Abstract
The partial (370 nucleotides) fusion gene sequences of 55 avian paramyxovirus type 1 (APMV-1) isolates were obtained. Included were 41 published sequences, of which 16 were from strains of APMV-1 of previously determined lineages included as markers for the data analysed and 25 were from APMV-1 viruses isolated from game birds of the order Galliformes. In addition, we sequenced a further 14 game bird isolates obtained from the repository at the Veterinary Laboratories Agency. The game bird isolates had been obtained from 17 countries, and spanned four decades. Earlier studies have shown that class II APMV-1 viruses can be divided into at least 15 lineages and sub-lineages. Phylogenetic analysis revealed that the 39 game bird isolates were distributed across 12 of these sub-lineages. We conclude that no single lineage of Newcastle disease viruses appears to be prevalent in game birds, and the isolates obtained from these hosts reflected the prevailing, both geographically and temporally, viruses in poultry, pigeons or wild birds.
Introduction
Newcastle disease (ND) or avian paramyxovirus type 1 (APMV-1) is regarded throughout the world as one of the two most important diseases of poultry and other birds, not only due to the serious disease and high flock mortality that may result from some ND virus (NDV) infections, but also the economic impact that may ensue due to trading restrictions and embargoes placed on areas and countries where outbreaks have occurred (Commission of the European Communities, Citation1992; Alexander, Citation2008).
Although the first isolations of NDV were from chickens, since that time the virus has been isolated from many domestically-reared flocks including turkeys (Alexander et al., Citation1998, Citation1999; Roy et al., Citation1999), ostriches (Huchzermeyer & Gerdes, Citation1993), pheasants, other game birds (Aldous & Alexander, Citation2008) and pigeons (Vindevogel & Duchatel, Citation1988). NDV can also infect many wild bird species including cormorants, wild ducks, starlings, penguins and wild waterfowl (Lipkind et al., Citation1987; Takakuwa et al., Citation1998; Kuiken, Citation1999). Kaleta & Baldauf (Citation1988) reviewed NDV isolations from all bird species and recorded that NDV infections have been established and detected in at least 241 species of birds, representing 27 of the 50 orders of the class Aves. It would seem probable that all birds are likely to be susceptible to infection with this virus (Alexander, Citation2009).
Although in its broadest definition the term ‘game birds’ includes any bird that is hunted for sport or for food (i.e. including ducks, geese, etc.), it is more usually applied to species of the families Phasianidae (pheasants and partridges), Tetraonidae (grouse) and Numididae (guinea fowl), and in the present study we have considered isolates from birds in this narrower definition. Some game bird species may be semi-domesticated (e.g. pheasants, partridges, etc.) or even intensively reared (quail), but generally rearing is extensive and/or the birds are released into the wild for hunting.
The two most recent outbreaks of ND in the UK were in game bird species. In 2005, pheasants presenting with mild ataxia and blindness were confirmed to be infected with virulent NDV, lineage 5b (Aldous et al., Citation2007); and in 2006, partridges showing clinical signs of loss of condition, diarrhoea, progressive neurological signs and mortality totalling about 30% were also confirmed to be infected with virulent NDV (Irvine et al., 2009). Infections of game birds with APMV-1 viruses are not uncommon, but these outbreaks, in the absence of infections in chickens or turkeys, were of considerable concern for control of this notifiable avian disease. In light of this new, elevated risk that may be presented by this group of birds, the viruses isolated from these host species that are held in the repository of the International Reference Laboratory for ND at the Veterinary Laboratories Agency (VLA) were obtained and sequenced. The data have been analysed to enable insights into any host restrictions that may occur within these species and the recognition of any potentially high-risk strains.
The genetic and antigenic relationships between many different isolates of NDV have been investigated and discussed (Ballagi-Pordany et al., Citation1996; Alexander et al., Citation1997b; Lomniczi et al., Citation1998; Aldous et al., Citation2003; Czegledi et al., Citation2006). In the present study, the phylogenetic relationships between 55 isolates, including 39 from game bird hosts, were investigated. The analysis was based on the nucleotide sequence of a 370-nucleotide fragment at the 3′ end of the fusion protein gene. As one of the major antigenic determinants of NDV, the fusion protein gene is likely to display greater genetic variation than the internal genes, an important characteristic for studying quite closely related virus populations, where a more conserved gene may show insufficient sequence variation to allow deduction of evolutionary hypotheses.
Materials and Methods
Isolates
Virus isolates were grown in the allantoic cavities of 9-day-old to 10-day-old embryonated fowls’ eggs originating from a commercial specific-pathogen-free flock and were supplied by the International Reference Laboratory at VLA-Weybridge. Details of the isolates, including their abbreviations and accession numbers are presented in . Intracerebral pathogenicity index values for the viruses (where available) were supplied by the International Reference Laboratory.
Table 1. Isolate details.
Reverse transcriptase-polymerase chain reaction and polymerase chain reaction
Viral RNA was extracted from infective allantoic fluid using the Qiagen QIAamp viral RNA kit. The preparation of cDNA was carried out as described (Collins et al., Citation1993) using an alternative cDNA primer (#MSF1), 5′-GACCGCTGACCACGAGGTTA. ‘Reddy Mix’ polymerase chain reaction (PCR) master mix (ABgene) was used for the PCR, using the cDNA forward primer and reverse primer (#2) 5′-AGTCGGAGGATGTTGGCAGC, to generate an amplicon of approximately 700 bp. The final concentrations of components in the PCR mix were 1.25 units Taq DNA polymerase, 75 mM Tris–HCl, 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% (v/v) Tween 20, 1 µM each primer and 0.2 mM dATP, dCTP, dGTP and dTTP. The cycling parameters were an initial denaturation step of 1 min at 95°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50°C and 3 min at 72°C, completed with a final extension step of 72°C for 10 min. The PCR products were separated by gel electrophoresis using 2% w/v agarose gel in Tris–acetate buffer, stained with ethidium bromide and visualized under ultraviolet light. The product was excised and purified using the QiaQuick (Qiagen) gel extraction kit.
Nucleotide sequencing
The forward primer used was (#7) 5′-TTAGAAAAAACACGGGTAGAA (Collins et al., Citation1996), the reverse primer was the same as for the PCR (#2). Nucleotide sequencing was carried out using ‘BigDye’ DNA sequencing kits (V3.1) and a 3130 genetic analyser (Applied Biosystems) according to the manufacturer's instructions.
Sequence analysis
Nucleotide sequence analysis and alignment was done using Lasergene DNASTAR software version 8. The sequences were aligned to begin at the start codon (ATG at position 47) of the fusion protein gene and finish just downstream from the cleavage activation site. The sequences were aligned using the Clustal V method in Megalign. Subsequent phylogenetic analysis was carried out using MEGA, v4.1. The results are presented as phylogenetic trees in which the branch lengths are proportional to the predicted number of substitutions.
Results and Discussion
The results obtained from the phylogenetic analysis are shown as a phylogenetic tree in , which was generated from a dataset based on the partial fusion gene nucleotide sequences of 56 APMV-1 isolates. Included in this dataset are 41 published sequences, of which 16 represent previously determined lineages of APMV-1 as markers for the data analysed and presented in this study (Ballagi-Pordany et al., Citation1996; Aldous et al., Citation2003); the remaining 25 published sequences are APMV-1 isolates from game birds. In addition to this, we sequenced a further 14 game bird isolates obtained from the NDV repository at VLA. The predicted virulence of these isolates is presented in .
Figure 1. Evolutionary relationships of 56 APMV-1 isolates. The evolutionary history was inferred using the neighbour-joining method (Saitou & Nei, Citation1987). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, Citation1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al., Citation2004) and are in the units of the number of base substitutions per site. Codon positions included were first+second+third. There were a total of 370 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., Citation2007). ▴, isolates sequenced in the present study; •, ‘reference strains’ to indicate genetic lineages according to Aldous et al. (2003). Host abbreviations: PH, pheasant; QU, quail; PT, partridge; GL, gull; CK, chicken; PI, pigeon; DK, duck. Country abbreviations are according to their ISO 3166-1 two-letter code (http://www.iso.org/iso/english_country_names_and_code_elements). The grey box indicates the NDV lineages (Aldous et al., 2003) and genotypes (Lomniczi et al., 1998) of the isolates in the present study.
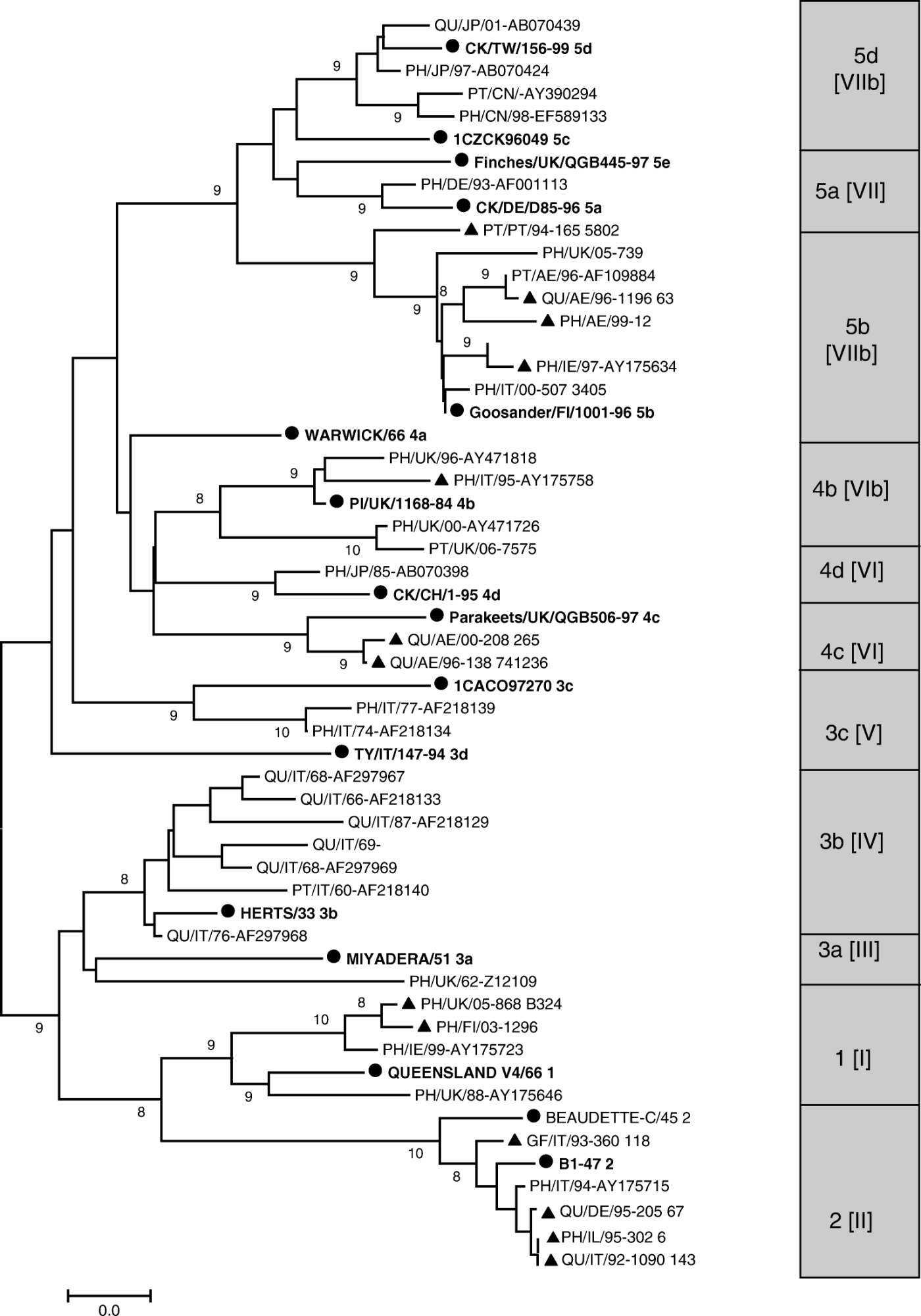
Table 2. Deduced F0 cleavage site motif and predicted virulence for chickens of game bird isolates sequenced in the present study.
Only class II viruses (Czegledi et al., Citation2006), which correspond to viruses of lineages 1 to 5 defined by Aldous et al. (Citation2003), were included in this study as there were no available data on isolations of class I (lineage 6; Aldous et al., Citation2003) viruses from game bird hosts. The five genetic lineages defined for class II viruses can be further divided into at least 15 sub-lineages. As can be seen ( and ), the 39 isolates from game birds analysed in this study are distributed across 12 of these sub-lineages. Of the main lineages for virulent NDVs (lineages 3, 4 and 5), the four sub-lineages with the greatest numbers of game bird viruses are 3b, 4b, 5b and 5d, although sub-lineages 3a, 3c, 4c, 4d and 5a are also represented. Nine isolates fall into lineages 1 and 2, the majority of which are likely to be of low virulence and could be the result of spread from wild birds, deliberate infection with live vaccines or accidental infection with live vaccines as a result of spread from poultry.
Four game bird isolates were placed in sub-lineage 4b, one from Italian pheasants, the other three from pheasants (two isolates) and partridges in the UK. These isolations covered a period from 1995 to 2006 and all appeared to be the result of spread from pigeons as there were no outbreaks in other poultry at the time they occurred (Alexander et al., Citation1997a; Irvine et al., Citation2009).
Ten of the isolates were placed in lineage 3 in sub-lineages 3a (one isolate), 3c (two isolates) and 3b (seven isolates). Nine of these viruses were isolated between 1960 and 1977 and reflect the prevailing ND strains in poultry at that time. The 10th virus placed in sub-lineage 3b was isolated from quail in Italy in 1987, and is consistent with the finding that viruses with specific genetic characteristics do not necessarily disappear as other variants arise and may continue to be isolated many years after their first appearance (Lomniczi et al., Citation1998).
The main conclusion from the present study is that no single lineage of ND viruses appears to be prevalent in game birds. This is unlike the situation in racing, farmed and feral pigeons, in which a panzootic caused exclusively by ND viruses of lineage 4b has continued for about 30 years (Vindevogel & Duchatel, Citation1988; Aldous et al., Citation2004; Alexander, Citation2009). The viruses isolated from game birds appear generally to represent geographically and temporally contemporaneous viruses affecting chickens, turkeys and/or other poultry. There is no way of knowing whether chickens and other poultry affected game birds or the game birds were responsible for spread to chickens and other poultry. Presumably transmission has occurred in both directions. However, infections of game birds, especially pheasants, with viruses virulent for chickens do not always result in severe disease (Aldous et al., Citation2007; Aldous & Alexander, Citation2008). There is therefore a real concern that, because of the international nature of trade in such birds and, when they are to be hunted, their release into the wild, they may represent a reservoir host for the spread and dissemination of virulent NDVs.
Acknowledgements
The present work was supported financially by a Department of Environment, Food and Rural Affairs research grant (SE0779). The authors thank Ruth Manvell for supplying the virus isolates.
References
- Aldous , E.W. and Alexander , D.J. 2008 . Newcastle disease in pheasants (Phasianus colchicus): a review . The Veterinary Journal , 175 : 181 – 185 .
- Aldous , E.W. , Fuller , C.M. , Mynn , J.K. and Alexander , D.J. 2004 . A molecular epidemiological investigation of isolates of the variant APMV-1 virus (PPMV-1) responsible for the 1978–2002 panzootic in pigeons . Avian Pathology , 33 : 258 – 269 .
- Aldous , E.W. , Manvell , R.J. , Cox , W.J. , Ceeraz , V. , Harwood , D.G. Shell , W. 2007 . Outbreak of Newcastle disease in pheasants (Phasianus colchicus) in south-east England in July 2005 . The Veterinary Record , 160 : 482 – 484 .
- Aldous , E.W. , Mynn , J.K. , Banks , J. and Alexander , D.J. 2003 . A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene . Avian Pathology , 32 : 239 – 357 .
- Alexander , D.J. 2008 . “ Newcastle disease In ” . In World Organisation for Animal Health: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals , 6th edn , 576 – 589 . Paris : OIE .
- Alexander , D.J. 2009 . “ Ecology and epidemiology of Newcastle disease ” . In Diagnosis of Avian Influenza and Newcastle Disease: A Field and Laboratory Manual , Edited by: Capua , I. and Alexander , D.J. 19 – 27 . Milan : Springer-Verlag .
- Alexander , D.J. , Manvell , R.J. , Banks , J. , Collins , M.S. , Parsons , G. Cox , B. 1999 . Experimental assessment of the pathogenicity of the Newcastle disease viruses from outbreaks in Great Britain in 1997 for chickens and turkeys, and the protection afforded by vaccination . Avian Pathology , 28 : 501 – 511 .
- Alexander D.J. Manvell R.J. Frost K.M. Pollitt W.J. Welchman D. Perry K. 1997a An outbreak of Newcastle disease in pheasants in Great Britain in May 1996 The Veterinary Record 140 20 22
- Alexander D.J. Manvell R.J. Lowings J.P. Frost K.M. Collins M.S. Russell P.H. Smith J.E. 1997b Antigenic diversity and similarities detected in avian paramyxovirus type 1 (Newcastle disease virus) isolates using monoclonal antibodies Avian Pathology 26 399 418
- Alexander , D.J. , Morris , H.T. , Pollitt , W.J. , Sharpe , C.E. , Eckford , R.L. Sainsbury , R.M. 1998 . Newcastle disease outbreaks in domestic fowl and turkeys in Great Britain during 1997 . The Veterinary Record , 143 : 209 – 212 .
- Ballagi-Pordany , A. , Wehmann , E. , Herczeg , J. , Belak , S. and Lomniczi , B. 1996 . Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene . Archives of Virology , 141 : 243 – 261 .
- Collins , M.S. , Bashiruddin , J.B. and Alexander , D.J. 1993 . Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity . Archives of Virology , 128 : 363 – 370 .
- Collins , M.S. , Strong , I. and Alexander , D.J. 1996 . Pathogenicity and phylogenetic evaluation of the variant Newcastle disease viruses termed' pigeon PMV-1 viruses' based on the nucleotide sequences of the fusion protein gene . Archives of Virology , 141 : 635 – 647 .
- Commission of the European Communities 1992 Council directive 92/66/EEC of 14th July 1992 introducing Community measures for the control of Newcastle disease Official Journal of the European Communities L260 1 20
- Czegledi , A. , Ujvari , D. , Somogyi , E. , Wehmann , E. , Werner , O. and Lomniczi , B. 2006 . Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications . Virus Research , 120 : 36 – 48 .
- Felsenstein , J. 1985 . Confidence limits on phylogenies: an approach using the bootstrap . Evolution , 39 : 783 – 791 .
- Huchzermeyer , F. and Gerdes , G. 1993 . Newcastle disease virus isolated from ostriches in South Africa [letter] . Journal of the South African Veterinary Association , 64 : 160
- Irvine , R.M. , Aldous , E.W. , Manvell , R.J. , Cox , W.J. , Fuller , C.M. Wood , A.M. 2009 . Outbreak of Newcastle disease due to pigeon paramyxovirus type 1 in grey partridgees (Perdix perdix) in Scotland in October 2006 . The Veterinary Record , 165 : 531 – 535 .
- Kaleta E.F. Baldauf C. 1988 Newcastle disease in free-living and pet birds In D.J. Alexander Newcastle Disease 197 246 MA Kluwer Academic Publishers
- Kuiken , T. 1999 . Review of Newcastle disease in cormorants . Waterbirds , 22 : 333 – 347 .
- Lipkind , M. , Rivetz , B. and Shihmanter , E. 1987 . The first isolation of Newcastle disease virus (NDV) from free flying birds in Israel: comparative studies on NDV strain isolated from migrating starlings (Sturnus vulgaris) wintering in Israel . Comparative Immunology and Microbiology of Infectious Disease , 10 : 65 – 70 .
- Lomniczi , B. , Wehmann , E. , Herczeg , J. , Ballagi-Pordany , A. , Kaleta , E.F. Werner , O. 1998 . Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII) . Archives of Virology , 143 : 49 – 64 .
- Roy , P. , Koteeswaran , A. , Venugopalan , A.T. and Chandran , N.D.J. 1999 . Newcastle disease in turkeys and ducks . Indian Veterinary Journal , 76 : 1121 – 1122 .
- Saitou , N. and Nei , M. 1987 . The neighbor-joining method: a new method for reconstructing phylogenetic trees . Molecular Biology and Evolution , 4 : 406 – 425 .
- Takakuwa , H. , Ito , T. , Takada , A. , Okazaki , K. and Kida , H. 1998 . Potentially virulent Newcastle disease viruses are maintained in migratory waterfowl populations . Japanese Journal of Veterinary Research , 45 : 207 – 215 .
- Tamura , K. , Dudley , J. , Nei , M. and Kumar , S. 2007 . MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 . Molecular Biology and Evolution , 24 : 1596 – 1599 .
- Tamura , K. , Nei , M. and Kumar , S. 2004 . Prospects for inferring very large phylogenies by using the neighbor-joining method . Proceedings of the National Academy of Sciences of the USA , 101 : 11030 – 11035 .
- Vindevogel H. Duchatel J.P. 1988 Panzootic Newcastle disease in pigeons In D.J. Alexander Newcastle Disease 184 196 MA Kluwer Academic Publishers