ABSTRACT
Pigeons are considered as one of the major natural reservoirs in the epidemiology of Newcastle disease (ND). In this study, the partial sequence of fusion protein gene of 17 pigeon-origin ND viruses (NDVs) isolated during 2012–2013 in Iran was analysed. Since the studied isolates showed F0 protein cleavage sites compatible with velogenic NDVs, all were considered as virulent NDVs. Two isolates carried 112RRQKRF117 as the cleavage site motif, whereas the rest demonstrated 112KRQKRF117 motif which just recently has been reported among Iranian virulent NDVs. Phylogenetic analysis divided all these diverse isolates in two distinct clusters within class II genotype VI. Based on the partial fusion protein gene sequence, 15 out of 17 isolates showed the highest genetic identity to subgenotype VIb/2 and the other two isolates were placed in a distinct genetic group of genotype VI. Based on recent findings, at least two different sublineages of genotype VI are causing the ND outbreaks in the pigeon population and are circulating simultaneously along with virulent NDVs of genotype VII in various species in Iran. The continuing circulation of a diverse group of virulent NDVs as an enzootic in widespread species such as pigeon can cause outbreaks in commercial poultry flocks and also failure in controlling programmes. Therefore, the constant monitoring and awareness of the virus characteristics should be considered in controlling programmes against ND in Iran.
Introduction
Newcastle disease (ND) is a detrimental worldwide disease having had significant impact not only on the poultry industry but also on the world economy. This economical effect is due to the enzootic or sporadic outbreaks in birds, subsequent trade restrictions and the cost of control programmes (Alexander et al., Citation2012).
Newcastle disease viruses (NDVs) are a diverse group of viruses that all belong to serotype 1 of avian paramyxovirus (APMV-1) along with 12 other serotypes constitute genus of Avulavirus of the Paramyxoviridae family (Alexander et al., Citation2012; Terregino et al., Citation2013; Yamamoto et al., Citation2015).
NDV isolates have been characterized based on pathogenicity, antigenicity, the genome size and the nucleotide sequence of the fusion protein (F) gene (Alexander et al., Citation1985; Collins et al., Citation1993; Ujvári et al., Citation2006; Diel et al., Citation2012). Different lineages or genotypes systems based on the phylogenetic analysis of the partial or complete nucleotide sequences of the F gene have been developed to classify NDV isolates (Aldous et al., Citation2004; Miller et al., Citation2010). The genotypes system, which is the mostly used system, classifies NDVs in two major groups of class I and II (Miller et al., Citation2010). However, both systems have led to much confusion in NDVs classification. Diel et al. (Citation2012) attempted to prepare a unified nomenclature and classification system of NDV genotypes based on the phylogenetic analyses of all complete F gene sequences available on GenBank. They proposed that class I viruses contain only a single genotype, whereas class II NDV isolates are divided into 18 genotypes. However, the suggestion of novel genetic group is still occurring (Courtney et al., Citation2013; De Almeida et al., Citation2013; Snoeck et al., Citation2013).
The third panzootic of ND was caused by variant strains of NDV, designated as pigeon paramyxovirus type 1 (PPMV-1) (Alexander et al., Citation1985; Aldous et al., Citation2004). Despite the initial occurrence in pigeon flocks, multiple outbreaks of PPMV-1 have been reported in chickens since the early 1980s and this situation remains a perpetual threat to the poultry industry (Alexander, Citation2011; Aldous et al., Citation2014).
Having been reported in multiple species such as wild birds, various domestic poultry and as frequent outbreaks in commercial chicken flocks since the first confirmation in 1951, ND is presently considered to be enzootic in Iran (Sohrab, Citation1974; Bozorgmehri-fard & Keyvanfar, Citation1979; Momayez et al., Citation2007; Ghiamirad et al., Citation2010; Madadgar et al., Citation2013; Hosseini et al., Citation2014). In the ND control programmes in enzootic regions, constant monitoring and identification of the causative agent are required (Miller et al., Citation2013). Yet, the characterization or detailed molecular studies, even on poultry origin isolates, is limited so far in Iran (Ebrahimi et al., Citation2012). Recently, NDV was isolated from pigeons in Iran; however, there is still a paucity of information on molecular characterization of Iranian pigeon-origin PPMV-1 (Madadgar et al., Citation2013).
In the present study, the partial sequence of the F gene of 17 NDVs isolated from pigeons in Iran was analysed to provide a better understanding of the epidemiological relationships in this variant group of APMV-1.
Materials and methods
Viruses
From January 2012 to November 2013, pigeons from distinct NDV-affected flocks with high mortality rates referred to the Avian Clinic of the University of Tehran, as a veterinary reference centre for the country, were sampled. Brain tissues were collected aseptically from dead pigeons with a history of ND including nervous signs and immediately stored at −70°C. In total, 55 brains were separately inoculated into eggs. Inoculation of 200 µl aliquots of each tissue suspensions into the allantoic cavity of five 9–10-day-old embryonated specific-pathogen-free chicken eggs was performed according to standard procedures (OIE, Citation2012). All the inoculated embryos were incubated at 37°C and monitored for 7 days. The allantoic fluids of 17 samples that caused embryo mortality within 3–4 days post-inoculation were collected, each were examined separately for haemagglutination activity (HA) and shown to be HA positive (OIE, Citation2012). Then, all 17 HA positive samples were identified as APMV-1 through a haemagglutination inhibition test by using an APMV-1 specific antiserum provided by Razi Vaccine and Serum Research Institute (Karaj, Iran). The allantoic fluid of the embryos which survived until seven days post-inoculation was extracted and shown to be negative in the HA test. All 17 pigeon-origin NDV of the present study were considered as mesogenic or lentogenic NDV based on mean death time results. Half of the studied isolates were from the flocks in Tehran province but the other 50% were from the neighbouring provinces.
Reverse transcription-polymerase chain reaction (RT-PCR)
Viral RNAs were extracted from all haemagglutination inhibition-positive allantoic fluids using High Pure Viral RNA isolation kit (Roche Molecular Biochemicals, Mannheim, Germany), as described by the manufacturer. The extracted RNAs were stored at −70°C until use. cDNA synthesis from the viral RNA was carried out by using a specific primer (Aldous et al., Citation2004), 5′-GACCGCTGACCACGAGGTTA and with the RevertAid® reverse transcriptase kit (Fermentas-Thermo Fisher Scientific, Burlington, Canada) in a 20 µl reaction volume containing 1 ng viral RNA, 60 pM final concentration of specific primer, 20 mM each dNTPs, 80 U Mmulv enzyme Revertaid®, 5 U Riboluck® RNase inhibitor, 4 µl 5 × reaction buffer and up to 20 µl DEPC treated water. The reaction mixture was incubated for 60 min at 45°C followed by 10 min at 70°C. Overlapping PCR products as a partial region of approximately 700 base pairs (bp) of the F gene, including the cleavage site, were accomplished using a pair of specific primers, as previously described by Aldous et al. (Citation2004). Primers sequences were 5′-GACCGCTGACCACGAGGTTA (Forward) and 5′-AGTCGGAGGATGTTGGCAGC (reverse). The PCR reaction mixture for each sample consisted of 50 mM MgCl2, 2.5 µl 10 × PCR buffer, 4 mM each dNTPs, 1.5 pM concentration of each primer, 1.25 U Taq DNA polymerase and 5 µl cDNA in a final volume of 25 µl. Amplification was programmed in a thermocycler (Peqstar 2X®, Peqlab, Erlangen, Germany) as follows: 95°C for 2 min followed by 35 cycles of 94°C for 1 min, 52°C for 1 min, 72°C for 3 min and a final extension at 72°C for 10 min. The amplified products were detected on SYBR® Green-stained (Invitrogen, Carlsbad, CA, USA) 1% agarose gel after electrophoresis and ultraviolet illumination.
Sequence and phylogenetic analysis
Seventeen PCR products (700 bp) were purified using the AccuPrep® DNA Gel Purification Kit (Bioneer, Daejeon, Korea) according to the manufacturer’s recommendation and submitted for automated sequencing in both directions at the Sequetech company (Mountain View, CA, USA) using PCR primers as sequencing primers. The 375-nucleotide (nt) sequences of fusion gene were aligned and analysed by BioEdit software (Hall, Citation1999) with selected sequences available in GenBank, including APMV-1 strains representing different genotypes and subgenotypes (Diel et al., Citation2012), the PPMV-1 obtained from other parts of the world representing different genetic groups (Ujvári et al., Citation2003; Aldous et al., Citation2004; Pchelkina et al., Citation2013) and the reported NDVs isolated in Iran (Ebrahimi et al., Citation2012; Hosseini et al., Citation2104; Samadi et al., Citation2014). Phylogenetic trees were constructed using the maximum likelihood method based on the Tamura-Nei model (Tamura & Nei, Citation1993) with MEGA 6 software (Tamura et al., Citation2013).
GenBank accession numbers
The sequence data from the present study were submitted to the GenBank database with the accession numbers given in .
Table 1. Seventeen PPMV-1 strains characterized in this study.
Results
The degree of nucleotide sequence divergence among all isolates of this study ranged from 0.3% to 11.5% (). A high degree of genetic diversity (11.5%) was observed between two isolates (NDP21 and NDP22) and the rest of 15 isolates, among which a diversity between 0.3% and 3% was found. The genetic similarity between NDP21 and NDP22 was 100%.
Table 2. Identity matrix of F gene partial nucleotide (nucleotides 47–420) (lower triangle) and amino acid (upper triangle) sequences of pigeon-origin APMV-1 characterized in the present study. The sequence identity matrix was generated by BioEdit software (Hall, Citation1999).
The phylogenetic trees based on the partial sequence of the F gene are shown in and . The latter shows a more detailed view of the origin of the pigeon NDV isolates. All 17 isolates belonged to genotype VI in class II APMV-1. Within genotype VI, as demonstrated in , the sequence analysis of our NDVs placed 15 isolates in subgenotype VIb but the other two isolates, NDP21 and NDP22, were placed in a distinct cluster. As shown in , VIb subgenotype has been divided in two separate groups of VIb/1 and VIb/2.
Figure 1. Phylogenetic tree based on the variable region (nucleotides 47–420) of the fusion protein gene representing different genotypes of class II, including reference strains, the Iranian strains (AY928937, AY928933, AY928934, JQ267579) and the isolates obtained in current study (bullet points). Genotype designation based on Diel et al. (Citation2012) is mentioned before accession numbers. Bootstrap values (500 replications) are shown next to the branches.
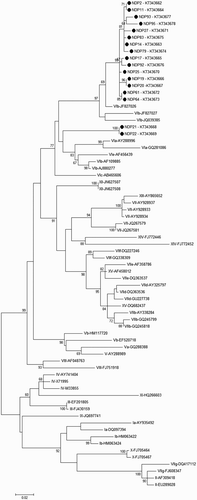
Figure 2. Phylogenetic analysis of 64 PPMV-1 strains based on the variable region (nucleotides 47–420) of the fusion protein gene. Bootstrap values (500 replications) are shown next to the branches. Genotype designation according to Ujvári et al. (Citation2003) and Diel et al. (Citation2012) is given to the right. Dots indicate isolates of the present study.
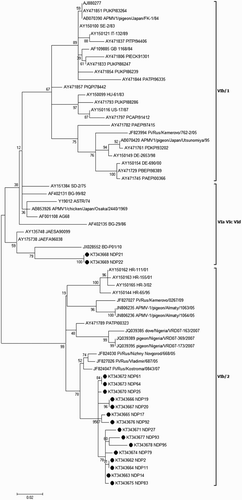
All 17 NDV isolates in different clusters shared only 88.2–91.9% similarity at the nucleotide level with the classical PPMV-1, VIb/1, which was the causative agent of the third ND panzootic. NDP21 and NDP22 isolates were close to middle-eastern fowl isolates (VIa) but separate from the classical PPMV-1 (). These two isolates, despite being obtained from ND occurrences in distinct pigeon flocks, showed 100% similarity with each other and were placed along with two NDV strains obtained from raptors in United Arab Emirates (UAE), JAESA90099 (Accession number AY135748) and JAEFA96038 (Accession number AY175738), and also with a recent pigeon-origin strain from Bangladesh, BD-P01/10 (Accession number JX028552) with 95% similarity. The other 15 NDV isolates of this study showed the highest similarity, up to 99%, to a distinct genetic cluster of subgenotype VIb, that is, VIb/2.
Nucleotide sequence of the variable region in the F gene (47–421 nt), including the precursor fusion protein (F0) cleavage site, was analysed in all 17 isolates. The deduced amino acid sequences of the cleavage site (residues 112–117) in all 17 isolates had multiple basic amino acids (MBAA) indicating that all 17 isolates belonged to velogenic Newcastle disease viruses (vNDVs) as defined by the World Organization for Animal Health (OIE). Except isolates NDP21 and NDP22, in which 112 K was replaced with R, the other 15 isolates demonstrated 112KRQKRF117 cleavage sequence.
Comparison of the partial F gene sequences among the 17 studied isolates and the Iranian chicken-origin NDV strains which were available in GenBank showed 77.8–87.1% similarity. Both NDP21 and NDP22 isolates showed identical cleavage site motif and the highest nucleotide sequence similarity with Iranian NDVs submitted to GenBank.
Discussion
ND is considered to be an enzootic disease in Iran and ND outbreaks are frequently occurring even in vaccinated poultry flocks (Madadgar et al., Citation2013; Hosseini et al., Citation2014). In the present study, we obtained 17 NDV isolates from pigeons with clinical disease and characterized each isolate based on its variable region of F gene sequence.
Since the initiation of the third ND panzootic in the 1980s, the isolation of its causative agent, PPMV-1, has been continuing in most regions of the world (Alexander, Citation2011). Ujvári et al. (Citation2003) studied a fragment of the F gene in numerous PPMV-1 isolates suggested the presence of four separate sublineages. In a phylogenetic study of 208 strains, PPMV-1 viruses were separated into six clades within two different genetic groups (Aldous et al., Citation2004). Despite using two different classification systems in the above-mentioned two studies and the variable nature of PPMV-1 strains, all isolates were situated in one group of class II APMV-1 either in 4b or in VIb. Not surprisingly, the divergence among our isolates was 11.5% for the F gene. However, the members of subgenotype VIb showed the highest similarity to our isolates. Considering this sequence divergence which indicates a continual circulation and the fact that all our 17 NDVs have been isolated from several non-related outbreaks within two years, it can be presumed that ND in pigeon population, like other countries, has been enzootic in Iran (Miller et al., Citation2010; Alexander, Citation2011; Borm et al., Citation2012; Aldous et al., Citation2014). The analysis of the nucleotide sequence of the variable region of the F gene (47–421 nt), including the cleavage site, is a standard tool for molecular characterization of NDVs and also a determinant to recognize the potential virulence of the isolates (Qin et al., Citation2008; OIE, Citation2012). One of the two criteria to recognize vNDV based on OIE definition is the presence of MBAA at the residue 112–116 of the F protein, the cleavage activation site and phenylalanine at residue 117 (OIE, Citation2012). In the present study, two different cleavage site motifs belonging to vNDV were observed. However, despite the presence of MBAA in the cleavage site of PPMV-1 strains, the lack of pathogenicity for chickens is a common finding in these strains (Meulemans et al., Citation2002; Guo et al., Citation2013) although the virulence of PPMV-1 strains for chickens after several serial passages has been already observed (Dortmans et al., Citation2011).
Circulation of vNDV and occurrence of ND in Iranian poultry flocks has already shown that all isolated NDVs belonged to genotype VII (Ebrahimi et al., Citation2012; Hosseini et al., Citation2014; Samadi et al., Citation2014). We, therefore, suggest that at least two NDV genotypes of VI and VII are concurrently enzootic in various species in Iran. This is consistent with the remarkable roles of genotypes VI and VII, respectively, in the continuing third and fourth ND panzootics (Miller et al., Citation2010; Alexander, Citation2011; Chong et al. Citation2013).
Ujvári et al. (Citation2003) reported the presence of lysine (K) in residue 112 of the fusion protein gene for the first time. There is also a report showing the motif 112KRQKRF117 in a pigeon-origin NDV isolates from China dating back to 1996 (Qin et al., Citation2008). This motif, which was dominant in the present study, has been previously reported in relation to a distinct group of pigeon-origin isolates known as VIb/2 (Śmietanka & Minta, Citation2011; Bogoyavlenskiy et al., Citation2012; Van Borm et al., Citation2012; Pchelkina et al., Citation2013). Isolates NDP21 and NDP22 of this study which showed significant nucleotide divergence from the typical PPMV-1 contained the characteristic cleavage site motif (112RRQKRF117) of PPMV-1 (Meulemans et al., Citation2002). Interestingly, the amino acid sequence of 112RRQKRF117 is the most commonly reported motif among Iranian vNDVs, especially in recent years (Ebrahimi et al., Citation2012; Hosseini et al., Citation2014).
The variable region of the F gene (47–420 nt) has been widely used in genotypic studies of NDVs (Qin et al., Citation2008; Aldous et al., Citation2014). The phylogenetic analysis of our NDVs was based on partial sequence of the F genes and the criteria used by Diel et al. (Citation2012). Our findings revealed that almost all strains remained close to cluster VIb/2 in genotype VI, which recently has been suggested as novel subgenotype VIg (Ujvári et al., Citation2003; De Almeida et al., Citation2013). The two other isolates, NDP21 and NDP22, were grouped in a distinct cluster together with PPMV-1 strains from UAE and Bangladesh (Aldous et al., Citation2004; Nooruzzaman et al., Citation2015). The degree of nucleic acid variation among these two genetic clusters is further than the suggested cut-off value to be considered as an identical subgenotype (Diel et al., Citation2012). In some studies, the classification of NDVs at the level of subgenotypes has been based on the partial sequence of the F gene (Ujvári et al., Citation2003; Aldous et al., Citation2004, Citation2014). However, the findings of recent studies have reinforced the need for the full sequence of the F gene of more distinct NDV isolates in order to propose a novel subgenotyping reliably (Diel et al., Citation2012; De almeida et al., Citation2013).
Since the first comprehensive studies on PPMV-1, a number of atypical PPMV-1 strains which are distinct from the main genetic group have been reported (Ujvári et al., Citation2003; Aldous, et al., Citation2004; Śmietanka & Minta, Citation2011). Aldous et al. (Citation2004) reported the presence of two pigeon-origin strains distinct from all other PPMV-1 in sublineage 4b and named them as 4a. In addition, the circulation of an uncertain diverse group of pigeon-origin isolates, sublineage VIb/2, between 1995 and 2002, has been reported in Croatia (Ujvári et al., Citation2003). It has been recently proved that the sublineage VIb/2 has had a prominent prevalence in the first decade of year 2000 in Russia (Pchelkina et al., Citation2013). In the present study, it was found that most of the isolates from pigeon outbreaks within years 2012–2013 in Iran had the highest genetic identity (99%) in their partial sequence of F gene with the members of subgenotype VIb/2. This data along with the evidence of pigeon NDV isolations from Nigeria, Kazakhstan and Poland suggest a more than expected extended geographical region of subgenotype VIb/2 (Śmietanka & Minta, Citation2011; Bogoyavlenskiy et al., Citation2012; Borm et al. Citation2012).
The most similar NDVs to 15 of 17 isolates of this study were a cluster of pigeon-origin isolates from Russia (Pchelkina et al., Citation2013). This is consistent with the results of previous studies about the close relation of chicken-origin genotype VII strains between Iran and Russia (Kianizadeh et al., Citation2002; Esmaelizad et al., Citation2012; Langeroudi et al., Citation2014). The presence of two different genotypes of vNDV in two distinct species of the birds in Iran and Russia suggested the occurrence of multiple introduction events. The high similarity of Iranian vNDVs to Russian strains may suggest the role of migratory wild bird populations (Kianizadeh et al., Citation2002; Esmaelizad et al., Citation2012; Langeroudi et al., Citation2014). It is not surprising to see a close relationship among APMV-1 isolates obtained from Iran with those of Russia, Kazakhstan and even the region around the Bay of Bengal. The reasons are the location of Iran on the main migration corridor to connect the wintering grounds in south (Indian subcontinent and Ethiopian Region) to the northern breeding grounds of Palaearctic and Eurasia and also the existence of some lagoons, wetlands and refuges where numerous wild birds are hosted. This North–South corridor through Iran has not only been taken by waterfowl but also is an access route for various species of raptor to reach Siberia and eastern north region of Central Asia from Arabian Peninsula such as UAE and even South Africa (Sehhatisabet & Khaleghizadeh, Citation2013). ND also affects these species in a way that the most similar isolates to NDP21 and NDP22 were two viruses of raptor origin. One of the main food resources for raptors, even in captivity, are pigeons or doves which might cause the hazard of disease transmission, in particular ND (Chitty, Citation2008). In a recent study, a phylogenic relationship was found between NDVs isolated from infected preys and raptors and a viral transmission possibility was suggested among these species (Jindal et al., Citation2010). Considering the extensive distribution of Columbidae species in Iran in the wild and around the poultry farms, the encounter of infected pigeons with raptors is not far-fetched. As mentioned above, there are some raptor origin isolates similar to both genetic clusters found in this research, which indicates the potential for intra-species transmission.
In summary, we observed the circulation of two distinct genetic groups of vNDV concurrently in Iran, causing mortality in pigeons. The achieved result together with the isolation of subgenotype VIIb and VIId from outbreaks of ND in poultry flocks (Ebrahimi et al., Citation2012; Hosseini et al., Citation2014; Langeroudi et al., Citation2014; Samadi et al., Citation2014) suggest co-circulation of at least four distinct genetic groups of vNDV, which is in agreement with earlier evidence of the simultaneous circulation of vNDVs (Aldous et al., Citation2004, Citation2014; Miller et al., Citation2010). The continuing circulation of a diverse group of vNDVs as an enzootic infection in widespread species such as pigeons can cause outbreaks in poultry and also failure of ND control programmes. Therefore, constant monitoring and awareness of the virus characteristics should be considered as important tools in any control programme against ND in Iran.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aldous, E.W., Fuller, C.M., Mynn, J.K. & Alexander, D.J. (2004). A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons. Avian Pathology, 33, 258–269. doi: 10.1080/0307945042000195768
- Aldous, E.W., Fuller, C.M., Ridgeon, J.H., Irvine, R.M., Alexander, D.J. & Brown, I.H. (2014). The evolution of pigeon paramyxovirus type 1 (PPMV-1) in Great Britain: a molecular epidemiological study. Transboundary and Emerging Diseases, 61, 134–139. doi: 10.1111/tbed.12006
- Alexander, D.J., RussellParsons, G., Abu Elzein, E.M.E., Ballouh, A., Cernik, K., Engstrom, B., Fevereiro, M., Fleury, H.J.A., Guittet, M., Kaleta, E.F., Kihm, U., Kosters, J., Lomniczi, B., Meister, J., Meulemans, G., Nerome, K., Petek, M., Pokomunski, S., Polten, B., Prip, M., Richter, R., Saghy, E., Samberg, Y., Spanoghe, L. & Tumova, B. (1985). Antigenic and biological characterization of avian paramyxovirus type 1 isolates from pigeons — an international collaborative study. Avian Pathology, 14, 365–376. doi: 10.1080/03079458508436238
- Alexander, D.J. (2011). Newcastle disease in the European Union 2000 to 2009. Avian Pathology, 40, 547–558. doi: 10.1080/03079457.2011.618823
- Alexander, D.J., Aldous, E.W. & Fuller, C.M. (2012). The long view: a selective review of 40 years of Newcastle disease research. Avian Pathology, 41, 329–335. doi: 10.1080/03079457.2012.697991
- Bogoyavlenskiy, A., BerezinPrilipov, A., Usachev, E., Korotetskiy, I., Zaitceva, I., Kydyrmanov, A. & Sayatov, M. (2012). Characterization of pigeon paramyxoviruses (Newcastle disease virus) isolated in Kazakhstan in 2005. Virologica Sinica, 27, 93–99. doi: 10.1007/s12250-012-3234-0
- Borm, S.V., Obishakin, E., Joannis, T., Lambrecht, B. & Berg, T.V.D. (2012). Further evidence for the widespread co-circulation of lineages 4b and 7 velogenic Newcastle disease viruses in rural Nigeria. Avian Pathology, 41, 377–382. doi: 10.1080/03079457.2012.696311
- Bozorgmehri-fard, M.H. & Keyvanfar, H. (1979). Isolation of Newcastle disease virus from Teals (Anas crecca) in Iran. Journal of wildlife Diseases, 15, 335–337. doi: 10.7589/0090-3558-15.2.335
- Chitty, J. (2008). Raptor: nutrition. In J. Chitty & M. Lierz (Eds.), BSAVA Manual of Raptors, Pigeons and Passerine Birds (pp. 195–200). Quedgeley: British Small Animal Veterinary Association.
- Chong, Y.L., Lam, T.T., Kim, O., Lu, H., Dunn, P. & Poss, M. (2013). Successful establishment and global dispersal of genotype VI avian paramyxovirus serotype 1 after cross species transmission. Infection, Genetics and Evolution, 17, 260–268. doi: 10.1016/j.meegid.2013.04.025
- Collins, M.S., Bashiruddin, J.B. & Alexander, D.J. (1993). Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Archives of Virology, 128, 363–370. doi: 10.1007/BF01309446
- Courtney, S.C., Susta, L., Gomez, D., Hines, N.L., Pedersen, J.C., Brown, C.C., Miller, P.J. & Afonso, C. (2013). Highly divergent virulent isolates of Newcastle disease virus from the Dominican Republic are members of a new genotype that may have evolved unnoticed for over 2 decades. Journal of Clinical Microbiology, 51, 508–517. doi: 10.1128/JCM.02393-12
- De Almeida, R.S., HammoumiGil, P., Briand, F., Molia, S., Gaidet, N., Cappelle, J., Chevalier, V., Balanc, G., Traore, A., Grillet, C., Maminiaina, O.F., Guendouz, S., Dakouo, M., Samake, K., El Mamy Bezeid, O., Diarra, A., Chaka, H., Goutard, F., Thompson, P., Martinez, D., Jestin, V. & Albina, E. (2013). New avian paramyxoviruses type I strains identified in Africa provide new outcomes for phylogeny reconstruction and genotype classification. PloS One, 8, e76413. doi: 10.1371/journal.pone.0076413
- Diel, D.G., Da Silva, L.H., Liu, H., Wang, Z., Miller, P.J. & Afonso, C.L. (2012). Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infection, Genetics and Evolution, 12, 1770–1779. doi: 10.1016/j.meegid.2012.07.012
- Dortmans, J.C.F.M., Rottier, P.J.M., Koch, G., Peeters, B.P.H. (2011). Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. Journal of General Virology, 92, 336–345. doi: 10.1099/vir.0.026344-0
- Ebrahimi, M.M., Shahsavandi, S., Moazenijula, G. & Shamsara, M. (2012). Phylogeny and evolution of Newcastle disease virus genotypes isolated in Asia during 2008–2011. Virus Genes, 45, 63–68. doi: 10.1007/s11262-012-0738-5
- Esmaelizad, M., Ashtiani, M.P., Jelokhani-Niaraki, S. & Hashemnejad, K. (2012). Identification of 23 specific nucleotide patterns in the HN gene of Newcastle disease viruses isolated from Iran. Turkish Journal of Biology, 36, 135–142.
- Ghiamirad, M., Pourbakhsh, A., Keyvanfar, H., Momayaz, R., Charkhkar, S. & Ashtari, A. (2010). Isolation and characterization of Newcastle disease virus from ostriches in Iran. African Journal of Microbiology Research, 23, 2492–2497.
- Guo, H., Liu, X., Han, Z., Shao, Y., Chen, J., Zhao, S., Kong, X. & Liu, S. (2013). Phylogenetic analysis and comparison of eight strains of pigeon paramyxovirus type 1 (PPMV-1) isolated in China between 2010 and 2012. Archives of Virology, 6, 1121–1131. doi: 10.1007/s00705-012-1572-8
- Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
- Hosseini, H., Langeroudi, A.G. & Torabi, R. (2014). Molecular characterization and phylogenetic study of Newcastle disease viruses isolated in Iran, 2010–2012. Avian Diseases, 58, 373–376. doi: 10.1637/10743-120713-Reg.1
- Jindal, N., Chander, Y., Primus, A., Redig, P.T. & Goyal, S.M. (2010). Isolation and molecular characterization of Newcastle disease viruses from raptors. Avian Pathology, 39, 441–445. doi: 10.1080/03079457.2010.517249
- Kianizadeh, M., Aini, I., Omar, A.R., Yusoff, K., Sahrabadi, M. & Kargar, R. (2002). Sequence and phylogenetic analysis of the fusion protein cleavage site of Newcastle disease virus field isolates from Iran. Acta Virologica, 46, 247–251.
- Langeroudi, A.G., Hosseini, H., Karimi, V., Hashemzadeh, M., Estabragh, A.S. & Madadgar, O. (2014). Phylogenetic study base on matrix gene of Iranian Newcastle disease virus isolates, 2011–2012. Comparative Clinical Pathology, 23, 77–81. doi: 10.1007/s00580-012-1573-8
- Madadgar, O., Karimi, V., Nazaktabar, A., Kazemimanesh, M., Ghafari, M.M., Azimi Dezfouli, S.M. & Hojjati, P. (2013). A study of Newcastle disease virus obtained from exotic caged birds in Tehran between 2009 and 2010. Avian Pathology, 42, 27–31. doi: 10.1080/03079457.2012.752791
- Meulemans, G., VanDen Berg, T.P., Decaesstecker, M. & Boschmans, M. (2002). Evolution of pigeon Newcastle disease virus strains. Avian Pathology, 31, 515–519. doi: 10.1080/0307945021000005897
- Miller, P.J., Decanini, E.L. & Afonso, C.L. (2010). Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infection, Genetics and Evolution, 10, 26–35. doi: 10.1016/j.meegid.2009.09.012
- Miller, P.J., Afonso, C.L., El Attrache, J., Dorsey, K.M., Courtney, S.C., Guo, Z. & Kapczynski, D.R. (2013). Effects of Newcastle disease virus vaccine antibodies on the shedding and transmission of challenge viruses. Developmental and Comparative Immunology, 41, 505–513. doi: 10.1016/j.dci.2013.06.007
- Momayez, R., Gharahkhani, P., Pourbakhsh, S.A., Toroghi, R., Shoushtari, A.H. & Banani, M. (2007). Isolation and pathogenicity identification of avian paramyxovirus serotype 1 (Newcastle disease) virus from a Japanese quail flock in Iran. Archives of Razi Institute, 62, 39–44.
- Nooruzzaman, M., Mazumder, A.C., Khatun, S., Chowdhury, E.H., Das, P.M. & Islam, M.R. (2015). Pathotypic and genotypic characterization of two Bangladeshi isolates of Newcastle disease virus of chicken and pigeon origin. Transboundary and Emerging Diseases, 62, 102–107. doi: 10.1111/tbed.12086
- OIE (World Animal Health Organization). (2012). Newcastle Disease. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: Mammals, Birds and Bees (pp. 555–573). Paris: Office International des Epizooties.
- Pchelkina, I.P., ManinKolosov, S.N., Starov, S.K., Andriyasov, A.V., Chvala, I.A., Drygin, V.V., Yu, Q., Miller, P.J. & Suarez, D.L. (2013). Characteristics of pigeon paramyxovirus serotype-1 isolates (PPMV-1) from the Russian Federation from 2001 to 2009. Avian Diseases, 57, 2–7. doi: 10.1637/10246-051112-Reg.1
- Qin, Z.M., Tan, L.T., Xu, H.Y., Ma, B.C., Wang, Y.L., Yuan, X.Y. & Liu, W.J. (2008). Pathotypical characterization and molecular epidemiology of Newcastle disease virus isolates from different hosts in China from 1996 to 2005. Journal of Clinical Microbiology, 46, 601–611. doi: 10.1128/JCM.01356-07
- Samadi, S., KianizadehNajafi, M.F., Nasab, S.D., Davatgar, A.M., Royaee, A. & Pilvar, P. (2014). Molecular characterization and phylogenetic study of velogenic Newcastle disease virus isolates in Iran. Virus Genes, 48, 290–295. doi: 10.1007/s11262-013-1015-y
- Sehhatisabet, M.E. & Khaleghizadeh, A. (2013). A review of current knowledge of radio-tracking of waterbirds and raptors in Iran. Podoces, 8, 22–30.
- Śmietanka, K. & Minta, Z. (2011). Isolation of an atypical pigeon paramyxovirus type 1 in Poland. Polish Journal of Veterinary Sciences, 14, 141–143. doi: 10.2478/v10181-011-0021-z
- Snoeck, C.J., Owoade, A.A., Couacy-Hymann, E., Alkali, B.R., Okwen, M.P., Adeyanju, A.T., Komoyo, G.F., Nakouné, E., Le Faou, A. & Muller, C. (2013). High genetic diversity of Newcastle disease virus in poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. Journal of Clinical Microbiology, 51, 2250–2260. doi: 10.1128/JCM.00684-13
- Sohrab, V. (1974). Newcastle disease in Iran. Bulletin de l′office International des Épizooties, 81, 899–903.
- Tamura, K. & Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10, 512–526.
- Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729. doi: 10.1093/molbev/mst197
- Terregino, C., Aldous, E.W., Heidari, A., Fuller, C.M., De Nardi, R., Manvell, R.J., Beato, M.S., Shell, W.M., Monne, I., Brown, I.H., Alexander, D.J. & Capu, I. (2013). Antigenic and genetic analyses of isolate APMV/wigeon/Italy/3920-1/2005 indicate that it represents a new avian paramyxovirus (APMV-12). Archives of Virology, 158, 2233–2243. doi: 10.1007/s00705-013-1735-2
- Ujvári, D., Wehmann, E., Kaleta, E.F., Werner, O., Savić, V., Nagy, E., Czifra, G. & Lomniczi, B. (2003). Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Research, 96, 63–73. doi: 10.1016/S0168-1702(03)00173-4
- Ujvári, D., Wehmann, E., Herczeg, J. & Lomniczi, B. (2006). Identification and subgrouping of pigeon type Newcastle disease virus strains by restriction enzyme cleavage site analysis. Journal of Virological Methods, 131, 115–121. doi: 10.1016/j.jviromet.2005.07.012
- Yamamoto, E., Ito, H., Tomioka, Y. & Ito, T. (2015). Characterization of novel avian paramyxovirus strain APMV/Shimane67 isolated from migratory wild geese in Japan. The Journal of Veterinary Medical Science, 77, 1079–1085. doi: 10.1292/jvms.14-0529
