Abstract
Tembusu virus (TMUV) belongs to the genus Flavivirus that may cause severe egg drop in ducks. In order to evaluate the most efficient TMUV detection method, the performances of a conventional RT-PCR (C-RT-PCR), a semi-nested PCR (SN-RT-PCR), a reverse-transcriptase real-time quantitative PCR (Q-RT-PCR), and a reverse-transcription loop-mediated isothermal amplification (RT-LAMP) targeting the TMUV virus-specific NS5 gene were examined. In order to compare the sensitivity of these four techniques, two templates were used: (1) plasmid DNA that contained a partial region of the NS5 gene and (2) genomic RNA from TMUV-positive cell culture supernatants. The sensitivities using plasmid DNA detection by C-RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP were 2 × 104 copies/μL, 20 copies/μL, 2 copies/μL, and 20 copies/μL, respectively. The sensitivities using genomic RNA for the C-RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP were 100 pg/tube, 100, 10, and 100 fg/tube, respectively. All evaluated assays were specific for TMUV detection. The TMUV-specific RNA was detected in cloacal swabs from experimentally infected ducks using these four methods with different rates (52–92%), but not in the control (non-inoculated) samples. The sensitivities of RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP performed with cloacal swabs collected from suspected TMUV infected ducks within 2 weeks of severe egg-drop were 38/69 (55.1%), 52/69 (75.4%), 57/69 (82.6%), and 55/69 (79.7%), respectively. In conclusion, both RT-LAMP and Q-RT-PCR can provide a rapid diagnosis of TMUV infection, but RT-LAMP is more useful in TMUV field situations or poorly equipped laboratories.
Introduction
Tembusu virus (TMUV) belongs to the genus Flavivirus of the family Flaviviridae, comprising one long open reading frame that includes four 5′ structural genes (nucleocapsid [NC], pre-membrane [prM], membrane [M] and envelope [E]) and seven 3′ non-structural [NS] genes (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Lindenbach & Rice, Citation2003; Mukhopadhyay et al., Citation2005). TMUV was first isolated from mosquitoes of the genus Culex in the 1970s in Malaysia, but the disease associated with TMUV infection was not known at that time (Platt et al., Citation1975). A chick-origin TMUV strain, originally named Sitiawan virus, can cause encephalitis and impede growth in broiler chicks (Kono et al., Citation2000). Outbreaks of TMUV infection with severe egg-drop in many egg-laying and breeder duck farms were observed in China (Cao et al., Citation2011; Su et al., Citation2011; Tang et al ., Citation2011a; Yan et al., Citation2011b; Liu et al., Citation2012). The affected ducks showed significant egg-drop. Hyperaemia, haemorrhage, degeneration, distortion and lymphocyte infiltration in the ovaries and interstitial inflammation in the portal areas of livers were the main pathological changes consistently observed in almost all diseased ducks, which had caused serious economic losses (Cao et al., Citation2011).
Rapid detection methods are the key for successful disease control. Immunological tests, such as immunoperoxidase test, enzyme-linked immunosorbent assay (ELISA) (Gould et al., Citation1985; Koraka et al., Citation2002; Yin et al., Citation2013), and the indirect immunofluorescence assay (IFA) (Nagarkatti & Nagarkatti, Citation1980), are currently in use widely for Flaviviruses. Not only are these tests complicated to apply for clinical diagnosis, but they also have low sensitivities in the early stage of Flavivirus infection due to insufficient production of antibodies, and often require further testing to validate results. The detection of a specific gene by RT-PCR assay has been used for the rapid detection of Flaviviruses (Scaramozzino et al., Citation2001). Nested RT-PCR (N-RT-PCR) or semi-nested RT-PCR (SN-RT-PCR) is widely used to improve the sensitivity of conventional RT-PCR (C-RT-PCR) in Flavivirus detection (Yao & Tavis, Citation2005; Bhatnagar et al., Citation2007). In addition, real-time quantitative RT-PCR (Q-RT-PCR) and reverse-transcription loop-mediated isothermal amplification (RT-LAMP) targeting a specific gene permit the detection of TMUV within 2 h and have high sensitivity and specificity (Yan et al., Citation2011a; Yun et al., Citation2012). As few studies have compared the abilities of the four aforementioned methods (RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP) for detecting the same TMUV-specific target gene, the aim of the study was to (1) develop the NS5-based semi-nested RT-PCR and real-time quantitative RT-PCR method for the diagnosis of TMUV and (2) assess the efficiency of these methods on plasmid DNA, genomic RNA, experimentally infected samples, and naturally infected samples.
Materials and Methods
Samples and preparation of RNA template
The samples of the duck's yolk membranes and cloacal swabs were obtained from ducks that suffered from severe cases of egg production drop or oophoritis from various duck farms in Shandong, China. The tissue samples were homogenized or eluted in sterile phosphate-buffered saline (PBS, pH 7.2) to give a 20% suspension (w/v). After centrifugation at 6000 g for 30 min, the supernatants were filtered through a 0.2 μm syringe-driven filter. The filtered suspension was used for direct RNA extraction and virus isolation (Tang et al., 2011). TMUV Shandong strain was obtained from the Poultry Disease Institute of Shandong Agricultural University (PDSDAU). RNA extraction was carried out with a RNA extraction Kit (TIANGEN, Beijing, China) following the manufacturer's instructions. The genomic RNA was determined by DU 800 (Beckman Coulter, Brea, CA, USA) and used to make standard dilutions.
Cloning of partial TMUV-NS5 gene
To determine the sensitivity of the TMUV detection assays, a 1084 bp specific genomic fragment was amplified by RT-PCR using the primers FU1 and cFD3 (Kuno et al., Citation1998). The reverse transcription step was performed using the cFD3 primer and carried out in 10 μL volume containing 5 μL of extracted RNA, 2 μL of 5 × RT Buffer, 0.5 μL of dNTP mixture (10 mM each dNTP), 20U RNase Inhibitor (TAKARA, Dalian, China), 1 μM cFD3 primer and 5U AMV Reverse Transcriptase (TAKARA). The reaction mixture was heated at 42 °C for 20 min, 99°C for 5 min, and 5°C for 5 min. Following cDNA synthesis, a PCR reaction mix containing 5 μL of 10 × EX Taq Buffer (Mg2+ Plus), 1 μL of dNTP mixture (10 mM each dNTP), 5U EX Taq polymerase (TAKARA) and 2 μL of each of the two primers (FU1/cFD3) was brought to 40 μL with RNase-free water and added directly to the 10 μL RT tubes. The thermal cycling programme consisted of an initial denaturation step at 94°C for 4 min, followed by 35 cycles of denaturation (30 s at 94°C), annealing (30 s at 50°C) and extension (60 s at 72°C), with a final extension step for 5 min at 72°C. The PCR product was purified and cloned into the pMD18-T vector (TAKARA) following the manufacturer's instructions. The concentration of the recombinant plasmid, designated pMD18-T-NS5, was determined by DU 800 (Beckman Coulter) and used to make standard dilutions to test the lower detection limit of the different assays.
Design of primers
The primers used in this study are summarized in . A diagram of the locations of the primers is shown in . The RT-LAMP primers and RT-PCR primers were designed as described by our previous study (Tang et al., 2011). The SN-RT-PCR primer (SNR) and Q-RT-PCR primers (QF/QR) were manually designed according to the sequence of NS5 gene on TMUV-YY5 genomic sequence (GenBank accession no. HQ641390) (Cao et al., Citation2011). Their respective melting temperatures (Tm) and potential secondary structures were evaluated using PRIMERSELECT® in LASERGENE® gene analysis software (DNASTAR Inc., Madison, WI, USA) in order to select suitable primer pairs.
Figure 1. Locations of the selected primers in the NS5 gene (strainYY5, GenBank accession number HQ641390). Primers FU1 and cFD3 were used to amplify a 1084 bp segment of the NS5 gene. Primers F3 and B3 were used to amplify a 211 bp segment of the NS5 gene in C-RT-PCR and also used as external primers in RT-LAMP method. Primers FIP and BIP were used in RT-LAMP as internal primers. Primers F3 and SNR were used to amplify a 139 bp segment of the NS5 gene in the second stage of SN-RT-PCR. Primers QF and QR were used in Q-RT-PCR.
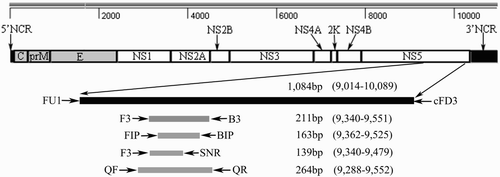
Table 1. Primers used for TMUV detection methods.
The RT-LAMP conditions
The RT-LAMP reaction was carried out by a previously utilized method (Tang et al., Citation2011b). Briefly, the reaction volume was 25 μL and contained 1.6 μM each of FIP and BIP primer, 0.2 μM each of F3 and B3 primer, 1.6 mM dNTPs, 1 M betaine (Solarbio, Beijing, China), 4 mM MgSO4, 10 × ThermoPol reaction buffer, 8 U of Bst DNA polymerase (New England Biolabs. Inc., MA, USA), 5U of AMV reverse transcriptase (TAKARA) and 2 μl of RNA template. The reaction mixture was incubated at 63°C for 60 min in a water bath, and then heated to 80°C for 10 min to terminate the reaction. The LAMP products were electrophoresed on a 2% (w/v) agarose gel in order to determine detection results.
The RT-PCR and semi-nested RT-PCR conditions
The C-RT-PCR/first-stage SN-RT-PCR for the amplification of the NS5 gene was performed with cDNA or plasmid DNA using 20 pmol of each LAMP outer primer (F3/B3). The PCR reaction mix containing 5 μL of 10 × EX Taq Buffer (Mg2+ Plus), 1 μL of dNTP mixture (10 mM each dNTP), 5U EX Taq polymerase (TAKARA), and 1 μL template DNA was brought to 50 μL with RNase-free water. The thermal cycling programme involved an initial denaturation at 94°C for 5 min, followed by 35 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, with a final extension for 10 min at 72°C. The second stage of SN-RT-PCR was performed with 2 μL of the product resulting from the first stage. The reaction mix contained 5 μL of 10 × EX Taq Buffer (Mg2+ Plus), 1 μL of dNTP mixture (10 mM each dNTP), 5U EX Taq polymerase, and RNase-free water added to give a final volume of 50 μL. The thermal cycling programme conditions were identical to those used in the first stage. The PCR products were electrophoresed on a 2% (w/v) agarose gel in order to determine detection results.
Real-time quantitative RT-PCR conditions
The real-time quantitative RT-PCR for the NS5 gene was conducted in a 20 μL reaction system containing the following ingredients: 10 μL of 2 × SYBR® premix EX Taq buffer (TAKARA) (reaction buffer, EX Taq DNA polymerase, MgCl2, SYBR® Green I, and dNTPs), 1 μL of each primer (10 μM of QF and QR), 2 μL of diluted plasmid, and 6 μL of sterile water. The reaction mixture was applied to 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). The data for the Q-RT-PCR were analyzed using the SDS software program (version 1.4). The thermal profile of Q-RT-PCR proceeded as follows: an initial denaturation step at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s and annealing and elongation at 60°C for 60 s, followed by the dissociation programme for measuring the Tm of PCR products. Samples with a cycle threshold (CT) of 35 or lower showing a single TM peak were classified as positive for Q-RT-PCR.
Sensitivity and specificity
To compare the sensitivity of the four assays using plasmid DNA and genomic RNA, RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP were performed with serial dilutions of plasmid DNA from 2 × 108 copies/μL to 2 × 101 copies/μL and serial dilutions of genomic RNA from 100 ng/tube to 10 fg/tube. In addition, three strains of TMUV and one of each of avian reovirus (ARV), infectious bursal disease virus (IBDV), avian infectious bronchitis virus (IBV), avian influenza virus (AIV) subtype H9N2, Newcastle disease virus (NDV), Egg-drop syndrome 1976 virus (EDS-76), Duck circovirus (DuCV), Duck reovirus (DRV), Duck parvovirus (DPV) and Duck hepatitis A virus (DHAV) were also tested to show the specificity of these assays. The list of strains is shown in .
Table 2. Viruses used in this study.
Comparative analysis of the detection methods using clinical samples
To evaluate the performances of the four assays in detecting TMUV in clinical samples, 25 cloacal swabs of experimental TMUV infected ducks (25 ducks were simultaneously infected by intramuscular injection with 0.8 mL of the TMUV with titre of 104.9 ELD50/mL, the cloacal swabs were collected at day 4 post-infection) and 69 cloacal swabs from suspected TMUV infected ducks collected from different areas of Shandong province were tested in all of four evaluated methods and virus isolation. In order to determine that the samples testing positive were true positives, amplification products of RT-PCR, SN-RT-PCR and Q-RT-PCR were purified using a gel extraction kit (TransGen, Beijing, China) and submitted to the commercial facility (BGI, Shenzhen, Guangdong, China) for sequencing. Ten cloacal swab samples from different healthy ducks worked as the negative controls in the test. The duck infection experiments were conducted with the approval of Animal Ethics Committee of Shandong Agriculture University.
Results
Sensitivity
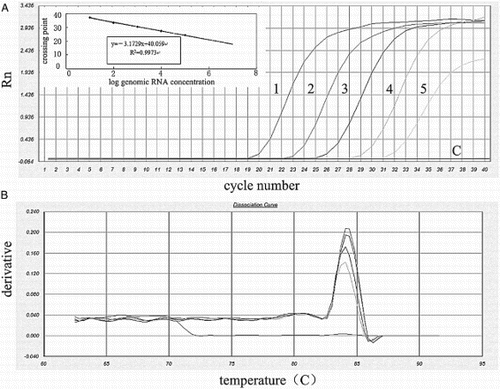
When plasmid DNA was used as a template, the sensitivity of C-RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP were 2 × 104, 20, 2 and 20 plasmid copies/μL, respectively (). When genomic RNA was used as a template, the sensitivity of C-RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP were 100 pg, 100, 10, and 100 fg per reaction, respectively (). The detection limit of the SN-RT-PCR and RT-LAMP methods was 103 times lower than that of the C-RT-PCR in both diluted plasmid and genomic RNA samples, although the same locus of F3 and B3 primers was targeted in all of three assays. The Q-RT-PCR assay was demonstrated to be 10 times more sensitive than the SN-RT-PCR and RT-LAMP assays ().
Figure 2. Detection sensitivity of C-RT-PCR, SN-RT-PCR, and RT-LAMP. Lane M, DL2000 DNA marker (TransGen); lane 1 and lane 8, negative control (sterile distilled water); lane 2–7, C-RT-PCR, SN-RT-PCR, and RT-LAMP conducted using 10-fold serial dilutions of standard plasmid DNA, pMD18-T-NS5: 2 × 105,2 × 104,2 × 103,2 × 102,2 × 101, and 2 × 100 copies/μL, respectively; lane 9–14, C-RT-PCR, SN-RT-PCR, and RT-LAMP conducted using 10-fold serial dilutions of TMUV genomic RNA: 100, 10, 1 pg/tube, 100 fg, 10 fg and 1 fg/tube, respectively.
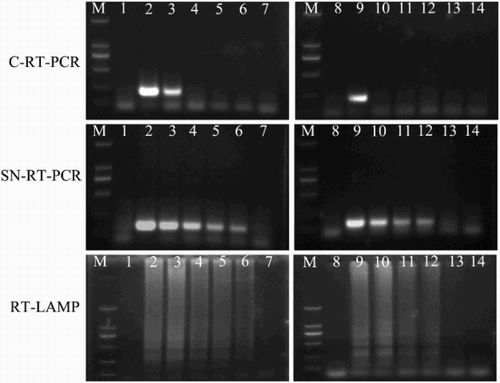
Figure 3. Detection sensitivity of Q-RT-PCR using 10-fold serial dilutions of genomic RNA. (A) Detection sensitivity of Q-RT-PCR. line 1, 100 pg/tube genomic RNA (CT, 19.1); line 2, 10 pg/tube genomic RNA (CT, 22.5); line 3, 1 pg/tube genomic RNA (CT, 25.9); line 4, 100 fg/tube genomic RNA (CT, 29.3); line 5, 10 fg/tube genomic RNA (CT, 31.4); and line C, D/W. The standard curve (insert) was generated using logs of genomic RNA concentration versus cycle threshold of each sample. (B) Representative SYBR Green dissociation curves of amplified products to determine the melting temperature values.
Specificity
To determine the specificity of C-RT-PCR, SN-RT-PCR, Q-RT-PCR, and RT-LAMP methods for the detection of TMUV NS5 gene, different reactions were performed using DNA or RNA from ARV, IBDV, IBV, AIV (H9), NDV, DuCV, DRV, DPV and DHAV as templates. Agarose gel electrophoresis analysis showed no visible band when any of these viruses were used as templates in C-RT-PCR, SN-RT-PCR, and RT-LAMP. The Q-RT-PCR result showed that the high CT values were above the cut-off point (CT > 35) for most of the test viruses (ARV, IBDV, IBV, AIV-H9, NDV, EDS-76, DuCV, DRV, DPV, and DHAV), except for three strains of TMUV (CT < 23.2). The results of specificity tests are shown in .
Comparison of the four assays in samples from artificially and naturally infected ducks
When cloacal swabs from experimentally TMUV infected ducks were used, C-RT-PCR (targeting the NS5 gene) had a detection rate of 52% (13/25 samples). In contrast, SN-RT-PCR, Q-RT-PCR, and RT-LAMP (also targeting NS5 gene) had detection rates of 92% (23/25), 88% (22/25), and 88% (22/25), respectively. One positive sample was negative by Q-RT-PCR and RT-LAMP methods. No virus could be detected in any of the cloacal swabs collected before day 4 post-inoculation. Of the 69 cloacal swabs from suspected TMUV infected ducks, 57 samples (82.6%) tested positive by Q-RT-PCR assay, 52 (75.4%) by SN-RT-PCR, and 55 (79.7%) by RT-LAMP, 38 (55.1%) by RT-PCR and 33 (47.8%) by virus isolation. Only five of the 69 tissue samples tested differently in SN-RT-PCR, Q-RT-PCR, and RT-LAMP. By sequencing amplified products of the C-RT-PCR, SN-RT-PCR and Q-RT-PCR, all the positive samples were confirmed to be TMUV NS5-related sequence. Because all of the C-RT-PCR, SN-RT-PCR and RT-LAMP assays were based on the same primers F3 and B3 (no primer mismatches) and all normal ducks tested negative, the result of the evaluated assays were shown to be specific for TMUV clinical samples. The detection results of clinical samples are listed in .
Table 3. Comparison of C-RT-PCR, SN-RT-PCR, Q-RT-PCR, RT-LAMP, and virus isolation for detection of TMUV in clinical samples and experimental infection samples.
Discussion
TMUV has spread very fast among ducks causing high morbidity and mortality in East China since 2010 (Yan et al., Citation2011b). Rapid diagnostic methods are crucial for successful TMUV infection control and prevention, such as emergency vaccination and infected duck quarantine. Several studies have reported molecular methods for the detection of TMUV (Yan et al., Citation2011a; Jiang et al., Citation2012; Liu et al., Citation2013). Nevertheless, each study used a single method and different types of samples; to our knowledge, only one comparative study has been carried out for TMUV detection (Yan et al., Citation2012), making the choice of a suitable assay inadequate.
Most of the molecular assays have their own strengths and limitations. The N-RT-PCR or SN-RT-PCR amplification requires only several hundreds of picogram (pg) of RNA and 104 to 102 times more sensitive than C-RT-PCR but doubles the reaction time and increases the risk of cross-contamination (Lee et al., Citation1998). Despite the high cost, Q-RT-PCR is useful for rapid detection of viruses because it takes less than 2 h and is helpful in evaluating the response to treatment and clinical outcome (Bustin, Citation2000; Lang et al., Citation2009). The RT-LAMP can amplify RNA sequences without reverse-transcriptase step (Notomi et al., Citation2000) with high sensitivity and requires little time (within 1 h). However, RT-LAMP requires primers with high specificity (mutations in the primer region, especially for virus, will abolish detection) (Nagamine et al., Citation2002). Also due to the production of a large number of amplifications, cross-contamination is more likely in LAMP reaction (Hong et al., Citation2004).
In this study, the sequence of the genomic regions (approximately 1 kb long) at the 3’ terminus of the NS5 gene was used to design primers. NS5 is the largest of the Flavivirus proteins (105 kDa). The RNA-dependent RNA polymerase (RdRp) domain in C-terminal of NS5 protein is highly conserved (Tan et al., Citation1996; Ackermann & Padmanabhan, Citation2001). For this reason, most Flavivirus detection methods have targeted the conserved region of NS5 gene (Fulop et al., Citation1993; Kuno et al., Citation1996). The four assays in this study also targeted NS5 gene sequence that was known to be conserved in the SDHS, YY5, BYD, FX2010, and JS804 strains of TMUV.
The detection limits for both genomic RNA and plasmid DNA of Q-RT-PCR, SN-RT-PCR, and RT-LAMP were low (from 2 copies/μL to 20 copies/μL), but C-RT-PCR showed a significantly high limit. With clinical cloacal swabs, more than 20% samples were tested negative by C-RT-PCR, but tested positive in the other three assays. This is significant considering the amount of TMUV in cloacal swabs (>30%) ranged from 2 copies/μL to 2 × 104 copies/μL, that could remain undetected by both C-RT-PCR and virus isolation. Indeed, the Tm values of productions of cloacal swabs and other two nucleic acid templates were identical in melting curve analysis, which indicated that the established Q-RT-PCR was specific and stable in TMUV clinical cloacal swabs detection.
In conclusion, TMUV RT-LAMP assay requires simple equipment and results that can be obtained within 1 h. The Q-RT- PCR is more sensitive than RT-LAMP assay but is also more time-consuming (about 2 h) and would require advanced equipment. Both of these two assays can provide the rapid diagnosis of TMUV infection, but RT-LAMP is more useful in TMUV field situations or poorly equipped laboratories.
Funding
This research was supported by the China Agriculture Research System [grant number CARS-43-10], National Natural Science Foundation of China [grant numbers 31272583 and 31472199], and Natural Science Foundation of Shandong [grant number ZR2012CM002].
References
- Ackermann, M. & Padmanabhan, R. (2001). De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. Journal of Biological Chemistry, 276, 39926–39937.
- Bhatnagar, J., Guarner, J., Paddock, C.D., Shieh, W.J., Lanciotti, R.S., Marfin, A.A., Campbell, G.L. & Zaki, S.R. (2007). Detection of West Nile virus in formalin-fixed, paraffin-embedded human tissues by RT-PCR: a useful adjunct to conventional tissue-based diagnostic methods. Journal of Clinical Virology, 38, 106–111.
- Bustin, S.A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology, 25, 169–193.
- Cao, Z., Zhang, C., Liu, Y., Ye, W., Han, J., Ma, G., Zhang, D., Xu, F., Gao, X., Tang, Y., Shi, S., Wan, C., Zhang, C., He, B., Yang, M., Lu, X., Huang, Y., Diao, Y., Ma, X. & Zhang, D. (2011). Tembusu virus in ducks, China. Emerging Infectious Diseases Journal, 17, 1873–1875.
- Fulop, L., Barrett, A.D., Phillpotts, R., Martin, K., Leslie, D. & Titball, R.W. (1993). Rapid identification of flaviviruses based on conserved NS5 gene sequences. Journal of Virological Methods, 44, 179–188.
- Gould, E.A., Buckley, A. & Cammack, N. (1985). Use of the biotin-streptavidin interaction to improve Flavivirus detection by immunofluorescence and ELISA tests. Journal of Virological Methods, 11, 41–48.
- Hong, T.C., Mai, Q.L., Cuong, D.V., Parida, M., Minekawa, H., Notomi, T., Hasebe, F. & Morita, K. (2004). Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. Journal of Clinical Microbiology, 42, 1956–1961.
- Jiang, T., Liu, J., Deng, Y.Q., Su, J.L., Xu, L.J., Liu, Z.H., Li, X.F., Yu, X.D., Zhu, S.Y., Gao, G.F., Qin, E.D. & Qin, C.F. (2012). Development of RT-LAMP and real-time RT-PCR assays for the rapid detection of the new duck Tembusu-like BYD virus. Archives of Virology, 157, 2273–2280.
- Kono, Y., Tsukamoto, K., Abd Hamid, M., Darus, A., Lian, T.C., Sam, L.S., Yok, C.N., Di, K.B., Lim, K.T., Yamaguchi, S. & Narita, M. (2000). Encephalitis and retarded growth of chicks caused by Sitiawan virus, a new isolate belonging to the genus Flavivirus. American Journal of Tropical Medicine and Hygiene, 63, 94–101.
- Koraka, P., Zeller, H., Niedrig, M., Osterhaus, A.D. & Groen, J. (2002). Reactivity of serum samples from patients with a Flavivirus infection measured by immunofluorescence assay and ELISA. Microbes and Infection Journal, 4, 1209–1215.
- Kuno, G., Chang, G.J., Tsuchiya, K.R., Karabatsos, N. & Cropp, C.B. (1998). Phylogeny of the genus Flavivirus. Journal of Virology, 72, 73–83.
- Kuno, G., Mitchell, C.J., Chang, G.J. & Smith, G.C. (1996). Detecting bunyaviruses of the Bunyamwera and California serogroups by a PCR technique. Journal of Clinical Microbiology, 34, 1184–1188.
- Lang, J.E., Magbanua, M.J., Scott, J.H., Makrigiorgos, G.M., Wang, G., Federman, S., Esserman, L.J., Park, J.W. & Haqq, C.M. (2009). A comparison of RNA amplification techniques at sub-nanogram input concentration. BMC Genomics, 10, 326.
- Lee, S.E., Kim, S.Y., Kim, S.J., Kim, H.S., Shin, J.H., Choi, S.H., Chung, S.S. & Rhee, H.H. (1998). Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. Journal of Clinical Microbiology, 36, 2887–2892.
- Lindenbach, B.D. & Rice, C.M. (2003). Molecular biology of Flaviviruses. Advances in Virus Research, 59, 23–61.
- Liu, M., Chen, S., Chen, Y., Liu, C., Chen, S., Yin, X., Li, G. & Zhang, Y. (2012). Adapted Tembusu-like virus in chickens and geese in China. Journal of Clinical Microbiology, 50, 2807–2809.
- Liu, Z., Fu, Y., Ji, Y., Wei, J., Cai, X. & Zhu, Q. (2013). Development and validation of one-step SYBR green real-time RT-PCR for the rapid detection of newly emerged duck Tembusu virus. Avian Diseases, 57, 595–601.
- Mukhopadhyay, S., Kuhn, R.J. & Rossmann, M.G. (2005). A structural perspective of the Flavivirus life cycle. Nature Reviews Microbiology, 3, 13–22.
- Nagamine, K., Hase, T. & Notomi, T. (2002). Accelerated reaction by loop-mediated isothermal amplification using loop primers. Molecular and Cellular Probes, 16, 223–229.
- Nagarkatti, P.S. & Nagarkatti, M. (1980). Comparison of haemagglutination inhibition (HI) and indirect fluorescent antibody (IFA) techniques for the serological diagnosis of certain Flavivirus infections. Journal of Tropical and Medical Hygiene, 83, 115–117.
- Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N. & Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 28, E63.
- Platt, G.S., Way, H.J., Bowen, E.T., Simpson, D.I., Hill, M.N., Kamath, S., Bendell, P.J. & Heathcote, O.H. (1975). Arbovirus infections in Sarawak, October 1968-February 1970 Tembusu and Sindbis virus isolations from mosquitoes. Annals of Tropical Medicine and Parasitology, 69, 65–71.
- Scaramozzino, N., Crance, J.M., Jouan, A., DeBriel, D.A., Stoll, F. & Garin, D. (2001). Comparison of Flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. Journal of Clinical Microbiology, 39, 1922–1927.
- Su, J., Li, S., Hu, X., Yu, X., Wang, Y., Liu, P., Lu, X., Zhang, G., Hu, X., Liu, D., Li, X., Su, W., Lu, H., Mok, N.S., Wang, P., Wang, M., Tian, K. & Gao, G.F. (2011). Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related Flavivirus. PLoS One, 6, e18106.
- Tan, B.H., Fu, J., Sugrue, R.J., Yap, E.H., Chan, Y.C. & Tan, Y.H. (1996). Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology, 216, 317–325.
- Tang, Y., Diao, Y., Gao, X., Yu, C., Chen, L. & Zhang, D. (2011a). Analysis of the complete genome of Tembusu virus, a Flavivirus isolated from ducks in China. Transboundary and Emerging Diseases, 59, 336–343.
- Tang, Y., Diao, Y., Yu, C., Gao, X., Chen, L. & Zhang, D. (2011b). Rapid detection of Tembusu virus by Reverse-Transcription, Loop-mediated Isothermal Amplification (RT-LAMP). Transboundary and Emerging Diseases, 59, 208–213.
- Yan, L., Peng, S., Yan, P., Zhou, J., Teng, Q., Li, G., Li, X. & Li, Z. (2012). Comparison of real-time reverse transcription loop-mediated isothermal amplification and real-time reverse transcription polymerase chain reaction for duck Tembusu virus. Journal of Virological Methods, 182, 50–55.
- Yan, L., Yan, P., Zhou, J., Teng, Q. & Li, Z. (2011a). Establishing a TaqMan-based real-time PCR assay for the rapid detection and quantification of the newly emerged duck Tembusu virus. Virology Journal, 8, 464.
- Yan, P., Zhao, Y., Zhang, X., Xu, D., Dai, X., Teng, Q., Yan, L., Zhou, J., Ji, X., Zhang, S., Liu, G., Zhou, Y., Kawaoka, Y., Tong, G. & Li, Z. (2011b). An infectious disease of ducks caused by a newly emerged Tembusu virus strain in mainland China. Virology, 417, 1–8.
- Yao, E. & Tavis, J.E. (2005). A general method for nested RT-PCR amplification and sequencing the complete HCV genotype 1 open reading frame. Virology Journal, 2, 88.
- Yin, X., Lv, R., Chen, X., Liu, M., Hua, R. & Zhang, Y. (2013). Detection of specific antibodies against Tembusu virus in ducks by use of an E protein-based enzyme-linked immunosorbent assay. Journal of Clinical Microbiology, 51, 2400–2402.
- Yun, T., Ni, Z., Hua, J., Ye, W., Liu, C., Zhang, S., Zhang, Y. & Zhang, C. (2012). Development of a one-step real-time RT-PCR assay using a minor-groove-binding probe for the detection of duck Tembusu virus. Journal of Virology Methods, 85, 370–377.
