ABSTRACT
A distinctive infectious bursal disease (IBD) virus genotype (ITA) was detected in IBD-live vaccinated broilers in Italy without clinical signs of IBD. It was isolated in specific-pathogen-free eggs and molecularly characterized in the hypervariable region of the virus protein (VP) 2. Phylogenetic analysis showed that ITA strains clustered separately from other homologous reference sequences of IBDVs, either classical or very virulent, retrieved from GenBank or previously reported in Italy, and from vaccine strains. The new genotype shows peculiar molecular characteristics in key positions of the VP2 hypervariable region, which affect charged or potentially glycosylated amino acids virtually associated with important changes in virus properties. Characterization of 41 IBDV strains detected in Italy between 2013 and 2014 showed that ITA is emergent in densely populated poultry areas of Italy, being 68% of the IBDV detections made during routine diagnostic activity over a two-year period, in spite of the immunity induced by large-scale vaccination. Four very virulent strains (DV86) and one classical strain (HPR2), together with eight vaccine strains, were also detected. The currently available epidemiological and clinical data do not allow the degree of pathogenicity of the ITA genotype to be defined. Only in vivo experimental pathogenicity studies conducted in secure isolation conditions, through the evaluation of clinical signs and macro/microscopic lesions, will clarify conclusively the virulence of the new Italian genotype.
Introduction
Infectious bursal disease virus (IBDV) belongs to the family Birnaviridae, within the genus Avibirnavirus. IBDV has a non-enveloped virion with icosahedral symmetry and a diameter varying from 55 to 65 nm. Two serotypes of IBDV are known: serotype 1, further categorized into classical, attenuated, very virulent (vvIBDV) and antigenic variant strains, and serotype 2, including only avirulent strains (Kibenge et al., Citation1988). Antigenic variant strains, recognized in the USA in the early 1980s, are able to cause disease in the face of immunity to classical viruses (Rosenberger & Cloud, Citation1986).
Independent of the pathogenicity of the strain and the severity of clinical signs, IBDV infection is always associated with damage to the bursa of Fabricius and immunosuppression, which is more severe if the infection occurs during the first three weeks of age in birds with low levels of maternally derived antibodies.
The virus has a bi-segmented double-stranded RNA genome. Segment A (3300 base pairs) encodes a polyprotein that is self-cleaved to yield virus protein (VP) 4 protease, the structural proteins VP2 (major capsid protein) and VP3, and the non-structural protein VP5. Genome segment B (2800 base pairs) encodes VP1, the RNA dependent-RNA polymerase, which complexes with VP3 to initiate capsid formation (Eterradossi & Saif, Citation2013).
Inside the amino acid sequence of the VP2 protein, a hypervariable region is recognized between aa 206 and 350. This sequence includes two major hydrophilic peaks, A and B, at positions 212–224 and 314–324, respectively, and two minor hydrophilic peaks, C and D, at positions 249–254 and 279–289, respectively. Most of the amino acid changes observed among IBDV strains, which are associated with antigenic and virulence variations, are clustered in these regions (Nagaraian & Kibenge, Citation1997).
IBDV affects chickens and is widespread in nearly all commercial poultry-producing countries around the world. The first report of IBD in Italy dates back to 1965 (Rinaldi et al., Citation1965); since then the disease has become endemic in the country and live vaccination of young birds was introduced (Coletti et al., Citation1983; Asdrubali & Franciosini, Citation1993). In the early 1990s the appearance of vvIBDVs in Europe challenged the poultry industry and traditional vaccine schemes became insufficient to control the disease (Chettle et al., Citation1989; Eterradossi et al., Citation1990; Asdrubali & Franciosini, Citation1993; Tosi et al., Citation1997).
More recently, the use of sequence analysis has allowed genotyping of Italian IBDV isolates and gave a clearer epidemiological picture. Predominant circulation of very virulent strains has been confirmed, and the presence of atypical classical strains reported (Moreno et al., Citation2007, Citation2010a), despite the application of intermediate and intermediate plus live vaccines, and inactivated booster in breeders.
In this study, we report on the isolation of a genetically distinctive IBDV in Italy and on its diffusion in Italian broiler farms.
Materials and methods
Samples
Longitudinal studies
Two longitudinal studies were performed in broiler flocks with a history of poor growth performance and secondary Escherichia coli infections suspected to be related to immunosuppression by IBDV without overt clinical signs. They belonged to the same integrated poultry company and were located in densely populated poultry areas (DPPA) of two different Northern Italian regions, Veneto and Lombardia. Flock 1 had been vaccinated against IBD with a commercial intermediate live vaccine Nobilis Gumboro D78 Live (MSD Animal Health, Milton Keynes, UK) at 16 and 24 days of life (d.o.l.). Flock 2 received the vaccines Bursine 2 (Zoetis, Campus Drive, NJ, USA) and Nobilis Gumboro D78 Live at 15 and 20 d.o.l., respectively. Samples were collected twice in both groups: at 21 and 33 d.o.l. in flock 1 and at 17 and 35 d.o.l. in flock 2.
On each sampling time, from eight birds, bursae of Fabricius were collected aseptically and processed in pools of four each for IBDV-RNA detection by RT-PCR and virus isolation in specific-pathogen-free (SPF) eggs.
Bursal homogenates were prepared from pools as a 20% (w/v) suspension in Minimal Essential Medium (MEM) containing antibiotics and antimycotics (penicillin, streptomycin and amphotericin B) (Applied Biosystems, CA, USA). After a two-hour incubation period at 4°C the suspension was centrifuged at 3000 × g for 20 min. Supernatants were stored at −80°C until processing for RNA extraction, RT-PCR, DNA sequencing and virus isolation as described below.
IBDV field isolates
41 IBDV strains were obtained in 2013–2014 from broiler flocks located in Northern Italian DPPA during routine molecular diagnostic activity. All detections originated from birds vaccinated for IBD (), with poor growth performances without overt IBDV-like clinical signs. RNA extraction, RT-PCR and sequence analysis of the hypervariable region in the VP2 gene were performed as described below. Molecular typing of the IBDV sequences was determined using the nucleotide BLAST algorithm with GenBank database (http://www.ncbi.nlm.nih.gov) (Johnson et al., Citation2008).
RNA extraction and RT-PCR for IBDV
RNA was extracted from supernatants using the QIAamp® Viral RNA Mini kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instructions. RT-PCR was performed using the forward primer 2 (5′-GCCCAGAGTCTACACCAT) and the reverse primer 5 (5′- CCCGGATTATGTCTTTGA) that amplify a 742-bp fragment from nt 737 to 1479 of the hypervariable region of the VP2 gene (Jackwood et al., Citation2008). RT was performed using ImProm-IITM Reverse Transcriptase (Promega, Milan, Italy) in a 20 μl final volume containing 4 μl of 5X reaction buffer, 2.4 μl of MgCl2 (3 mM), 1 μl of dNTP (0.5 mM), 1 μl of the reverse primer, 3.1 μl of nuclease free water and 7 μl of RNA. The RT mix was incubated at 42°C for 60 min then heated at 70°C for 15 min. PCR was carried out using Go Taq® DNA Polymerase (Promega, Milan, Italy) in a final 50-μl volume containing 10 μl of 5X Go Taq® Flexi Buffer (Promega, Milan, Italy), 3.5 μl of MgCl2, 1 μl of dNTP (0.2 mM), 1 μl of each primer, 25 μl of nuclease free water and 8 μl of cDNA. The PCR cycling parameters were: a precycle step at 95°C for two min followed by 35 cycles consisting of 95°C for 1 min, 55°C for 1 min, 72°C for 1 min. The last cycle was followed by a final extension step of 72°C for 5 min.
DNA sequencing and sequence analysis
The Wizard® SV Gel and PCR Clean-Up System (Promega, Milan, Italy) was used to purify RT-PCR products, according to the manufacturer’s instructions. Sequencing was performed in both directions using the PCR primers at Bio-Fab Research (Rome, Italy), with the 96-capillary 3730xl DNA Analyzer (Applied Biosystems, CA, USA).
Nucleotide sequences were edited using Bioedit software, aligned and compared with sequences of IBDV strains previously published (Moreno et al., Citation2007) or retrieved from GenBank database (), using ClustalW software (Larkin et al., Citation2007).
Table 1. Accession numbers and molecular type of IBDV sequences retrieved from GenBank used in the analysis.
JModelTest 2.1.2 (Darriba et al., Citation2012) and ProtTest 3.2 (Darriba et al., Citation2011) software were used to select, according to the Bayesian Information Criterion, the most appropriate evolution model to generate the phylogenetic tree for nucleotide and deduced amino acid sequences, respectively. Phylogenetic trees were then generated by the Maximum Likelihood method using the PhyML 3.0 software (Guindon et al., Citation2010). Bootstrap values, obtained with 1000 replicates, were considered significant when >70.
Virus isolation in SPF eggs
Virus isolation was attempted from one selected bursal homogenate that tested positive for IBDV by RT-PCR. The supernatant (0.2 ml/egg) was used to inoculate ten 12-day-old SPF embryonated chicken eggs via the chorioallantoic membrane (CAM) route (Senne, Citation2008). The eggs were incubated at 37.7°C until embryo death was observed or up to 7 days. From the eggs suspected to be infected due to embryo death or embryo lesions, the CAMs were aseptically harvested, homogenized, pooled and prepared as 20% (w/v) suspensions as described above. IBDV isolation was confirmed by the RT-PCR described above.
Accession numbers
The partial sequences of the VP2 gene of four strains detected in longitudinal studies were deposited under the GenBank accession numbers JN852985.1, JN852986.1, JN852987.1, JN852988.1.
Results
Detection and molecular characterization of the unique IBDV genotype ITA
RT-PCR resulted positive for IBDV in four out eight pools analysed; strains were named ITA01 (flock 1, 21 d.o.l.) ITA02 (flock 2, 17 d.o.l.), ITA03 and ITA04 (flock 2, 36 d.o.l.). Virus isolation of strain ITA01 was attempted in SPF embryonated eggs and resulted in embryo death on day 5–7 post-inoculation, associated with oedema and congestion.
The VP2 sequences (nucleotide positions 737–1479) of the detected strains were determined and the amino acid sequences deduced (). Sequence analysis showed that the detected strains had >99.9% nucleotide, and 100% amino acid identity between themselves and unique molecular characteristics compared to vaccines and reference strains. None of the sequences of the VP2 hypervariable region had the 253-H and 284-T mutations typically found in attenuated vaccine strains (Mundt, Citation1999; Brandt et al., Citation2001). Furthermore, the comparison of our sequences to known sequences of vaccine strains showed <93% identity with the vaccines, respectively, used in each sampled flocks.
Figure 1. Deduced amino acid sequences of the VP2 hypervariable region of IBDV strains detected in the study (grey box) and reference strains. Only residues that differ from the sequence of the F52/70 are shown.
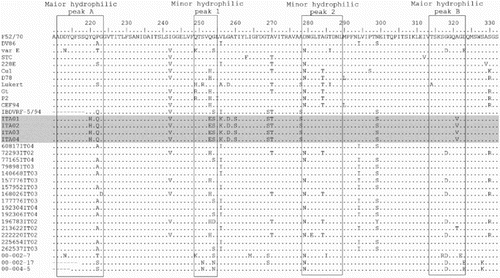
Compared to classic and very virulent reference IBDVs, amino acid sequences of ITA strains showed substitution mutations in three out of four hydrophilic peaks: A, B and minor peak 2. The most notable mutations were observed at amino acid positions 220, 222, 253, 254 and 321. At position 222, in hydrophilic peak A, the strains had glutamine (hydrophilic and negative charge) instead of alanine (hydrophobic) observed in vvIBDV strains and proline (hydrophobic and neutral) in classic IBDV strains. A substitution from tyrosine (hydrophobic) to histidine (hydrophilic and positive charge) was observed at position 220. At positions 253 and 254 the mutations were from glutamine (hydrophilic and negative charged) and glycine (aliphatic) to glutamate (hydrophilic and negative charged) and serine (hydrophilic and neutral), respectively. The substitution in position 254 was reported for other Italian and European IBDV strains (Jackwood et al., Citation2006; Moreno et al., Citation2007). In position 321, an alanine is replaced by a valine.
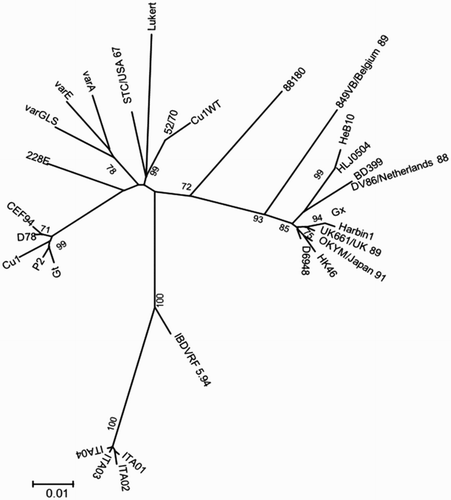
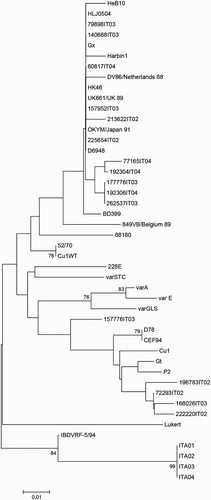
Phylogenetic trees based on nucleotide () and amino acid () sequences both showed that ITA strains clustered together on a completely separated branch (bootstrap 100) from the classical, very virulent, variant and vaccine IBDV strains included in the analysis.
Molecular characterization of IBDVs circulating in Italy
Characterization of IBDV strains detected in Italy between 2013 and 2014 is displayed in . The majority of the genotyped strains (28 out of 41) showed the highest similarity (>99%) to ITA strains; four to the very virulent strain DV86 (>97%); one to the classical strain HPR2 (>96%).
Table 2. Year and age of birds at detection, vaccination plan applied to the flock and molecular type of the IBDV strains characterized in the study.
Eight strains were vaccine detections, showing 100% nucleotide identity to published vaccine sequences.
Discussion
A distinctive IBDV genotype (ITA), detected in Italy in live vaccinated broilers without clinical signs of IBD, has been molecularly characterized in the hypervariable region of the VP2 protein. It seems to be emergent in Italian DPPA being the 68% of the IBDV detections made during routine diagnostic activity over a two-year period.
Phylogenetic analysis showed that ITA strains clustered separately from other homologous reference sequences of IBDVs, either classical or very virulent, retrieved from GenBank or previously reported in Italy (Moreno et al., Citation2007) and from vaccine strains. Mutations characteristic of these latter attenuated strains, such as a glutamine at position 253 and a threonine at position 284 (Mundt, Citation1999; Brandt et al., Citation2001) have indeed not been observed in our ITA strains.
A previous European survey reported the circulation, in France and Spain, of IBDV strains molecularly similar to the USA variants (Jackwood et al., Citation2006). None of the nucleotide substitutions characterizing those strains have been observed in the ITA genotype.
The new genotype shows peculiar molecular characteristics, which have never been observed before, in key positions of the VP2 hypervariable region, where substitution mutations responsible for antigenic drift or virulence occur. In particular the ITA genotype, possess amino acid mutations at positions 222 and 254 which have been reported to allow the virus to circumvent induced immunity (Heine et al., Citation1991; Jackwood et al., Citation2006; Jackwood & Sommer-Wagner, Citation2011; Jackwood & Stoute, Citation2013). Most of the observed mutations affected charged or potentially glycosylated amino acids and therefore could be associated with important changes in virus properties. In the case of virus changes involving charged amino acids, mutations are likely to affect the ability of antibodies and other immune cells to recognize the virus antigen.
Our results show that ITA strains are widespread in Italy in spite of the immunity induced by large-scale vaccination, thus allowing us to hypothesize an evolution of the virus in response to a selective pressure exerted by vaccines. A similar scenario of avoidance of vaccine-induced immunity, involving an endemic viral infection controlled by extensive vaccination plans, has been observed in Italian DPPA for avian metapneumovirus, another RNA virus (Catelli et al., Citation2010; Cecchinato et al., Citation2010). To confirm this hypothesis, both antigenic characterization of the new IBDV genotype and in vivo cross-protection studies using existing vaccines should be carried out in secure isolation conditions.
The currently available epidemiological and clinical data do not allow definition of the degree of pathogenicity of the ITA genotype, since the viruses have been detected in birds vaccinated for IBD, and hence provided with some kind of immune protection which could have masked or altered clinical signs and post-mortem lesions.
Molecular sequences typical of virulent strains (very virulent or classical) are observed in fragments of the ITA sequences: the serine-rich heptapeptide region in position 326–332, close to the major hydrophilic peak B, reported in very virulent strains by Nagarajan and Kibenge (Citation1997) and specific amino acid-residues at positions 249-Q, 286-T and 318-G, hallmarks of classical IBDV strains, reported by Jackwood (Citation2012). These molecular characteristics, together with the observation of high mortality when virus was inoculated in embryonated SPF eggs (Eterradossi & Saif, Citation2013), suggest classification of ITA strains among classical IBDVs.
Only in vivo experimental pathogenicity studies conducted in secure isolation conditions, through the evaluation of clinical signs and macro/microscopic lesions, will clarify conclusively the virulence of the new Italian genotype.
Very virulent strains, which have been dominantly present in Italy between early 1990s and the 2000s (Asdrubali & Franciosini, Citation1993; Tosi et al., Citation1997; Moreno et al., Citation2007, Citation2010a) seem to circulate currently to a much less extent.
Live vaccine genotypes represent 20% of the detections, demonstrating vaccine virus persistence and circulation in broiler flocks. Although 100% identity between detections and vaccines was always observed in our survey, closely related but not identical vaccine strains have been previously reported to be circulating in Italy (Moreno et al., Citation2007) pointing out the possibility that reversion to virulence is always around the corner for vaccine viruses (Raue et al., Citation2004; Catelli et al., Citation2006; Oldoni et al., Citation2009; Moreno et al., Citation2010b; Lupini et al., Citation2011; Cecchinato et al., Citation2014; Ndegwa et al., Citation2014).
The reconstruction of the actual origin of ITA genotype is challenging. The phylogenetically closest strain is IBDVRF 5.94 (95% identity), which was isolated in Russia in 1994 (Shcherbakova et al., Citation1998); however clear epidemiological links are lacking. Further investigations based on the comparison of broader regions of the genome and taking advantage of the analysis of recombination patterns will provide more information on the origin of ITA genotype.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Asdrubali, G. & Franciosini, M.P. (1993). Recenti acquisizioni sulla malattia di Gumboro. Rivista di avicoltura, 7, 43–46.
- Brandt, M., Yao, K., Liu, M., Heckert, R.A. & Vakharia, V.N. (2001). Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. Journal of Virology, 75, 11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001
- Catelli, E., Cecchinato, M., Savage, C.E. Jones, R.C. & Naylor, C.J. (2006). Demonstration of loss of attenuation and extended field persistence of a live avian metapneumovirus vaccine. Vaccine, 24, 6476–6482. doi: 10.1016/j.vaccine.2006.06.076
- Catelli, E., Lupini, C., Cecchinato, M., Ricchizzi, E., Brown, P. & Naylor, C.J. (2010). Field avian metapneumovirus evolution avoiding vaccine induced immunity. Vaccine, 28, 916–921. doi: 10.1016/j.vaccine.2009.10.149
- Cecchinato, M., Catelli, E., Lupini, C., Ricchizzi, E., Clubbe, J., Battilani, M. & Naylor, C.J. (2010). Avian metapneumovirus (AMPV) attachment protein involvement in probable virus evolution concurrent with mass live vaccine introduction. Veterinary Microbiology, 146, 24–34. doi: 10.1016/j.vetmic.2010.04.014
- Cecchinato, M., Catelli, E., Lupini, C., Ricchizzi, E., Prosperi, S. & Naylor, C.J. (2014). Reversion to virulence of a subtype B avian metapneumovirus vaccine: is it time for regulators to require availability of vaccine progenitors? Vaccine, 32, 4660–4664. doi: 10.1016/j.vaccine.2014.06.030
- Chettle, N., Stuart J.C. & Wyeth P.J. (1989). Outbreak of virulent infectious bursal disease in East Anglia. Veterinary Record, 125, 271–272. doi: 10.1136/vr.125.10.271
- Coletti, M., Asdrubali, G., Amico, V., Covarelli, G. & Mechelli, L. (1983). Ricerche sierologiche ed istologiche in polli vaccinati contro la malattia di Gumboro (bursite infettiva). Rivista zootecnica e veterinaria, 11, 304–307.
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics, 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
- Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. doi: 10.1038/nmeth.2109
- Eterradossi, N., Picault, J.P., Drouin, P., Lamandé, J., Alleé, C., Pichoire, M., Le-Coq, H., Morin, Y., Guittet, M. & Bennejean, G. (1990). Maladie de Gumboro: etude étiologique de cas aigus survenus en France en 1989. L’aviculteur, 511, 54–58.
- Eterradossi, N. & Saif, Y.M. (2013) Infectious bursal disease. In D.E. Swayne, J.R. Glisson, L.R. McDougald, L.K. Nolan, D.L. Suarez & N. Venugopal (Eds.). Diseases of Poultry 13th edn (pp. 219–246). Ames: Iowa State Press.
- Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. doi: 10.1093/sysbio/syq010
- Heine, H.G., Haritou, M., Failla, P., Fahey, K. & Azad, A. (1991). Sequence analysis and expression of the host-protective immunogen VP2 of a variant strain of infectious bursal disease virus which can circumvent vaccination with standard type I strains. Journal of General Virology, 72, 1835–1843. doi: 10.1099/0022-1317-72-8-1835
- Jackwood, D.J. (2012). Molecular epidemiologic evidence of homologous recombination in infectious bursal disease viruses. Avian Diseases, 56, 574–577. doi: 10.1637/10053-010912-ResNote.1
- Jackwood, D.J., Cookson, K.C., Sommer-Wagner, S.E., Le Galludec, H. & de Wit, J.J. (2006). Molecular characteristics of infectious bursal disease viruses from asymptomatic broiler flocks in Europe. Avian Diseases, 50, 532–536. doi: 10.1637/7528-032006R1.1
- Jackwood, D.J. & Sommer-Wagner, S.E. (2011). Amino acids contributing to antigenic drift in the infectious bursal disease Birnavirus (IBDV). Virology, 409, 33–37. doi: 10.1016/j.virol.2010.09.030
- Jackwood, D.J., Sreedevi, B., LeFever, L.J. & Sommer-Wagner, S.E. (2008). Studies on naturally occurring infectious bursal disease viruses suggest that a single amino acid substitution at position 253 in VP2 increases pathogenicity. Virology, 377, 110–116. doi: 10.1016/j.virol.2008.04.018
- Jackwood, D.J. & Stoute, S.T. (2013). Molecular evidence for a geographically restricted population of infectious bursal disease viruses. Avian Diseases, 57, 57–64. doi: 10.1637/10303-071212-Reg.1
- Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S. & Madden, T.L. (2008). NCBI BLAST: a better web Interface. Nucleic Acids Research, 36, W5–W9. doi: 10.1093/nar/gkn201
- Kibenge, F.S., Dhillon, A.S. & Russell, R.G. (1988). Biochemistry and immunology of infectious bursal disease virus. Journal of General Virology, 69, 1757–1775. doi: 10.1099/0022-1317-69-8-1757
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. & Higgins, D.G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
- Lupini, C., Cecchinato, M., Ricchizzi, E., Naylor, C.J. & Catelli, E. (2011). A turkey rhinotracheitis outbreak caused by the environmental spread of a vaccine-derived avian metapneumovirus. Avian Pathology, 40, 525–530. doi: 10.1080/03079457.2011.607428
- Moreno, A.M., Barbieri, I., Ceruti, R., Morandini, E. & Cordioli P. (2010a). Caratterizzazione genomica di ceppi del virus della malattia di Gumboro isolati in Italia nel periodo 2006–2009. Atti della Società Italiana di Patologia Aviare, 2010, 199–203.
- Moreno, A.M., Fallacara, F., Barbieri, I., Tosi, G., Rivallan, G., Eterradossi, N., Ceruti, R. & Cordioli P. (2007). Genetic and antigenic characterization of infectious bursal disease viruses isolated in Italy during the period 2002–2005. Avian Diseases, 51, 863–872. doi: 10.1637/7904-020107-REGR.1
- Moreno, A.M., Piccirillo, A., Mondin, A., Morandini, E., Gavazzi, L. & Cordioli, P. (2010b). Epidemic of infectious laryngotracheitis in Italy: characterization of virus isolates by PCR-restriction fragment length polymorphism and sequence analysis. Avian Diseases, 54, 1172–1177. doi: 10.1637/9398-051910-Reg.1
- Mundt, E. (1999). Tissue culture infectivity of different strains of infectious bursal disease virus is determined by distinct amino acids in VP2. Journal of General Virology, 80, 2067–2076. doi: 10.1099/0022-1317-80-8-2067
- Nagarajan, M.M. & Kibenge, F.S. (1997). Infectious bursal disease virus: a review of molecular basis for variations in antigenicity and virulence. Canadian Journal of Veterinary Research, 61, 81–88.
- Ndegwa, E.N., Toro, H. & van Santen, V.L. (2014). Comparison of vaccine subpopulation selection, viral loads, vaccine virus persistence in trachea and cloaca, and mucosal antibody responses after vaccination with two different Arkansas Delmarva Poultry Industry–derived infectious bronchitis virus vaccines. Avian Diseases, 58, 102–110. doi: 10.1637/10609-070613-Reg.1
- Oldoni, I., Rodríguez-Avila, A., Riblet, S.M., Zavala, G. & García, M. (2009). Pathogenicity and growth characteristics of selected infectious laryngotracheitis virus strains from the United States. Avian Pathology, 38, 47–53. doi: 10.1080/03079450802632031
- Raue, R., Islam, M.R., Islam, M.N., Islam, K.M., Badhy, S.C., Das, P.M. & Muller H. (2004). Reversion of molecularly engineered, partially attenuated, very virulent infectious bursal disease virus during infection of commercial chickens. Avian Pathology, 33, 181–189. doi: 10.1080/03079450310001652112
- Rinaldi, A., Cervio G. & Mandelli G. (1965). Comparsa di una nuova forma morbosa dei polli negli allevamenti avicoli della Lombardia verosimilmente identificabile con la cosiddetta Malattia di Gumboro. Selezione Veterinaria, 6, 207–209.
- Rosenberger, J.K. & Cloud, S.S. (1986, July 20–24). Isolation and characterization of variant infectious bursal disease viruses. Abstracts of the 123rd meeting of the American Veterinary Medical Association (p. 357). Atlanta, Georgia, USA.
- Senne, D.A. (2008). Virus propagation in embryonating eggs. In L. Dufour-Zavala, D.E. Swayne, J.R. Glisson, J.E. Pearson, W.M. Reed, M.W. Jackwood & P.R. Woolcock (Eds.). A Laboratory Manual for the Isolation, Identification and Characterization of Avian Pathogens. 5th edn (pp. 204–208). Madison, WI: American Association of Avian Pathologists.
- Shcherbakova, L.O., Lomakin, A.I., Borisov, A.V., Drygin, V.V. & Gusev, A.A. (1998). Comparative analysis of the VP2 variable region of the gene from infectious bursal disease virus isolates. Molekuliarnaia genetika, mikrobiologiia i virusologiia, 1, 35–40.
- Tosi, G., Pavesi, M., Massi, P. & Meini A. (1997). Osservazioni sull’andamento della Bursite Infettiva del pollo (IBD) in Italia e preliminare tipizzazione di alcuni ceppi di campo. La Selezione Veterinaria, 8, 631–636.