ABSTRACT
A peptide enzyme linked immunosorbent assay (ELISA) based on an epitope in the haemagglutinin (HA) of avian influenza virus H5N1, amino acid positions 274–288 (HA274–288) was evaluated for detection of H5N1-specific antibodies. An optimized ELISA based on the tetrameric form of the HA274–288 epitope designated MP15 gave low background with non-immune chicken sera and detected vaccinated and infected birds. The HA274–288 epitope was highly conserved in Indonesian H5N1 strains and antibody responses were detected in the majority of the vaccinated chickens regardless of the H5N1 strain used for vaccination. The HA274–288 epitope was also conserved in the majority of H5N1 strains from the neighbouring Asian region, and other H5 subtypes potentially allowing for a wider use of the MP15 ELISA in H5N1 vaccinated and infected flocks. The MP15 ELISA results correlated significantly with haemagglutination inhibition (HI) test results and test sensitivity and specificity were 87% and 92%, respectively. The MP15 ELISA titres were significantly higher than the HI titres in all immune sera allowing for sera to be tested at a single dilution of 1:400 which is of advantage in routine surveillance. The study indicated that the MP15 ELISA is potentially useful for serological detection of H5N1 vaccinated or infected poultry and to have some advantages over the standard HI test for routine monitoring of flocks’ immunity after vaccination.
Introduction
Highly pathogenic avian influenza (HPAI) H5N1 remains a global concern due to the devastating economic losses that it continues to cause in poultry worldwide and its zoonotic potential. There have been 840 confirmed human cases of avian influenza (AI) due to H5N1 between 2003 and 2015, including 447 deaths (WHO, Citation2015). H5N1 was first reported in Indonesian poultry in 2003 in Java and has spread to neighbouring islands soon after (Hartaningsih et al., Citation2015). Subsequently, more than 150 million of poultry were culled or died and this resulted in the introduction of vaccination for poultry in 2004 in areas where H5N1 was prevalent (Spackman & Swayne, Citation2013).
As vaccination for HPAI may not provide complete protection, flock immunity can be monitored following vaccination by measuring either the virus neutralizing or haemagglutination inhibiting antibodies (Swayne et al., Citation2015). Virus neutralizing antibodies are the most accurate reflection of the host protective immune status and the virus neutralization test is the “gold standard” for AI sero-diagnosis (Desvaux et al., Citation2012). The haemagglutination inhibition (HI) test is the most commonly used method for detecting antibody responses following AI infection or vaccination and also for virus sub-typing (Lee et al., Citation2004; Alexander, Citation2006). In H5N1 vaccinated poultry, HI antibody titres have been correlated with protection (Swayne et al., Citation1999) and the aim of an effective vaccination is to maintain these protective titres throughout the bird’s life. Therefore, frequent sero-monitoring of vaccinated flocks is beneficial. However, the set-up for HI test can be laborious (Zhou et al., Citation1998), as the production of haemagglutinating (HA) antigen requires amplification of potentially hazardous virus and thus requires appropriate biosafety conditions and technical expertise (Postel et al., Citation2011; Zhao et al., Citation2011). It also has some other disadvantages, including the necessity to use antigenically homologous HA antigen for accurate detection of immunity (Meulemans et al., Citation1987; Koel et al., Citation2014), low sensitivity (Lee et al., Citation2006; Postel et al., Citation2011), presence of non-specific agglutinins in some serum samples causing false-negative results and the need to standardize the virus concentration each time a test is performed. Therefore, alternative serological tests that can overcome the limitations of HI test (Chen et al., Citation2008; Prabakaran et al., Citation2009; Postel et al., Citation2011), and target subtype-specific antigenic epitopes of H5N1, would be advantageous.
Influenza A virus contains two surface proteins, haemagglutinin (HA) and neuraminidase (NA). Based on antigenic differences between their HA and NA proteins, AI viruses are subdivided into 16 different HA subtypes (H1–H16) and nine different NA subtypes (NA1–NA9). The HA is synthetized as HA0 precursor that is post-translationally cleaved into two polypeptides HA1 and HA2 (or subunits) which are, in the infectious virus particle, linked via disulphide bonds into trimers. In the trimer configuration, the HA1 subunit forms the globular head that has receptor binding properties and is able to attach to sialic acid receptors located on the host cell surface. The HA2 subunit forms the stem in the trimer and acts as a fusion peptide and anchor for the HA1 subunit (Wiley et al., Citation1981; Yoshida et al., Citation2009). The HA1 is the major antigen of the virus and contains most of antigenic epitopes that are involved in virus neutralization, protection, subtype specificity and serological detection (Okuno et al., Citation1993; Sahini et al., Citation2010; Wang et al., Citation2010). Several enzyme linked immunosorbent assays (ELISAs) have been developed using HA1 subunit antigenic epitopes that are subtype-specific. Most are competitive ELISAs using monoclonal antibodies specific to an antigenic epitope (Prabakaran et al., Citation2009; Yoshida et al., Citation2009; Postel et al., Citation2011) or indirect ELISAs using recombinant HA5 subtype glycoprotein (Rowe et al., Citation1999) and a few are peptide ELISAs (Velumani et al., Citation2011; Zhao et al., Citation2011). With most ELISAs inter-subtype cross-reactivity is unavoidable due to the degree of amino acid conservation between HA subtypes (Boer et al., Citation1990; Postel et al., Citation2011; Yoshida et al., Citation2009). ELISAs also might lack sensitivity in comparison to the HI test (Desvaux et al., Citation2012; Rowe et al., Citation1999) and most are qualitative, detecting seroconversion without indicating the level of immunity (Velumani et al., Citation2011). Some of the ELISAs using epitope-specific peptides exhibited good sensitivity and specificity in comparison with HI test and had no cross-reactivity (Zhao et al., Citation2011).
Previously, a blocking ELISA was developed for detection of H5N1 antibodies using monoclonal antibodies generated against an epitope in the HA1 subunit of H5N1, located between amino acid (aa) positions 274–281 and comprising the amino acid sequence CNTKCQTP (Prabakaran et al., Citation2009). In the subsequent study (Velumani et al., Citation2011), the blocking ELISA was replaced with a peptide ELISA using the same highly conserved HA1 epitope as an antigen that covered aa positions 274–288 and comprised the aa sequence CNTKCQTPMGAINSS. This HA274–288 epitope was immunogenic and an ELISA using CNTKCQTPMGAINSS peptide as antigen was specific for H5N1 subtype and sensitive for detecting antibodies in sera obtained from both humans and chickens following infection with H5N1 (Velumani et al., Citation2011).
In this study, we compared three different forms of HA274–288 epitope, peptide P8 comprising the sequence CNTKCQTP (Prabakaran et al., Citation2009), peptide P15 comprising the sequence CNTKCQTPMGAINSS (Velumani et al., Citation2011) and the tetrameric form of P15 (MP15), as antigens in an ELISA for detection of H5N1 antibodies in immune chicken sera. The MP15 ELISA was then used to detect H5N1 immune responses in vaccinated commercial layers and experimentally infected chickens and compared to the HI test to determine its sensitivity and specificity for detection of H5-specific antibodies.
Materials and methods
Peptides
Three peptides, namely P8 with the amino acid sequence CNTKCQTP comprising the N-terminal end of the epitope HA274–288, P15 with the sequence CNTKCQTPMGAINSS comprising the entire epitope HA274–288 and multimeric peptide (MP15) that consisted of four P15 peptides linked via a lysine backbone, were chemically synthesized by VCP BIO (Shenzhen City, China). The purity of P8, P15 and MP15 was 92.8%, 85.9% and 80.5%, respectively. The amino acid sequence of the P8 and P15 peptides corresponds to the amino acid residues 274–281 and 274–288, respectively, in the Indonesian H5N1 consensus sequence (). The position numbering refers to the mature HA of H5 subtype, that is, protein without the 16 amino acids signal peptide.
Table 1. Amino acid sequence similarities of HA274–288 epitope in H5N1 strains.
Comparison of HA274–288 amino acid sequence for H5N1 strains
The HA1 amino acid sequences of 113 Indonesian H5N1 strains isolated between 2003 and 2014 and deposited in the GenBank were aligned using the Molecular Evolutionary Genetics Analysis version 6 application (MEGA v.6) (Tamura et al., Citation2013) and a consensus sequence for the HA274–288 region obtained. All of the 113 individual HA274–288 sequences were then aligned to the HA274–288 consensus sequence and variations identified. The HA1 amino acid sequences of 301 H5N1 strains from other Asian countries (China, Malaysia, Myanmar and Vietnam), isolated between 1997 and 2014 and deposited in GenBank, were also aligned using MEGA v.6, after which the consensus sequence for the HA274–288 region was obtained and variations from the consensus sequence identified.
Antisera panels
Six panels of different chicken antisera were developed and used.
Panel 1 contained antisera against 10 H5 strains (): A/Ck/Konawe Selatan/BBV/2007 (KS), A/Ck/Indonesia/Wates1/2005 (Wt), A/Ck/Indonesia/CSLK-EB/2006 (CSLK), A/Ck/West Java/SMI-ENDRI2/2006 (SMI), A/Viet Nam/1203/2004 (Vt), A/Ck/West Java/Sbg-29/2007 (Sb29), A/Ck/Myanmar/1001/1/2006 (My06), A/Ck/Sagaing/295/2010 (My10), A/Mall/South Korea/12A/2007/12 (H5N2) and A/Dk/Victoria/1462/2008 (H5N3). Antisera were produced by inoculation of 6 to 8-week-old specific pathogen-free (SPF) chickens with 0.5 ml of inactivated virus mixed 1:1 with an adjuvant. Three weeks later the chicks were bled and challenged with 6 log10 of median egg infective doses of the homologous live virus by the ocular/nasal route. Two weeks later, sera were collected, pooled (post-infected sera) and stored at −20°C until testing. Each experiment was carried out in a separate bio security level 3 animal isolation room at the Australian Animal Health Laboratory (AAHL), Geelong. Experimental procedures were approved by the AAHL animal ethics committee.
Table 2. Amino acid sequence of HA274–288 epitope in AIV strains used to produce reference sera for the study.
Panel 2 sera comprised reference chicken antisera to H5N1, H4N6, H7N7 and H9N2 strains obtained from the Animal Health and Veterinary Laboratories Agency (AHVLA), UK. H5N1 serum was produced by immunization of SPF chickens with an inactivated virus followed by challenged with a homologous live virus (), whereas sera to H4N6, H7N7 and H9N2 were produced by immunization of chickens with live virus, followed by challenged with a homologous live virus.
Panel 3 sera comprised SPF chicken sera (n = 56), obtained from chicks hatched from SPF eggs (SPAFAS, Woodend) and reared in isolation chambers in animal facility at the Faculty of Veterinary and Agricultural Sciences, University of Melbourne, Werribee. The sera were collected at between 8 and 48 weeks of age, and were negative for antibodies to all known avian pathogens when tested according to the European Pharmacopoeia.
Panel 4 were non-vaccinated commercial broiler (n = 45) and layer (n = 85) chicken sera, obtained from the diagnostic laboratory of Asia Pacific Centre for Animal Health, University of Melbourne. Birds from which the sera were collected originated from Australian commercial poultry flocks, were not vaccinated with any avian influenza virus (AIV) vaccines, but were vaccinated against other common pathogens of poultry, hence considered AIV antibody free.
Panel 5 sera (n = 300) were obtained from 15 commercial layer flocks from two districts in West Java, Indonesia, that practised vaccinations prior to lay (usually at 4 and between 16 and 18 weeks of age). Farm selection and sample collection was facilitated by the staff of local district animal health services who contacted the local farms in late 2009 and ascertain if (a) they were agreeable for sample collection at before and after point-of-lay booster vaccination, (b) the type of AI vaccine used and the date (flock age) of the point-of-lay vaccination. Subsequently, two visits were made in 2010 for sample collection. Four flocks were vaccinated with a vaccine registered for a A/Ck/Legok/2003 (Legok) H5N1 seed strain, one with a vaccine registered for A/Ck/West Java/PWT/2006 (PWT) H5N1, seven with vaccines registered for A/Tk/England/N28/73 (N28/73) H5N2 and three with a vaccine registered for A/Ck/Mexico/232/94 (Mex/232/94) H5N2 strain (). From each flock 10 or 11 birds were randomly selected and bled at approximately one week before the vaccination at point-of-lay (flocks that were vaccinated a variable number of times before collection of sera at 15–17 weeks old) and sera labelled as pre-booster vaccination (preBV) sera. Sera were also collected from the same flocks, from 10 or 11 randomly selected birds, at between 3 and 4 weeks following the point-of-lay vaccination and labelled as post-booster vaccination (postBV) sera. Following collection sera were stored at −20°C until use. Sera from flocks vaccinated with virus strains homologous to HA274–288 epitope (Legok, PWT and N28/73 vaccines) were labelled as H5 sera and those vaccinated with a heterologous Mex/232/94 were labelled as H5# sera.
Table 3. Amino acid sequence of HA274–288 epitope in vaccine strains used for vaccination of commercial layer flocks from which Panel 5 sera were obtained.
Panel 6 sera were collected from layer birds (n = 11) vaccinated in the laboratory at 8, 12 and 16 weeks of age with a commercial inactivated vaccine Medivac-AI® (Pt. Medion, Bandung, Indonesia) that contained H5N1 strain PWT () and challenged at 18 weeks of age with H5N1 strain Sbg29 (). The sera were collected prior to challenge at day 0 (Vaccinated) and at day 14 post-challenge (Challenged). Vaccinated birds were kept in a closed hen house on the floor and were moved to negative pressure isolators in biosecurity level 3 facilities at Indonesian Research Centre for Veterinary Science, Bogor, Indonesia for challenge (Tarigan et al., Citation2015). The experimental protocol was approved by the Committee on the Ethics of Animal Experiments of the Indonesian Research Institute for Veterinary Sciences (Registration number: BB/V/A/01/2013) and carried out in accordance with the World Organisation for Animal Health (OIE) standard procedure (OIE, Citation2014).
Optimized ELISA protocol
Maxisorp flat bottom ELISA plates (Nunc, Roskilde, Denmark) were coated with 12.5 µg/ml of either P8, P15 or MP15 peptide in coating buffer (0.1 M Na2CO3, 0.1 M NaHCO3 pH 9.6), 100 μl/well, at 4°C overnight. After washing two times with wash buffer (WB) [PBS-T: phosphate-buffered saline containing 0.05% (v/v) Tween 20], the wells were blocked with 200 µl/well of 1% bovine serum albumin (Sigma–Aldrich) in PBS for 2 h at room temperature (RT). Then, the wells were washed two times and incubated with serial twofold dilutions of chicken sera (starting dilution of 1:100) in dilution buffer (DB) (0.1 M Tris pH 7.4, 0.5 M NaCl, 1 mM Na2EDTA, 2% bovine serum albumin, 3% TritonX-100, 3% Tween 20) for 2 h at RT. After washing three times, horseradish peroxidase (HRP)-conjugated rabbit anti-chicken IgG (Merck Millipore, Temecula, CA, USA) in DB was added to the plates and incubated for 1 h at RT. Finally, the wells were washed five times with WB, 3,3′,5,5′-tetramethylbenzidine substrate (Millipore) was added, incubated for 10 min and the reaction was stopped by addition of 50 µl/well of 1 M HCl. The optical density (OD) was measured at 450 nm (OD450) using an ELISA reader (Multiskan Labsytems, Thermo Scientific, Waltham, MA USA). The cut-off OD value for the MP15 peptide ELISA was the mean OD of non-vaccinated commercial chicken serum and SPF chicken serum at 1:400 dilution plus two times standard deviation (SD). The titre of each serum was the last dilution of sera that produced an OD450 above the calculated cut-off value and expressed as a log2 value.
Peptide ELISA using previously described protocol
The peptide ELISA was carried out with the peptide P8 as the coating antigen as described in Velumani et al. (Citation2011) with homologous KS and SPF sera in twofold dilutions starting from 1:100 dilution. In brief, Maxisorp flat bottom ELISA plates (Nunc) were coated with 10 μg/well of P8 peptide at 4°C overnight. The plates were blocked with 1% bovine serum albumin in PBS-T (BB) for 1 h at 37°C and after washing with WB, incubated with serial twofold dilutions of chicken sera (starting at a dilution of 1:100) in DB for 45 min at 37°C. Then, they were washed with WB followed by incubation with HRP-conjugated rabbit anti-chicken antibody (Millipore) at 1:2000 in DB at 37°C for 45 min. After washing, the colour change was produced by addition of substrate tetramethylbenzidine and OD450 measured using ELISA reader (Multiscan Labsytems).
Haemagglutination inhibition test
HA antigens were prepared and the HI test done according to the standard procedure (OIE, Citation2014). All vaccinated commercial sera (Panel 5 sera) were tested using HA antigens homologous to the registered vaccine seed strains recorded as being used for flock vaccination and were prepared from H5N1 strain Legok and H5N2 strains N28/73 and Mex/232/94. H5N1 strain PWT was not available at the time of HA antigen preparation but instead a related Indonesian H5N1 strain, namely KS, was used as HA antigen with PWT sera, Panel 6 sera were tested using HA antigen prepared from Sb29 strain. For the HI test, serum to be tested was serially diluted in 25 µl of PBS in V-bottom microtitre plates and an equal volume of HA antigen containing 4 HA units was added. After incubation at 25°C for 30 min, 25 µl of 1% suspension of chicken red blood cells was added and incubated for 40 min at 25°C. The HI titre was expressed in log2 units of the highest dilution of sera that completely inhibited haemagglutination.
Statistical analysis
ODs of SPF, non-vaccinated commercial, preBV and postBV sera were compared using the Kruskal–Wallis test, and the preBV and postBV ODs of the H5N1, H5N2, H5# and challenged sera were analysed using the Mann–Whitney U test. The log2 ELISA and HI titres were analysed using an unpaired t-test with Welch’s correction. The optimum cut-off point to use for the sensitivity and specificity calculations of MP15 ELISA versus HI test was determined by receiver operating characteristic curve analysis (Greiner et al., Citation2000; Desvaux et al., Citation2012). Data from Panels 5 and 6 were used for sensitivity and specificity calculations. Sensitivity and specificity were calculated separately for data from Panel 5 excluding heterologous sera H5# and Panel 6 and heterologous sera H5# only. Pearson’s correlation test was used to determine the correlation between HI and MP15 ELISA test results using data from Panels 5 and 6 sera separately. Sera were considered positive in the M15 ELISA if the OD was ≥0.15 at 1:400 dilution, and positive in the HI test if log2 HI titre with homologous HA antigen was ≥4 log2. Additionally, Pearson’s correlation test was applied using MP15 ELISA and HI titres. All statistical analyses were carried out using GraphPad Prism® version 6.
Results
Amino acid sequence of the HA274–288 epitope is conserved in most Indonesian H5N1 and in many of H5N1 strains from the region
Alignment of available Indonesian H5N1 amino acid sequences (n = 113) for the HA274–288 epitope indicated that the epitope was fully conserved, except in seven strains (). Alignment of aa sequences from 301 H5N1 strains from neighbouring Asian countries indicated that in 182/301 (60%) of strains, the HA274–288 epitope was conserved (Group 1), 98/301 (32.6%) of strains had at least one aa mutation either N275S, K277R or M282I (Group 2), 18/301 (6%) of strains had two aa mutations K/M277/282R/I (Group 3) with only 3/301 (1%) of strains having three aa mutations N/K/M275/277/282S/R/I (Group 4) (). Consequently, two peptides P8 and P15 corresponding to the truncated and full length HA274–288 epitope, respectively, were used with aa sequences identical to the consensus sequence of Indonesian and Southeast Asian H5N1 strains ().
Comparison of aa sequences of the HA274–288 epitope in strains used to produce Panel 1 and Panel 2 sera showed that all H5N1 viruses had sequences homologous to the P15 peptide, that is to the consensus aa sequence “CNTKCQTPMGAINSS” () except for My10 and Scot, each having two aa mutations: K/M277/282R/I or N/M275/282D/V, respectively. Strain H5N2 had the same sequence as the consensus H5N1 sequence, whereas H5N3 had one aa mutation M282 V (). The aa sequence of the HA274–288 epitope differed in H4N6, H7N7 and H9N2 strains from the H5N1 consensus sequence by 10, 10 and 9 aa, respectively (). Amino acid sequence of HA274–288 epitope in vaccine strains PWT, Legok and N28/73 used for vaccination of hens from which Panel 5 sera was collected had the same sequence to the consensus H5N1 and P15 peptide sequence, except for the H5N2 vaccine Mex/232/94 with 3 aa mutations N/T/M275/276/282D/A/V ().
MP15 ELISA had the best reactivity with homologous sera
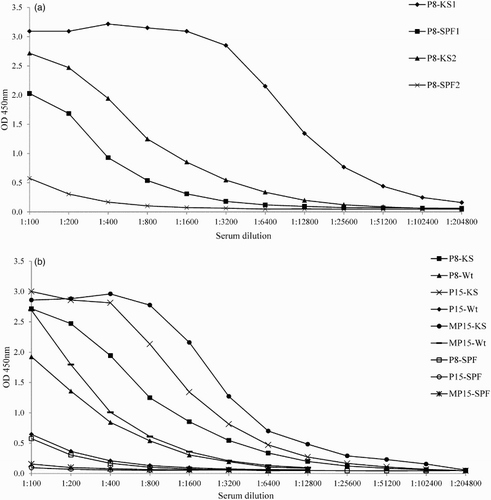
Initially, the P8 peptide ELISA was carried out as described in Velumani et al. (Citation2011). Homologous KS sera produced high OD and had a high titre of antibodies with P8; however, SPF negative sera also reacted strongly with the P8 peptide ((a); KS1 and SPF1). In an optimized ELISA, the high OD background of the SPF sera was reduced to 0.5 OD450 at the 1:100 dilution, but ODs with KS serum (P8-KS2) remained high. But P8 peptide ELISA still produced high background when Panel 4 sera (non-vaccinated commercial chicken sera) were used (data not shown). Therefore, P15 and multimeric form of P15 (MP15) peptides were tested as antigens in the ELISA ((b)). As shown, the use of the P15 peptide as the coating antigen produced less background than coating with the P8 peptide, but the MP15 peptide produced the highest ODs for both homologous sera KS and Wt, with low background for SPF sera (ODs of ≤0.2). Therefore, the MP15 peptide was selected as the antigen of choice for further testing in ELISA.
Figure 1. HA274–288 peptide ELISA with homologous KS and SPF chicken sera (a) Binding of homologous KS and SPF sera to P8 peptide (KS1 and SPF1), using the protocol according to Velumani et al. (Citation2011); KS2 and SPF2, using the optimized ELISA protocol. (b) Binding of homologous KS and Wt sera to P8, P15 and MP15 peptides using the optimized ELISA protocol. P8-KS, KS serum on P8 antigen; P15-KS, KS serum on P15 antigen; MP15-KS, KS serum on MP15 antigen; P8-SPF; SPF serum on P8 antigen; P15-SPF, SPF serum on P15 antigen; MP15-SPF; SPF serum on MP15 antigen.
Subsequently, the reactivity of the MP15 peptide with sera from AIV with diverse HA274–288 epitope sequences was tested. When five sera against H5N1 strains with homologous HA274–288 epitopes were tested in the MP15 ELISA, the antibody titres were 12,800 for KS sera, 3200, 1600 and 800 for Wt, CSLK and Vt sera, respectively, and 200 for SMI ( and (a)). Of the sera against H5N1 strains with heterologous sequence to P15, My10 and Scot, sera had titres of 1600 and 800, respectively, whereas sera to My06 and Sb29 with unpublished HA sequences had titres of 800 in the MP15 ELISA. Out of the two sera raised against the H5 subtypes other than H5N1, H5N2 sera had titres of 1600 and H5N3 had titres of 100. Sera from subtypes other than H5, H4N6 and H9N2 had titres of 200, whereas H7N7 had titres of 400 ( and (b)).
Figure 2. Titration of homologous and heterologous sera in MP15 ELISA (a) Sera from H5 subtypes homologous and heterologous to MP15 peptide titrated in twofold dilutions starting at 1:100. (b) Reference sera from different AI subtypes on MP15 peptide titrated in twofold dilutions starting at 1:100 in MP15 ELISA.
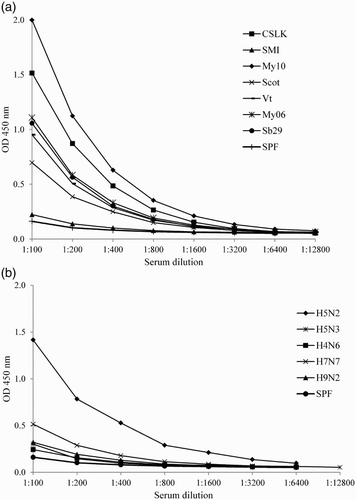
Table 4. Antibody titres to HA274–288 epitope in reference sera determined by MP15 peptide ELISA.
Background OD for non-vaccinated commercial chicken sera and cut-off OD values
Australian commercial layer (n = 85) and broiler (n = 45) sera, negative for antibodies to AIV, as well as the SPF (n = 56) sera were tested in MP15 peptide ELISA at a 1:400 dilution and cut-off was calculated as the mean OD + 2 × SD to be 0.15 [0.085 + 2 (0.034)].
MP15 ELISA clearly differentiated H5 vaccinated or infected flocks from those non-vaccinated
Ability of MP15 ELISA to detect H5-specific antibodies was evaluated using sera from commercial AIV non-vaccinated (Panel 4) and vaccinated (Panel 5) flocks. The ODs of sera from commercial layers vaccinated an unknown number of times, with different H5N1 and H5N2 vaccines prior to serum collection (Panel 5, preBV) were significantly higher (P < 0.0001) than ODs of chicken sera from non-vaccinated flocks (Panel 4 sera) with median OD of 0.31 (OD range 0.08–2.3) and 0.07 (OD range 0.05–0.25), respectively ( and (a)). Only 6/130 of Panel 4 sera were slightly above the cut-off OD and in preBV sera, 25/150 samples were below cut-off OD 0.15, and following booster vaccination 3–4 weeks later there was a significant increase (P < 0.001) in OD values with median OD of 0.4 (OD range 0.09–2.9) indicating that MP15 ELISA can detect antibody increase after booster vaccination ( and (a)). Among postBV sera 12/150 were below the cut-off OD of 0.15.
Figure 3. Performance of MP15 ELISA with non-vaccinated and vaccinated commercial chicken sera to detect antibodies against H5. SPF (Panel 3), birds (n = 56) reared in isolation and antibody free to all known avian pathogens; Non-vaccinated (Panel 4), commercial broilers (n= 45) and layers (n = 85) of different ages (Australian origin) not vaccinated with any AIV vaccines; preBV, commercial layers (n= 150) (Indonesian origin Panel 5) vaccinated unknown number of times before collection of sera at between 15 and 17 weeks; postBV, commercial layers (n= 150) (Indonesian origin Panel 5) vaccinated at between 16 and 18 weeks of age and sera collected at between 3 and 4 weeks after vaccination. All sera were tested at 1:400 dilution. Whiskers represent minimum and maximum OD values; line inside the bars represents the median OD values, dotted line represents the cut-off value of 0.15. ***P < 0.001 and ****P < 0.0001.
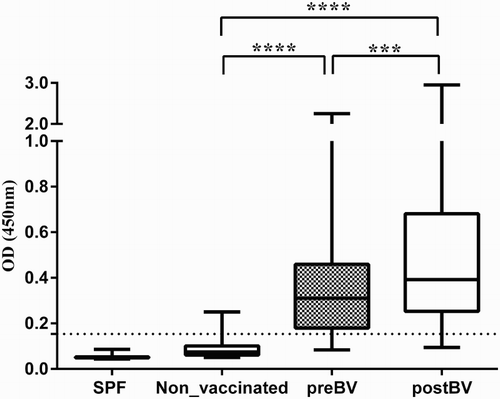
Table 5. Descriptive statistics of MP15 peptide ELISA and HI titres on sera.
Almost all layer flocks (11/15) had higher median OD values after re-vaccination (postBV), and regardless of the vaccine used, except for flocks A, D and M where the median OD in preBV and postBV sera did not differ significantly (P < 0.05) ().
Figure 4. Median ODs of pre- and post-booster vaccinated chicken sera (Panel 5 sera) from 15 farms. Columns represent the mean OD of 10 sera from each flock with whiskers representing the SD. All 15 flocks were commercial layer farms located in two districts in Indonesia, identified as A–O. preBV, flocks vaccinated unknown number of times before collection of sera at between 15 and 17 weeks of age; postBV, flocks vaccinated at between 16 and 18 weeks of age and sera collected at between 3 and 4 weeks after booster vaccination. All sera were tested at 1:400 dilution. Flocks vaccinated with vaccine derived from: A, PWT (H5N1); B–E, Legok (H5N1); F–L, N28/73 (H5N2) and M, N and O, Mex/232/94 (H5N2).
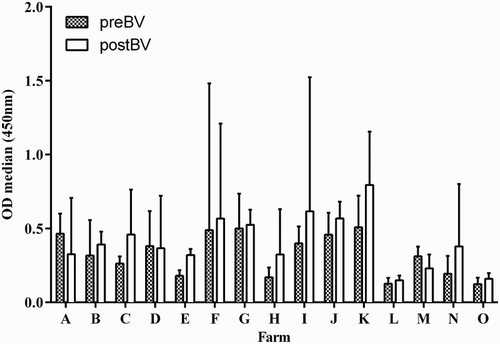
When panel 5 sera were separated according to the vaccine strain used for flock vaccination, that is vaccines that contained homologous HA274–288 epitope Legok, PWT (H5N1) and N28/73 (H5N2) or heterologous HA274–288 epitope Mex/232/94, the MP15 ELISA detected higher OD values in homologous (H5) vaccination ((a) and 5(b)) than in heterologous vaccination (H5#) ((c)). There was a significant increase (P < 0.03) in median postBV ODs of 0.35 (range 0.13–2.59) in comparison to preBV ODs of 0.31 (range 0.08–1.8) in H5N1 vaccinated flocks ( (flocks A–E) and (b)) and significant increase (P < 0.007) in median postBV OD of 0.52 (range 0.12–2.95) in comparison to preBV OD of 0.38 (range 0.084–2.3) in H5N2 vaccinated flocks ( (flocks F–L) and (b)). Flocks vaccinated with H5# vaccines did not show a significant increase in postBV OD 0.23 (range 0.09–2.5) ( (flocks M–O) and (b)) and 9/32 birds were negative in H5# preBV compared to 6/27 birds in H5# postBV.
Figure 5. MP15 ELISA was able to detect serological response to booster vaccination with commercial vaccines (Panel 5 sera) containing (a) H5N1 vaccines with homologous H5 sequence to P15; (b) H5N2 vaccines with homologous H5 sequence to P15 and (c) H5N2 vaccines with heterologous H5 sequence to P15 and (5) after experimental challenge with H5N1 virus (Panel 6 sera). preBV, vaccinated unknown number of times before collection of sera at between 15 and 17 weeks of age; postBV, vaccinated at between 16 and 18 weeks of age and sera collected at between 3 and 4 weeks after vaccination; Vaccinated, birds vaccinated experimentally with inactivated H5N1 commercial vaccine and sera collected at 2 weeks after the last vaccination (Panel 6 sera); Challenged, birds challenged experimentally with H5N1 strain Sb29 and sera collected 14 days post-challenge. All sera were tested at 1:400 dilution. Whiskers represent minimum and maximum OD values; line inside the bars represents the median OD values. *P < 0.05, **P < 0.01.
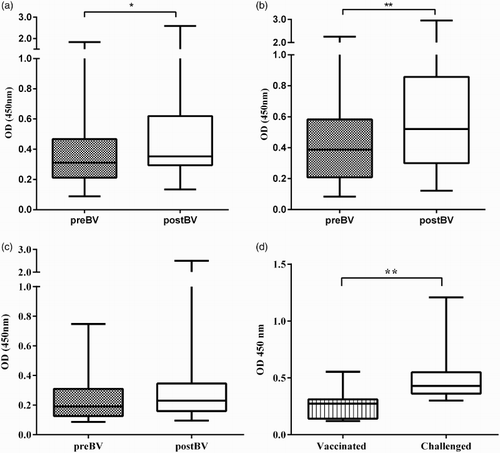
In sera obtained from vaccinated and infected birds (Panel 6 sera), the MP15 ELISA also detected an increase in ODs following experimental infection with H5N1 virus ((d)) with a significant increase (P < 0.01) in OD values of 0.43 at two weeks after challenge (range 0.3–1.2)] than after vaccination [OD of 0.27 (range 0.12–0.55)] ((c)). Only 3/11 had OD values below the cut-off after vaccination and all the birds after challenge (11/11) had ODs above the cut-off.
MP15 ELISA can be used as an alternative or confirmatory test for HI test results
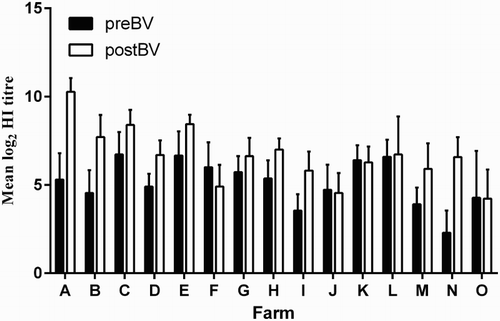
Since the level of AI protective immunity in vaccinated flocks is related to protective antibody titres (such as HI titres), HI antibody titres were also determined for Panels 5 and 6 sera ( and ). Mean homologous log2 HI titres in preBV sera (Panel 5) were above 4 log2 in the majority of flocks (12/15, the exceptions were I, M and N flocks) and were higher in postBV sera in all flocks (15/15) (). In four flocks J, K, L and O, preBV and postBV HI titres did not differ, and in flock F preBV titres were higher than postBV HI titres.
Figure 6. Mean log2 HI titres of pre- and post-booster vaccinated chicken sera (Panel 5 sera) from 15 flocks. Columns represent the mean log2 Hi titre of 10–11 sera from each flock with whiskers representing the SD. All 15 flocks were from commercial layer farms located in two districts in Indonesia, identified as A–O. preBV, flocks vaccinated unknown number of times before collection of sera at between 15 and 17 weeks of age; postBV, flocks vaccinated at between 16 and 18 weeks of age and sera collected at between 3 and 4 weeks after vaccination. Flocks vaccinated with vaccine derived from: A, PWT (H5N1); B–E, Legok (H5N1); F–L, AN28/73 (H5N2) and M, N and O, Mex/232/94 (H5N2). HI titres determined using homologous HA antigen except for flock A sera where KS HA antigen was used.
Comparison of HI and MP15 ELISA titres are shown in and . Mean log2 HI titre of all postBV sera, regardless of vaccine used (6.6 ± 1.9) were significantly higher (P < 0.0001) than mean log2 HI titre of preBV sera (5.05 ± 1.7) ((a) and (a)). Similarly, the mean log2 ELISA titre for all postBV sera regardless of vaccine used of 10.7 ± 1.8 was significantly higher (P < 0.001) than the mean log2 ELISA titre of the preBV sera (9.98 ± 1.5). Log2 HI titres of flocks vaccinated with either H5N1 or H5N2 vaccines showed significant increase (P < 0.0001) in HI titres after vaccination ((b)–(d) and (b)). Log2 ELISA titres of layers vaccinated with H5N1 strains containing homologous HA274–288 epitope sequences were significantly higher (P < 0.01) after vaccination (postBV) with 10.6 ± 1.4 titres, than at preBV (9.9 ± 1.16) ((b) and (b)) and also significantly higher (P < 0.01) in flocks vaccinated with H5N2 strains containing homologous HA274–288 epitope with postBV 11.2 ± 1.9 and preBV 10.44 ± 1.67 ((c) and (b)). But there was no significant increase in the log2 ELISA titre in the flocks vaccinated with Mex/232/94 H5N2 strain containing heterologous HA274–288 epitope sequence ((d) and (b)).
Figure 7. Comparison of serological responses in pre- and post-booster vaccination sera using HI and MP15 ELISA titres. (a) Combined results from all commercial layers (n = 150) (Indonesian origin sera) used in the study (Panel 5 sera) (b) Results from the flocks vaccinated with H5N1 vaccines with homologous H5 sequence to P15 (c) Results from the flocks vaccinated with H5N2 vaccines with homologous H5 sequence to P15. (d) Results from the flocks vaccinated with H5N2 vaccines with heterologous H5 sequence to P15. preBV, vaccinated unknown number of times before collection of sera at between 15 and 17 weeks of age; postBV, vaccinated at between 16 and 18 weeks of age and sera collected at between 3 and 4 weeks after vaccination; ELISA titre, log2 endpoint titres of MP15 ELISA; HI titre, log2 HI titres from the corresponding layer flocks. Columns represents the mean titres and whiskers represents the SD, *P < 0.05, ***P < 0.001, ****P < 0.0001.
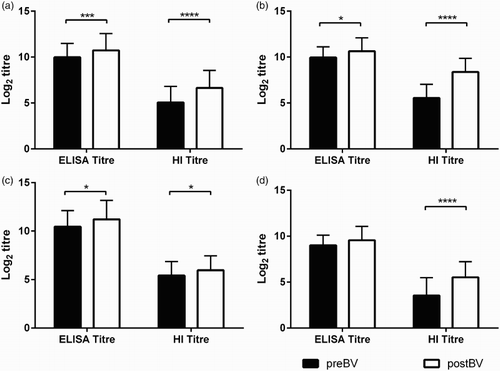
In the experimentally infected birds (Panel 6 sera), both mean log2 ELISA and HI titres after infection with Sb29 (11.1 ± 0.9 and 8.1 ± 0.9, respectively) were significantly higher than titres before challenge (9.5 ± 1.4 and 4.2 ± 0.6, respectively) ( and (c)).
Figure 8. Comparison of vaccinated and challenged sera (Panel 6 sera) using HI and MP15 ELISA titres. Vaccinated, sera from birds vaccinated with H5 inactivated vaccine (n = 11); Challenged, sera from birds 14 days after challenged with H5N1 strain (n = 11); ELISA titre, log2 endpoint titres from MP15 ELISA of sera from experimental birds; HI titre, log2 HI titres from the same birds. Columns represent the mean titres and whiskers represent the SD. **P < 0.01 and ****P < 0.0001.
Sensitivity and specificity of the MP15 ELISA
The sensitivity and specificity of the MP15 ELISA were determined using the HI test as reference method. All Panel 5 sera from 15 flocks vaccinated with vaccines strains with either homologous or heterologous HA274–288 (Flock A–O) (n = 300) and Panel 6 (n = 22) sera were used and, 35/300 and 1/22, respectively, tested HI negative (<4 log2 titre) (). As shown in , the sensitivity and specificity for the MP ELISA versus the HI test were 87% and 92%, respectively. The test sensitivity, however, was also dependent on the origin of sera, that is, on the type of vaccine used for vaccination. In flocks vaccinated with either H5N1 or H5N2 vaccine containing an AI strain with homologous sequence to the HA274–288 epitope, the sensitivity and specificity of the MP15 ELISA were 90% and 90% and in those vaccinated with Mex/232/94 vaccine (H5N2) containing heterologous sequence to HA274–288 epitope it was 74% and 100%, respectively (). The area under the curve (AUC) of all the receiver operating characteristic curves was greater than 0.9 indicating high accuracy of the MP15 ELISA when compared to the HI reference test.
Table 6. Comparison of MP15 ELISA and HI test.
Table 7. MP15 ELISA test performance at cut-off OD > 0.15 and HI test as the reference test.
Correlation between HI test and MP15 ELISA results
The degree of correlation between the HI and MP15 ELISA results (test positive or negative in both tests) was determined using Panel 3, Panel 4, Panel 5 [excluding results from farms vaccinated with the heterologous Mex/232/94 (H5#)] and Panel 6 sera. Pearson’s correlation coefficient was R = 0.79 [P = 0.0001] indicating highly significant correlation between the results obtained in MP15 ELISA and HI test. However, there was no correlation between the range of the antibody titres obtained by MP15 ELISA and HI test (results not shown).
Discussion
Our results show that the MP15 ELISA was able to detect H5–specific immune responses after H5N1 infection and/or vaccination in commercial chickens. The test requires a synthetic peptide thus the test set-up is more simple, specific and sensitive when compared to the HI test. As the detecting antigen is a H5-specific peptide based on the HA274–288 epitope located in the HA1 subunit, the test does not requires the use of live HPAI viruses, unlike the HI test. The HA274–288 epitope is specific for and highly conserved within the H5 subtype strains and therefore the MP15 ELISA can be used as a serological test to detect immune response after vaccination and/or infection with all H5N1 strains.
Three peptides, P8, P15 and MP15, with aa sequences CNTKCQTP (HA274–281), CNTKCQTPMGAINSS (HA274–288) and a tetramer of CNTKCQTPMGAINSS were all able to detect H5 antibodies in H5N1 immune sera, however, the MP15 peptide produced the highest ODs and titres with both homologous and heterologous immune sera, with low background in ELISA for non-immune commercial sera (ODs ≤ 0.2). In the initial studies of Prabakaran et al. (Citation2009), the epitope HA274–281 (corresponding to the P8) was identified as antigenic, but in the subsequent study (Velumani et al., Citation2011) a single peptide with sequence CNTKCQTPMGAINSS was used. In our study, the comparison of P8 and P15 peptides indicated that the P15 peptide is a better antigen and has higher sensitivity for detection of antibodies, therefore indicating that the epitope HA274–288 is more antigenic than the truncated epitope HA274–281. Further mapping studies would be necessary to determine more accurately the precise boundaries of this epitope.
Peptide ELISAs have been used for diagnosis of several viral pathogens (Gießauf et al., Citation2004; Gomara & Haro, Citation2007; Velumani et al., Citation2011) and the tetrameric form of the peptides also showed increased test sensitivity and low non-specific reactions with non-immune chicken sera in comparison to single peptide ELISA (Hemmatzadeh et al., Citation2013; Tarigan et al., Citation2015). The increased sensitivity in MP15 ELISA might be due to an increased number of antigenic epitopes available for antibody binding, or better antigenic presentation, as some studies have shown that most HA epitopes are conformation dependent (Yoshida et al., Citation2009; Zhao et al., Citation2011). Also, the optimized MP15 ELISA protocol was able to reduce the non-specific binding observed with the ELISA protocol used by Velumani et al. (Citation2011) without changing the sensitivity of antibody detection. Non-specific reactions occurring due to the use of serum from older chickens (Rowe et al., Citation1999) were also reduced.
The amino acid sequences of P8, P15 and MP15 were identical to the corresponding consensus sequence of Indonesian and other Asian H5N1 strains. Analysis of all Indonesian H5N1 strains for which the HA sequences were available indicated that the HA274–288 (P15) epitope was highly conserved, with only 7/113 strains being different. Five strains had one aa mutation each, at four different HA positions, 280, 283, 285 and 286, and two strains had two mutations at the same positions 277 and 283. High conservation of HA274–288 epitope is advantageous as it enables universal detection of H5N1-specific antibodies regardless of antigenic type of vaccines or challenge strains that elicited these antibodies. However, in only 60% (181/301) of H5N1 from other neighbouring Asian countries, was the HA274–288 epitope conserved, with 40% of strains (120/301) having at least one aa mutation at either position 275, 277 and/or 282. Two mutations K/M277/282R/I and N/M274/282D/V in H5N1 strains My10 and Scot, respectively, appeared not to have influenced the binding of their antisera to MP15 in ELISA, as titres of MP15 antibodies were high. However, H5N3 antisera, with a single aa mutation in HA274–288 M282 V, reacted poorly with MP15. Whether these aa mutations influence the detection of HA274–288-specific antibodies remains to be determined in a more comprehensive study that would require targeted mutation of the HA274–288 epitope.
In the studies of Prabakaran et al. (Citation2009) and Velumani et al. (Citation2011), antibodies to the HA274–288 epitope were found to be highly specific for the H5N1 subtype and antisera to other H subtypes did not react with the H5-specific HA274–288 peptide. In our study, eight out of nine antisera produced in our laboratory against different H5N1 strains were positive in the MP15 ELISA as well as the majority (46/48) of preBV and all postBV sera (47/47) from commercial flocks vaccinated with two H5N1 vaccines Legok and PWT. Five of these H5N1 strains had the HA274–288 epitope identical to the MP15, two (My10 and Scot), had two aa mutations each, whereas the sequence for strains My06 and Sb29 was not known. H5N1 strain SMI was not recognized by the MP15 ELISA regardless of its identical HA274–288 epitope to that of MP15. Antisera to H5N2 strain N28/73 with HA274–288 epitope identical to the MP15 were also recognized in the MP15 ELISA, whereas sera to H5N2 strain Mex/232/94, in which HA274–288 epitope had three aa mutations, were not. Also sera against an H5N3 virus with a single aa mutation in HA274–288 were not recognized. In agreement with the studies of Velumani et al. (Citation2011), sera to strains H9N2, H7N7 and H4N6 had a very low cross-reaction or did not react in the MP15 ELISA (titres of 200, 400 and 200, respectively). Overall, the results suggest that sera from poultry vaccinated or infected with H5N1 strains, as well as strains of other H5 subtype with conserved HA274–288, would be detected by the MP15 ELISA. However, testing field sera from birds infected with viruses of different H5N1 clades and other AIV strains would provide more insight into the subtype specificity and variation of the HA274–288 epitope-specific antibodies.
The MP15 ELISA was able to differentiate vaccinated/infected commercial poultry from those that were neither vaccinated nor infected. Almost all birds (46/48) in five commercial flocks vaccinated with H5N1 vaccines an unknown number of times and tested at 15–17 weeks of age were antibody positive in the MP15 ELISA, and the titres of antibodies ranged from 200 (below the cut-off titres of 1:400) to 2048. However, at four weeks after booster vaccination, all birds were antibody positive, giving high ODs and HA274–288 antibody titres that were significantly different from the preBV titres. In vaccinated and challenged birds, these differences were more pronounced and statistically significant. There were, however, two notable features of HA274–288 antibody response: (a) ODs and antibody titres were more widely distributed in sera/birds from the same flock and (b) in most sera, there were discrepancies between the ODs obtained in the MP15 ELISA and the titres of HA274–288 antibodies, that is, some sera with low OD had a high titre of HI antibodies or vice versa. At present, the features of antibody response to the HA274–288 epitope are unknown. For example, uniformity of antibody response following vaccination and infection, titres of antibodies induced in primary and booster vaccination and the time of appearance and duration of HA274–288 antibodies. Therefore, more experimental and field data would help to establish the detection interval of the HA274–288 antibodies.
Positive or negative results obtained with the MP15 ELISA correlated significantly [P = 0.0001] with the HI test positive or negative results, and the sensitivity and specificity of the MP15 ELISA, in comparison to the HI test, were 87% and 92%, respectively. Therefore, the MP15 ELISA demonstrated a considerable potential as test to determine if a flock is H5N1 antibody positive. One of the main advantages of the HI test is that titres are qualitative and might correlate well with protection; however, there was no correlation between the MP15 ELISA titres with the HI titres. At present, data were not available on factors that may influence HA274–288 epitope titres and further studies are needed. MP15 ELISA titres were significantly higher than the HI titres for all sera, preBV and postBV and post-challenge, and this has allowed for immune sera to be tested in the MP15 ELISA at 1:400 dilution, in comparison the HI test uses sera at 1:2 as a starting dilution. This is an advantage in routine surveillance, since a very small amount of sera are needed for testing in the MP15 ELISA.
As shown in this study, the HI test has several disadvantages. First, it was necessary to use four different HA antigens in order to obtain accurate (homologous) antibody titres. This is often not possible, as was the case in this study where one of the vaccine strains was not available for production of the HA antigen. Also, as has been the case in Indonesia, several new antigenic H5N1 variants have emerged since vaccination was introduced, all of which required change of the detecting HA antigen (Swayne et al., Citation2015). Selecting the optimal H5N1 vaccine strain and HA antigen from an array of emerging variants requires significant resources, which are often not available (Fouchier & Smith, Citation2010; Hartaningsih et al., Citation2015). Unlike the HI test, the MP15 ELISA did not significantly detect the antibody response after vaccination with Mex/232/94 which possess three aa substitutions in the HA274–288 epitope and had only 74% sensitivity but 100% specificity. These findings emphasize the importance of using epitopes relevant for the geographic region to achieve optimal results in serological tests (Freidl et al., Citation2014) and the use of vaccine seed strains selected from currently circulating strains to maintain vaccine efficacy (Lee et al., Citation2004). Vaccine pressure can cause antigenic drift especially in the HA1 region as it is highly antigenic (Lee et al., Citation2004; Escorcia et al., Citation2008), therefore, it is important to monitor the HA274–288 epitope for substitutions in the current circulating virus strains.
In summary, the MP15 ELISA provides a sensitive and specific detection of H5 subtype-specific immune responses in sera of AIV vaccinated and infected poultry. The MP15 ELISA compares well with the HI test and has an advantage for routine screening as it detects antibodies to an epitope, HA274–288, conserved in all tested H5N1 strains. Further studies are needed however to fully characterize the antigenicity of the epitope HA274–288 and validate the MP15 ELISA in experimental and commercial vaccinated and infected poultry.
Acknowledgements
The authors would like to thank the Directorate General of Animal and Livestock Health Services, Jakarta, for the permission to use the serum samples and Indrawati Sendow, John Allen and Trevor Taylor for help with shipment of samples. The assistance of District Animal Health Services from Sukabumi and Cianjur districts in West Java is gratefully acknowledged and special thanks are due to poultry farmers who gave permission and data for sample collection in Indonesia.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Peter A. Durr http://orcid.org/0000-0003-0851-5300
Additional information
Funding
References
- Alexander, D. (2006). Avian influenza viruses and human health. Developments in Biologicals, 124, 77–84.
- Boer, G.F., Back, W. & Osterhaus, A.D.M.E. (1990). An ELISA for detection of antibodies against influenza A nucleoprotein in humans and various animal species. Archives of Virology, 115, 47–61. doi: 10.1007/BF01310622
- Chen, Y.C., Chen, C.H. & Wang, C.H. (2008). H5 antibody detection by blocking enzyme-linked immunosorbent assay using a monoclonal antibody. Avian Diseases, 52, 124–129. doi: 10.1637/8076-071807-Reg
- Desvaux, S., Garcia, J.M., Nguyen, T.D., Reid, S.A., Bui, N.A., Roger, F., Fenwick, S., Peiris, J.S.M. & Ellis, T. (2012). Evaluation of serological tests for H5N1 avian influenza on field samples from domestic poultry populations in Vietnam: consequences for surveillance. Veterinary Microbiology, 156, 277–284. doi: 10.1016/j.vetmic.2011.11.010
- Escorcia, M., Vazquez, L., Mendez, S., Rodriguez-Ropon, A., Lucio, E. & Nava, G. (2008). Avian influenza: genetic evolution under vaccination pressure. Virology Journal, 5, 15. doi: 10.1186/1743-422X-5-15
- Fouchier, R.A.M. & Smith, D.J. (2010). Use of antigenic cartography in vaccine seed strain selection. Avian Diseases, 54, 220–223. doi: 10.1637/8740-032509-ResNote.1
- Freidl, G.S., de Bruin, E., van Beek, J., Reimerink, J., de Wit, S., Koch, G., Vervelde, L., van den Ham, H-J. & Koopmans, M.P.G. (2014). Getting more out of less – a quantitative serological screening tool for simultaneous detection of multiple influenza A hemagglutinin-types in chickens. PLoS ONE, 9, e108043. doi: 10.1371/journal.pone.0108043
- Gießauf, A., Letschka, T., Walder, G., Dierich, M.P. & Würzner, R. (2004). A synthetic peptide ELISA for the screening of rubella virus neutralizing antibodies in order to ascertain immunity. Journal of Immunological Methods, 287, 1–11. doi: 10.1016/j.jim.2003.12.011
- Gomara, M.J. & Haro, I. (2007). Synthetic peptides for immunodiagnosis of human diseases. Current Medicinal Chemistry, 14, 531–546. doi: 10.2174/092986707780059698
- Greiner, M., Pfeiffer, D. & Smith, R.D. (2000). Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Preventive Veterinary Medicine, 45, 23–41. doi: 10.1016/S0167-5877(00)00115-X
- Hartaningsih, N., Wibawa, H., Pudjiatmoko, H., Rasa, F.S.T., Irianingsih, S.H., Dharmawan, R., Azhar, M., Siregar, E.S., McGrane, J., Wong, F., Selleck, P., Allen, J., Broz, I., Torchetti, M.K., Dauphin, G., Claes, F., Sastraningrat, W. & Durr, P.A. (2015). Surveillance at the molecular level: developing an integrated network for detecting variation in avian influenza viruses in Indonesia. Preventive Veterinary Medicine, 120, 96–105. doi: 10.1016/j.prevetmed.2015.02.015
- Hemmatzadeh, F., Sumarningsih, S., Tarigan, S., Indriani, R., Dharmayanti, N.L.P.I., Ebrahimie, E. & Igniatovic, J. (2013). Recombinant M2e protein-based ELISA: a novel and inexpensive approach for differentiating avian influenza infected chickens from vaccinated ones. PLoS ONE, 8, e56801. doi: 10.1371/journal.pone.0056801
- Koel, B.F., van der Vliet, S., Burke, D.F., Bestebroer, T.M., Bharoto, E.E., Yasa, I.W.W., Herliana, I., Laksono, B.M., Xu, K., Skepner, E., Russell, C.A., Rimmelzwaan, G.F., Perez, D.R., Osterhaus, A.D.M.E., Smith, D.J., Prajitno, T.Y. & Fouchiera, R.A.M. (2014). Antigenic variation of clade 2.1 H5N1 virus is determined by a few amino acid substitutions immediately adjacent to the receptor binding site. mBio, 5, e01070-14.
- Lee, C.-W., Senne, D.A. & Suarez, D.L. (2004). Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. Journal of Virology, 78, 8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004
- Lee, C.-W., Senne, D.A. & Suarez, D.L. (2006). Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clinical and Vaccine Immunology, 13, 395–402. doi: 10.1128/CVI.13.3.395-402.2006
- Meulemans, G., Carlier, M.C., Gonze, M. & Petit, P. (1987). Comparison of hemagglutination-inhibition, agar gel precipitin, and enzyme-linked immunosorbent assay for measuring antibodies against influenza viruses in chickens. Avian Diseases, 31, 560–563. doi: 10.2307/1590740
- OIE. (2014). Manual of diagnostic tests and vaccines for terrestrial animals. Avian Influenza, Chapter 2.3.4., 1–23. Retrieved from http://www.oie.int/international-standard-setting/terrestrial-manual/access-online/
- Okuno, Y., Isegawa, Y., Sasao, F. & Ueda, S. (1993). A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. Journal of Virology, 67, 2552–2558.
- Postel, A., Ziller, M., Rudolf, M., Letzel, T., Ehricht, R., Pourquier, P., Dauber, M., Grund, C., Beer, M. & Harder, T.C. (2011). Broad spectrum reactivity versus subtype specificity – trade-offs in serodiagnosis of influenza A virus infections by competitive ELISA. Journal of Virological Methods, 173, 49–59. doi: 10.1016/j.jviromet.2011.01.006
- Prabakaran, M., Ho, H.T., Prabhu, N., Velumani, S., Szyporta, M., He, F., Chan, K.-P., Chen, L.-M., Matsuoka, Y., Donis, R.O. & Kwang, J. (2009). Development of epitope-blocking ELISA for universal detection of antibodies to human H5N1 influenza viruses. PLoS ONE, 4(2), e4566. doi: 10.1371/journal.pone.0004566
- Rowe, T., Abernathy, R.A., Hu-Primmer, J., Thompson, W.W., Lu, X., Lim, W., Fukuda, K., Cox, N.J. & Katz, J.M. (1999). Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. Journal of Clinical Microbiology, 37, 937–943.
- Sahini, L., Tempczyk-Russell, A. & Agarwal, R. (2010). Large-scale sequence analysis of hemagglutinin of influenza A virus identifies conserved regions suitable for targeting an anti-viral response. PLoS ONE, 5, e9268. doi: 10.1371/journal.pone.0009268
- Spackman, E. & Swayne, D.E. (2013). Vaccination of gallinaceous poultry for H5N1 highly pathogenic avian influenza: current questions and new technology. Virus Research, 178, 121–132. doi: 10.1016/j.virusres.2013.03.004
- Swayne, D.E., Beck, J.R., Garcia, M. & Stone, H.D. (1999). Influence of virus strain and antigen mass on efficacy of H5 avian influenza inactivated vaccines. Avian Pathology, 28, 245–255. doi: 10.1080/03079459994731
- Swayne, D.E., Suarez, D.L., Spackman, E., Jadhao, S., Dauphin, G., Kim-Torchetti, M., McGrane, J., Weaver, J., Daniels, P., Wong, F. & Selleck, P. (2015). Antibody titer has positive predictive value for vaccine protection against challenge with natural antigenic-drift variants of H5N1 high-pathogenicity avian influenza viruses from Indonesia. Journal of Virology, 89, 3746–3762. doi: 10.1128/JVI.00025-15
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. Retrieved from http://www.megasoftware.net/pdfs/TamuraStecher13.pdf
- Tarigan, S., Indriani, R., Durr, P.A. & Ignjatovic, J. (2015). Characterization of the M2e antibody response following highly pathogenic H5N1 avian influenza virus infection and reliability of M2e ELISA for identifying infected among vaccinated chickens. Avian Pathology, 44, 259–268. doi: 10.1080/03079457.2015.1042428
- Velumani, S., Ho, H.-T., He, F., Musthaq, S., Prabakaran, M. & Kwang, J. (2011). A novel peptide ELISA for universal detection of antibodies to human H5N1 influenza viruses. PLoS ONE, 6, e20737. doi: 10.1371/journal.pone.0020737
- Wang, G., Hu, S. & Yu, X. (2010). Development of a latex agglutination test for detecting antibodies against avian Influenza virus based on matrix 1 protein expressed in vitro. Avian Diseases, 54, 41–45. doi: 10.1637/9002-072309-Reg.1
- WHO. (2015). Cumulative number of confirmed human cases for avian influenza A (H5N1) reported to WHO, 2003–2015. Retrieved May 10, 2015, from http://www.who.int/influenza/human_animal_interface/EN_GIP_20150501CumulativeNumberH5N1cases.pdf?ua=1
- WHO/OIE/FAO H5N1 Evolution Working Group. (2014). Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza and Other Respiratory Viruses, 8, 384–388. doi: 10.1111/irv.12230
- Wiley, D.C., Wilson, I.A. & Skehel, J.J. (1981). Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature, 289, 373–378. doi: 10.1038/289373a0
- Yoshida, R., Igarashi, M., Ozaki, H., Kishida, N., Tomabechi, D., Kida, H., Ito, K. & Takada, A. (2009). Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathogens, 5, e1000350. doi: 10.1371/journal.ppat.1000350
- Zhao, R., Cui, S., Guo, L., Wu, C., Gonzalez, R., Paranhos-Baccalà, G., Vernet, G., Wang, J. & Hung, T. (2011). Identification of a highly conserved H1 subtype-specific epitope with diagnostic potential in the hemagglutinin protein of influenza A virus. PLoS One, 6, e23374. doi: 10.1371/journal.pone.0023374
- Zhou, E.M., Chan, M., Heckert, R.A., Riva, J. & Cantin, M.F. (1998). Evaluation of a competitive ELISA for detection of antibodies against avian influenza virus nucleoprotein. Avian Diseases, 42, 517–522. doi: 10.2307/1592678
