotavirus enterotoxin NSP4 binds to the extracellular matrix protein laminin-β3 to facilitate viral release (Boshuizen et al., Citation2004). In recent years, the role of collagen in virus infection has been increasingly recognized. The expression of collagen type I and other fibrosis-related molecules increased after the human hepatic stellate cell line (LX-2) was transfected with hepatitis B virus X protein gene (Guo et al., Citation2009). A collagen deposition was observed in lymphatic tissues that damaged the immune function during HIV-1 and SIV infections (Estes, Citation2009). In addition, there have been reports that the expression of extracellular matrix including laminin and collagens I, III and IV were up-regulated in HIV-1 that transfected human salivary gland cell lines, which related to the HIV pathogenesis (McArthur et al., Citation2001). In NDV infection, previous research has shown that the expression of α, β, and γ types of actin were altered, and suggested that host cells resist the viral invasion and budding by down-regulation of actin during infection (Munir et al., Citation2005). In the present study, we found that the expression of laminin and collagen were also impacted in NDV infection, and we speculate that high expression of laminin and collagen may facilitate the NDV invasion and budding.
Oxidation–reduction-related genes induced by NDV infection
The three oxidation–reduction-related genes TXN (thioredoxin protein), MSRB3 (methionine sulfoxide reductase B3), GCLC (gamma-glutamylcysteine synthetase) were up-regulated after NDV infection. Several viruses induce an imbalance of intracellular redox state toward pro-oxidant conditions, which contribute both to virus replication and to the pathogenesis of virus-induced disease (Nencioni et al., Citation2007). NDV infections induce a host cell oxidative stress and apoptosis has been confirmed in many experiments (Lam & Vasconcelos, Citation1994; Lam, Citation1995, Citation1996; Lam et al., Citation1996; Ravindra et al., Citation2008, Citation2009). but interestingly, some anti-oxidation-related genes were screened in our research. TXN is a small redox protein that can reduce the level of cell oxidation by self-oxidation–reduction reaction. TXN is recognized as an important anti-apoptosis factor because it can directly repress the apoptosis induced by TNF or reactive oxygen species (ROS) (Haendeler et al., Citation2002). MSRB3 is an enzyme with oxidation–reduction activity, which can repair oxidated methionine residues caused by ROS (Kim & Gladyshev, Citation2004). Methionine sulfoxide reductase is capable of protecting cells against oxidative damage and apoptosis by reducing the level of ROS (Prentice et al., Citation2008). GCLC is a rate-limiting enzyme that regulates the expression of glutathione, which controls the cellular redox status. Overexpression of GCLC can protect cells against oxidative damage and apoptosis by suppressing the activation of NF-κB and AP-1 induced by apoptosis (Manna et al., Citation1999). These findings showed that there exists a phenomenon of anti-oxidation at the time of NDV infection. Up-regulation of these anti-oxidation genes' expression seems to be a host-resistant mechanism that reduces the negative impact of oxidative and apoptotic damage caused by NDV infection. Therefore, the significance of anti-oxidation genes in NDV infection needs further study.
Metabolism-related genes induced by NDV infection
Some metabolism-related genes including energy, nucleic acid, protein, and so forth, were screened in the present studies. A typical representative of protein synthesis is the expression of RPS15A (ribosomal protein S15) and RPL32 (ribosomal protein L32); these ribosomal proteins increased during NDV infection. It was reported that the expression of ribosomal protein SA isoform 2 can be induced in infectious bursal disease virus infection, but the function in viral infection was unknown (Zheng et al., Citation2008). Energy metabolism-related genes include GLUT8 (solute carrier family 2 facilitated glucose transporter, member 8) and CYPR21 (cytochrome P450 2R1). GLUT8 is a novel member of the sugar transport facilitator family with glucose transport activity (Doege et al., Citation2000). The GLUT (glucose transporter) family was shown to exist in most immune cells (such as monocytes/macrophages, neutrophils, and B and T lymphocytes) (Chakrabarti et al., Citation1994; Malide et al., Citation1998; Fu et al., Citation2004), and a previous study demonstrated that the expression of GLUT3 and GLUT5 increased in the differentiation of monocytes to macrophages (Ercolani et al., Citation1985). CYPR21 is one component of the respiratory chain that generates energy through electron transport. Nucleic acid metabolism-related genes include DPYD (dihydropyrimidine dehydrogenase) and LOC417295 (UDP-N-acteylglucosamine pyrophosphorylase1-like 1). DPYD plays an important role in pyrimidine base degradation, and participates in the de novo pyrimidine base biosynthetic process. According to the results mentioned above, we find that host metabolism becomes active during NDV infection. It is known that generating an efficient and effective immune response involves large increases in cellular proliferative, biosynthetic, and secretory activities, processes that all require a mechanism support. In addition, when adaptive as well as innate immune cells shift from a quiescent phenotype to a highly active state after stimulation by pathogens, it is also associated with an alteration of metabolism (Wolowczuk et al., Citation2008). Therefore, we infer that the host metabolic activity will provide support for host resistance to the virus infection.
Taken together, our results demonstrate previously unrecognized changes in gene transcription that are altered during NDV infection in vivo, and provide useful information for future investigations of the molecular events in avian–NDV interactions.
Acknowledgements
The authors thank China Animal Health and Epidemiology Center for help with the animals and virus. The present work was supported by Nation Key Technology Research and Development Program of China (Grant 2006BAD06A11).
References
- Alexander , D.J. 2000 . Newcastle disease and other avian paramyxoviruses . Revue Scientifique et Technique de l'Office International des Epizooties , 19 : 443 – 462 .
- Arai , H. , Maki , K. , Waga , K. , Sasaki , K. , Nakamura , Y. , Imai , Y. Mitani , K. 2002 . Functional regulation of TEL by p38-induced phosphorylation . Biochemical and Biophysical Research Communications , 299 : 116 – 125 .
- Boshuizen , J.A. , Rossen , J.W. , Sitaram , C.K. , Kimenai , F.F. , Simons-Oosterhuis , Y. , Laffeber , C. Einerhand , A.W. 2004 . Rotavirus enterotoxin NSP4 binds to the extracellular matrix proteins laminin-beta3 and fibronectin . Journal of Virology , 78 : 10045 – 10053 .
- Chakrabarti , R. , Jung , C.Y. , Lee , T.P. , Liu , H. and Mookerjee , B.K. 1994 . Changes in glucose transport and transporter isoforms during the activation of human peripheral blood lymphocytes by phytohemagglutinin . The Journal of Immunology , 152 : 2660 – 2668 .
- Chakrabarti , S.R. and Nucifora , G. 1999 . The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A . Biochemical and Biophysical Research Communications , 264 : 871 – 877 .
- Chiu , W.L. , Lin , C.L. , Yang , M.H. , Tzou , D.L. and Chang , W. 2007 . Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin . Journal of Virology , 81 : 2149 – 2157 .
- Collins , P.L. and Hightower , L.E. 1982 . Newcastle disease virus stimulates the cellular accumulation of stress (heat shock) mRNAs and proteins . Journal of Virology , 44 : 703 – 707 .
- Culp , T.D. , Budgeon , L.R. , Marinkovich , M.P. , Meneguzzi , G. and Christensen , N.D. 2006 . Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells . Journal of Virology , 80 : 8940 – 8950 .
- Cvetanovic , M. , Rooney , R.J. , Garcia , J.J. , Toporovskaya , N. , Zoghbi , H.Y. and Opal , P. 2007 . The role of LANP and ataxin 1 in E4F-mediated transcriptional repression . EMBO Reports , 8 : 671 – 677 .
- De Leeuw , F. , Zhang , T. , Wauquier , C. , Huez , G. , Kruys , V. and Gueydan , C. 2007 . The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor . Experimental Cell Research , 313 : 4130 – 4144 .
- Doege , H. , Schurmann , A. , Bahrenberg , G. , Brauers , A. and Joost , H.G. 2000 . GLUT8, a novel member of the sugar transport facilitator family with glucose transport activity . The Journal of Biological Chemistry , 275 : 16275 – 16280 .
- Dohner , K. , Wolfstein , A. , Prank , U. , Echeverri , C. , Dujardin , D. , Vallee , R. and Sodeik , B. 2002 . Function of dynein and dynactin in herpes simplex virus capsid transport . Molecular Biology of the Cell , 13 : 2795 – 2809 .
- Ercolani , L. , Lin , H.L. and Ginsberg , B.H. 1985 . Insulin-induced desensitization at the receptor and postreceptor level in mitogen-activated human T-lymphocytes . Diabetes , 34 : 931 – 937 .
- Estes , J.D. 2009 . Role of collagen deposition in lymphatic tissues and immune reconstruction during HIV-1 and SIV infections . Current HIV/AIDS Reports , 6 : 29 – 35 .
- Finsterbusch , T. , Steinfeldt , T. , Doberstein , K. , Rodner , C. and Mankertz , A. 2009 . Interaction of the replication proteins and the capsid protein of porcine circovirus type 1 and 2 with host proteins . Virology , 386 : 122 – 131 .
- Fu , Y. , Maianu , L. , Melbert , B.R. and Garvey , W.T. 2004 . Facilitative glucose transporter gene expression in human lymphocytes, monocytes, and macrophages: a role for GLUT isoforms 1, 3, and 5 in the immune response and foam cell formation . Blood Cells, Molecules and Diseases , 32 : 182 – 190 .
- Galarneau , A. and Richard , S. 2005 . Target RNA motif and target mRNAs of the Quaking STAR protein . Nature Structural & Molecular Biology , 12 : 691 – 698 .
- Grimbert , P. , Valaciute , A. , Audard , V. , Lang , P. , Guellaen , G. and Sahali , D. 2004 . The Filamin-A is a partner of Tc-mip, a new adapter protein involved in c-maf-dependent Th2 signaling pathway . Molecular Immunology , 40 : 1257 – 1261 .
- Guo , G.H. , Tan , D.M. , Zhu , P.A. and Liu , F. 2009 . Hepatitis B virus X protein promotes proliferation and upregulates TGF-beta1 and CTGF in human hepatic stellate cell line, LX-2 . Hepatobiliary and Pancreatic Diseases International , 8 : 59 – 64 .
- Haendeler , J. , Hoffmann , J. and Zeiher , A.M. 2002 . Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69 . Circulation , 106 : 212 – 213 .
- Henry , T. , Gorvel , J.P. and Meresse , S. 2006 . Molecular motors hijacking by intracellular pathogens . Cellular Microbiology , 8 : 23 – 32 .
- Huang , Z.H. , Krishnamurthy , S. , Panda , A. and Samal , S.K. 2003 . Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist . Journal of Virology , 77 : 8676 – 8685 .
- Jain , S. , McGinnes , L.W. and Morrison , T.G. 2007 . Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein . Journal of Virology , 81 : 2328 – 2339 .
- Jain , S. , McGinnes , L.W. and Morrison , T.G. 2008 . Overexpression of thiol/disulfide isomerases enhances membrane fusion directed by the Newcastle disease virus fusion protein . Journal of Virology , 82 : 12039 – 12048 .
- Kaariainen , L. and Ranki , M. 1984 . Inhibition of cell functions by RNA-virus infections . Annual Review of Microbiology , 38 : 91 – 109 .
- Kawakami , A. , Tian , Q. , Duan , X. , Streuli , M. , Schlossman , S.F. and Anderson , P. 1992 . Identification and functional characterization of a TIA-1-related nucleolysin . Proceedings of the National Academy of Sciences of the USA , 89 : 8681 – 8685 .
- Kelkar , S.A. , Pfister , K.K. , Crystal , R.G. and Leopold , P.L. 2004 . Cytoplasmic dynein mediates adenovirus binding to microtubules . Journal of Virology , 78 : 10122 – 10132 .
- Kim , H.Y. and Gladyshev , V.N. 2004 . Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases . Molecular Biology of the Cell , 15 : 1055 – 1064 .
- Kozak , M. 1987 . An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs . Nucleic Acids Research , 15 : 8125 – 8148 .
- Kutney , S.N. , Hong , R. , Macfarlan , T. and Chakravarti , D. 2004 . A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Iβ in integrating chromatin hypoacetylation and transcriptional repression . The Journal of Biological Chemistry , 279 : 30850 – 30855 .
- Lam , K.M. 1995 . Apoptosis in chicken embryo fibroblasts caused by Newcastle disease virus . Veterinary Microbiology , 47 : 357 – 363 .
- Lam , K.M. 1996 . Growth of Newcastle disease virus in chicken macrophages . Journal of Comparative Pathology , 115 : 253 – 263 .
- Lam , K.M. and Vasconcelos , A.C. 1994 . Newcastle disease virus-induced apoptosis in chicken peripheral blood lymphocytes . Veterinary Immunology and Immunopathology , 44 : 45 – 56 .
- Lam , K.M. , Kabbur , M.B. and Eiserich , J.P. 1996 . Newcastle disease virus-induced functional impairments and biochemical changes in chicken heterophils . Veterinary Immunology and Immunopathology , 53 : 313 – 327 .
- Lee , J. , Beliakoff , J. and Sun , Z. 2007 . The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor . Nucleic Acids Research , 35 : 4523 – 4534 .
- Lyles , D.S. 2000 . Cytopathogenesis and inhibition of host gene expression by RNA viruses . Microbiology and Molecular Biology Reviews , 64 : 709 – 724 .
- Malide , D. , Davies-Hill , T.M. , Levine , M. and Simpson , I.A. 1998 . Distinct localization of GLUT-1, -3, and -5 in human monocyte-derived macrophages: effects of cell activation . American Journal of Physiology—Endocrinology and Metabolism , 37 : E516 – E526 .
- Manna , S.K. , Kuo , M.T. and Aggarwal , B.B. 1999 . Overexpression of gamma-glutamylcysteine synthetase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-kappa B and activator protein-1 . Oncogene , 18 : 4371 – 4382 .
- Mavrothalassitis , G. and Ghysdael , J. 2000 . Proteins of the ETS family with transcriptional repressor activity . Oncogene , 19 : 6524 – 6532 .
- McArthur , C.P. , Wang , Y. , Heruth , D. and Gustafson , S. 2001 . Amplification of extracellular matrix and oncogenes in tat-transfected human salivary gland cell lines with expression of laminin, fibronectin, collagens I, III, IV, c-myc and p53 . Archives of Oral Biology , 46 : 545 – 555 .
- Munir , S. and Kapur , V. 2003 . Regulation of host cell transcriptional physiology by the avian pneumovirus provides key insights into host–pathogen interactions . Journal of Virology , 77 : 4899 – 4910 .
- Munir , S. , Sharma , J.M. and Kapur , V. 2005 . Transcriptional response of avian cells to infection with Newcastle disease virus . Virus Research , 107 : 103 – 108 .
- Nanda , S. , Havert , M.B. , Calderon , G.M. , Thomson , M. , Jacobson , C. , Kastner , D. and Liang , T.J. 2008 . Hepatic transcriptome analysis of hepatitis C virus infection in chimpanzees defines unique gene expression patterns associated with viral clearance . PLoS One , 3 : e3442
- Nencioni , L. , Sgarbanti , R. , De Chiara , G. , Garaci , E. and Palamara , A.T. 2007 . Influenza virus and redox mediated cell signaling: a complex network of virus/host interaction . New Microbiologica , 30 : 367 – 375 .
- Nordentoft , I. and Jorgensen , P. 2003 . The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor . Biochemical Journal , 374 : 165 – 173 .
- Prentice , H.M. , Moench , I.A. , Rickaway , Z.T. , Dougherty , C.J. , Webster , K.A. and Weissbach , H. 2008 . MsrA protects cardiac myocytes against hypoxia/reoxygenation induced cell death . Biochemical and Biophysical Research Communications , 366 : 775 – 778 .
- Qiu , X.B. , Shao , Y.M. , Miao , S. and Wang , L. 2006 . The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones . Cellular and Molecular Life Sciences , 63 : 2560 – 2570 .
- Ravindra , P.V. , Tiwari , A.K. , Ratta , B. , Chaturvedi , U. , Palia , S.K. and Chauhan , R.S. 2009 . Newcastle disease virus-induced cytopathic effect in infected cells is caused by apoptosis . Virus Research , 141 : 13 – 20 .
- Ravindra , P.V. , Tiwari , A.K. , Sharma , B. , Rajawat , Y.S. , Ratta , B. , Palia , S. Chauhan , R.S. 2008 . HN protein of Newcastle disease virus causes apoptosis in chicken embryo fibroblast cells . Archives of Virology , 153 : 749 – 754 .
- Saccomanno , L. , Loushin , C. , Jan , E. , Punkay , E. , Artzt , K. and Goodwin , E.B. 1999 . The STAR protein QKI-6 is a translational repressor . Proceedings of the National Academy of Sciences of the USA , 96 : 12605 – 12610 .
- Sakurai , T. , Itoh , K. , Higashitsuji , H. , Nonoguchi , K. , Liu , Y. , Watanabe , H. Fujita , J. 2006 . Cirp protects against tumor necrosis factor-alpha-induced apoptosis via activation of extracellular signal-regulated kinase . Biochimica et Biophysica Acta—Molecular Cell Research , 1763 : 290 – 295 .
- Sasaki , H. , Nakamura , M. , Ohno , T. , Matsuda , Y. , Yuda , Y. and Nonomura , Y. 1995 . Myosin–actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells . Proceedings of the National Academy of Sciences of the USA , 92 : 2026 – 2030 .
- Seo , J. , Lozano , M.M. and Dudley , J.P. 2005 . Nuclear matrix binding regulates SATB1-mediated transcriptional repression . The Journal of Biological Chemistry , 280 : 24600 – 24609 .
- Sohn , S.Y. , Kim , S.B. , Kim , J. and Ahn , B.Y. 2006 . Negative regulation of hepatitis B virus replication by cellular Hsp40/DnaJ proteins through destabilization of viral core and X proteins . Journal of General Virology , 87 : 1883 – 1891 .
- Sousa , S. , Cabanes , D. , El-Amraoui , A. , Petit , C. , Lecuit , M. and Cossart , P. 2004 . Unconventional myosin VIIa and vezatin, two proteins crucial for Listeria entry into epithelial cells . Journal of Cell Science , 117 : 2121 – 2130 .
- Sullivan , C.S. and Pipas , J.M. 2002 . T antigens of simian virus 40: Molecular chaperones for viral replication and tumorigenesis . microbiology and Molecular Biology Reviews , 66 : 179 – 202 .
- Tomita , Y. , Mizuno , T. , Diez , J. , Naito , S. , Ahlquist , P. and Ishikawa , M. 2003 . Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis . Journal of Virology , 77 : 2990 – 2997 .
- Van Eldik , L.J. , Watterson , D.M. and Burgess , W.H. 1984 . Immunoreactive levels of myosin light-chain kinase in normal and virus-transformed chicken embryo fibroblasts . Molecular and Cellular Biology , 4 : 2224 – 2226 .
- Wilkinson , B. , Chen , J.Y. , Han , P. , Rufner , K.M. , Goularte , O.D. and Kaye , J. 2002 . TOX: an HMG box protein implicated in the regulation of thymocyte selection . Nature Immunology , 3 : 272 – 280 .
- Wolowczuk I. Verwaerde C. Viltart O. Delanoye A. Delacre M. Pot B. Grangette C. 2008 Feeding our immune system: impact on metabolism Clinical & Developmental Immunology 1 19
- Yasui , D. , Miyano , M. , Cai , S. , Varga-Weisz , P. and Kohwi-Shigematsu , T. 2002 . SATB1 targets chromatin remodelling to regulate genes over long distances . Nature , 419 : 641 – 645 .
- Yue , H. , Yu , N. , Tang , C. and Li , M.Y. 2007 . Development of real-time reverse-transcription PCR for detection of mesogenic and virulent Newcastle disease virus RNA . Chinese Journal of Veterinary Science , 27 : 300 – 304 .
- Zheng , X. , Hong , L. , Shi , L. , Guo , J. , Sun , Z. and Zhou , J. 2008 . Proteomics analysis of host cells infected with infectious bursal disease virus . Molecular & Cellular Proteomics , 7 : 612 – 625 .