ABSTRACT
A comparison of the unique long region 2 (UL2) gene sequences between 10 virulent and 11 attenuated duck plague virus (DPV) strains (including all DPV UL2 gene sequences registered in GenBank) showed that the UL2 genes in the attenuated DPV strains had a 528 bp deletion in the B fragment. Primers were designed based on the B fragment sequence of the UL2 gene in an attempt to establish a fluorescence quantitative polymerase chain reaction (PCR) and a conventional PCR detection method that could specifically detect virulent DPV strains (i.e. positive detection for virulent DPV strains and negative detection for attenuated DPV strains). Additionally, PCR products were cloned for sequence analysis. These two methods detected five attenuated DPV strains in addition to the virulent DPV strains. Sequence analysis of the PCR products showed that the amplified products were the B fragments of the UL2 gene. These results indicated that detection methods specific for virulent DPV strains could not be established using primers designed based on the UL2 gene B fragment.
Introduction
Duck plague, also known as duck viral enteritis, is an acute, contagious and lethal disease of ducks, geese, swans and many other species of waterfowl within the order Anseriformes (Willemse et al., Citation1994). This disease causes high mortality and decreased egg production in domestic and wild waterfowl, resulting in significant economic losses (Walker et al., Citation1969; Campagnolo et al., Citation2001; Sandhu & Shawky, Citation2003; Ji et al., Citation2009; Yang et al., Citation2014). The disease is caused by anatid herpesvirus 1 (duck plague virus, DPV). Attenuated or naturally apathogenic DPV strains are used in live vaccines to provide protection against clinical duck plague in industry settings (Islam et al., Citation2005; Liu et al., Citation2007; Li et al., Citation2009b; Lian et al., Citation2010). However, there have been reports that duck plague (caused by a virulent DPV strain) can also break out after immunization with live vaccines containing attenuated DPV strains (Diao et al., Citation2006).
The existing diagnostic methods for DPV (such as ELISA, electron microscopy, polymerase chain reaction (PCR), real time PCR and Loop-mediated isothermal amplification (LAMP) (Plummer et al., Citation1998; Kumar et al., Citation2004; Yang et al., Citation2005; Ji et al., Citation2009) cannot differentiate between virulent and attenuated DPV strains. In addition, virus isolation and identification of DPV are time consuming.
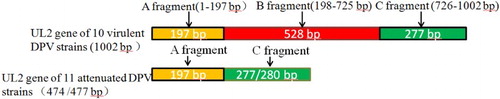
The DPV unique long region 2 (UL2) is a homologue of the herpes virus UL2 protein, which encodes a non-essential enzyme with uracil-DNA glycosylase activity (Li et al., Citation2009a). Sequence analysis of the UL2 gene of 10 virulent DPV strains and 11 attenuated DPV strains, including all the UL2 gene sequences released in GenBank, have revealed that there is a 528 bp (B fragment) deletion within the UL2 gene of the attenuated DPV strains. Therefore, based on differences in the UL2 gene between the virulent and attenuated DPV strains, this study aimed to design specific primers based on the UL2 gene-specific B fragment of the virulent strains in an attempt to establish a fluorescence quantitative PCR detection method for virulent DPV strains. However, this study was not successful in achieving its aims. We found that the attenuated DPV strains also contained the UL2 gene-specific B fragment from the virulent DPV strains, and the B fragment of the attenuated DPV strains was separate from the UL2 gene.
Materials and methods
Viruses and DNA/RNA extraction
The DPV strains used in this study are listed in . Genomic DNA were extracted from 200 μL of the viruses listed in using an EasyPure Viral DNA/RNA kit (Transgen, Beijing, China) according to the manufacturer’s protocol. DNA samples were immediately stored at −70°C until required.
Table 1. DPV strains used in the study.
Clone and sequence analysis of the UL2 gene from the DPV strains listed in Table 1
The PCR primers DPV-F: ACGAGGGAGACCCAAATGAC and DPV-R: TTTATACTGTTCCACAAGGAAGTTG were used to amplify the open reading frame (ORF) of the UL2 gene; the expected PCR amplification size was 474 bp for the attenuated strains and 1002 bp for the virulent strains. PCR amplification was performed using the conventional method. The DNA samples from the DPV strains listed in were used as templates. The PCR products were cloned into pMD18-T cloning vectors (TaKaRa Biotechnology, Dalian, China) according to the manufacturer’s instructions. The constructed recombinant plasmids were identified by PCR and then sequenced by Invitrogen (Guangzhou, China). The nucleotide sequence analysis was performed using Lasergene software (Version 7.0, Lasergene, USA). Some sequence data were submitted to GenBank.
Analysis of UL2 genes from virulent and attenuated strains
The UL2 genes of 10 virulent DPV strains were compared with those of 11 attenuated DPV strains, including all UL2 gene sequences released in GenBank (). The ORF of the UL2 gene of the attenuated DPV strains was 474 bp, but for virulent DPV strains it was 1002 bp. It was confirmed that there is a 528 bp (B fragment) deletion within the UL2 gene of the attenuated DPV strains, and that the deleted fragment was located at nucleotides 198-725 of the virulent strains (). This finding is consistent with previous results reported by Wang et al. (Citation2011) and Yang et al. (Citation2014) who compared a small number of virulent and attenuated strains. Therefore, the use of primers designed based on the B fragment sequences of the UL2 gene could theoretically establish a fluorescence quantitative PCR method for the specific detection of virulent DPV strains.
Table 2. The DPV strains used for sequence analysis.
Establishment of a quantitative fluorescence PCR method
One pair of specific primers (DPlague virulent F 507-528: TGGGCAAGATCCATATCATCAA; and DPlague virulent R 551-569: CCTCTCCGCACGCTGAAT; expected amplification size 63 bp) and one TaqMan probe (DPlague virulent T 531-549: FAM-CGGCCAGGCTCACGGGCTC-BHQ1) were designed according to the UL2 gene-specific B fragment sequences of virulent DPV strains using Primer Express 3.0 software. The primers and probe were synthesized by TaKaRa Biotechnology. The DNA samples from four virulent DPV strains (DPVGuangxi01, DPVGuangxi02, Yulin2016-60D and Yulin2016-30D), and five attenuated DPV strains (AV18 strain and four attenuated DPV vaccine strains) were used as templates.
The fluorescence quantitative PCR mix contained 1× real-time PCR Premix (Perfect Real Time PCR kit, TaKaRa Biotechnology); 0.1 μM primers of DPlague virulent F, DPlague virulent R and probe DPlague virulent T; and 2 μl of template. Distilled H2O was added to bring the final volume to 20 μl. PCR amplification consisted of an initial step at 95°C for 20 s, followed by 40 cycles at 95°C for 5 s and 60°C for 20 s.
PCR amplification of the UL2 gene B fragment
Because the fluorescence quantitative PCR method established using primers designed based on the B fragment sequences (previously expected to amplify only virulent DPV strains) also detected the attenuated strains, we suspected that the attenuated strains also contained free B fragments. Therefore, we designed conventional PCR primers (DPV F 228-246: TATAGCACAACAACCTCAG and DPV R 700-718: GCCCCTTTCGTACTGTGAG) based on the B fragment sequences to specifically amplify the B fragment (expected size 491 bp). PCR amplification was performed using the conventional method. The DNA samples from four virulent DPV strains (DPVGuangxi01, DPVGuangxi02, Yulin2016-60D and Yulin2016-30D) and five attenuated DPV strains (AV18 and four attenuated DPV vaccine strains) were used as the templates.
After a total of 50 µl of PCR product was subjected to electrophoresis, the PCR product was cloned into pMD18-T cloning vectors (TaKaRa Biotechnology) according to the manufacturer’s instructions. The constructed recombinant plasmids were identified by PCR and then sequenced by Invitrogen (Guangzhou, China). The nucleotide sequence analysis was performed using Lasergene software (Version 7.0, Madison, WI, USA).
Results
Clone and sequence analysis of the UL2 gene from the DPV strains listed in Table 1
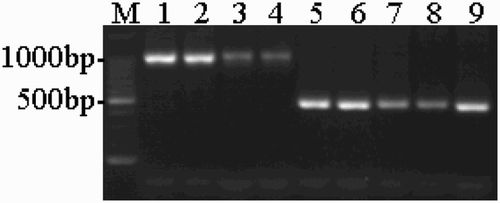
The PCR amplification results for the ORF of the UL2 gene of the DPV strains listed in are shown in . For the five attenuated DPV strains, the PCR amplification results only showed a 474 bp attenuated strain band (with its correctness also confirmed by sequencing), whereas a 1002 bp virulent strain band was not observed. The sequence analysis results confirmed that the ORF of the UL2 gene of the attenuated DPV strains (AV18 and the four vaccine strains listed in ) was 474 bp, but for the virulent DPV strains (DPVGuangxi01, DPVGuangxi02, Yulin2016-60D and Yulin2016-30D, listed in ), the length was 1002 bp. Sequence data for the virulent DPV strains Yulin2016-60D and Yulin2016-30D were submitted to GenBank under accession number KX925439 and KX925440, respectively.
Figure 2. PCR amplification of the ORF of the DPV UL2 gene. M: 100-bp DNA ladder; 1–4: virulent strains (DPV Guangxi01 strain, Guangxi02 strain, Yulin2016-60D strain and Yulin2016-30D strain); 5–8: attenuated vaccine strains (Harbin Pharmaceutical Group Bio-vaccine, Co., Ltd., Guangxi Liyuan Biological Co., Ltd., Shanghai Hile Bio-Technology, Co., Ltd. and Liaoning Yikang Biological, Co., Ltd.); 9: AV18 strain (attenuated strain).
Fluorescence quantitative PCR detection results
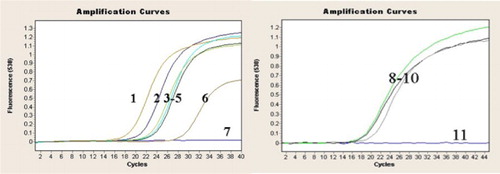
The detection results for the virulent and attenuated DPV strains using fluorescence quantitative PCR established with primers designed based on the UL2 gene B-fragment sequence are shown in . Theoretically, the fluorescence quantitative PCR method established using primers designed based on the UL2 gene B fragment sequence should have specifically detected the virulent DPV strains (i.e. shown negative detection results for the attenuated strains). However, the actual detection results showed that the established method also detected the attenuated DPV strains (curves 2–6 in ). These results indicated that the UL2 gene-specific B fragment of the virulent strains was also present in the attenuated strains and that the B fragment was separate from the UL2 gene in the attenuated strains.
Figure 3. Detection of DPV virulent and attenuated strains using fluorescence quantitative PCR. 1: Yulin2016-30D strain (virulent); 2–5: attenuated vaccine strains (Harbin Pharmaceutical Group Bio-vaccine Co., Ltd., Guangxi Liyuan Biological Co., Ltd., Shanghai Hile Bio-Technology, Co., Ltd. and Liaoning Yikang Biological, Co., Ltd.); 6: AV18 strain (attenuated); 7: Normal duck embryo allantoic fluid; 8–10: virulent strains (DPV Guangxi01strain, Guangxi02 strain and Yulin2016-60D strain); 11: Normal duck embryo allantoic fluid.
PCR amplification of the UL2 gene B fragment
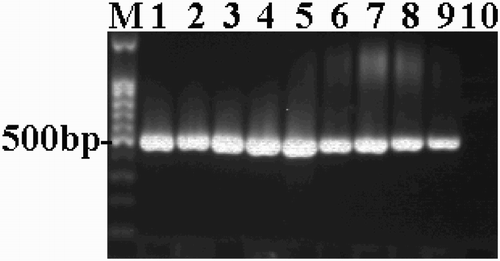
The results for the virulent and attenuated DPV strains detected using the conventional PCR method established using primers designed based on the UL2 gene B fragment are shown in . This method amplified 491 bp bands consistent with the experimental design based on the four virulent DPV strains (DPVGuangxi01, DPVGuangxi02, Yulin2016-60D and Yulin2016-30D) and five attenuated strains (AV18 and the attenuated vaccine strains listed in ). The results were in agreement with the fluorescence quantitative PCR results.
Figure 4. The PCR-amplified B fragments of the UL2 gene. M: 100-bp DNA ladder; 1–4: virulent strains (DPV Guangxi01strain, Guangxi02 strain, Yulin2016-60D strain and Yulin2016-30D strain); 5–9: attenuated vaccine strains (Harbin Pharmaceutical Group Bio-vaccine, Co., Ltd., Guangxi Liyuan Biological Co., Ltd., Shanghai Hile Bio-Technology, Co., Ltd. and Liaoning Yikang Biological, Co., Ltd.); 9: AV18 strain (attenuated strain); 10: Normal duck embryo allantoic fluid.
Sequencing analysis results
The sequencing analysis results confirmed that the amplified PCR products from the virulent and attenuated DPV strains were all 491 bp in size, which is consistent with the anticipated results. The sequences were subjected to Lasergene software analysis (). The results showed that the sequences for the five attenuated strains (AV18 and four attenuated vaccine strains) had consistent homology with the corresponding fragment (i.e. the B fragment of the UL2 gene) of the virulent DPV strains used for primer design.
Figure 5. Sequence analysis of the B fragment of the UL2 gene by Lasergene software. B fragment sequences for the five attenuated strains (AV18 and four attenuated vaccine strains) had consistent homology with the corresponding fragment (i.e. the B fragment of the UL2 gene) of the 10 virulent DPV strains. “-” represent nucleotide deletion.

Discussion
This study compared UL2 gene sequences between 11 attenuated and 10 virulent DPV strains (including all DPV UL2 gene sequences registered in GenBank). The UL2 genes in the attenuated DPV strains all contained a 528 bp deletion in the B fragment. These results were consistent with the results reported by Wang et al. (Citation2011) and Yang et al. (Citation2014), who compared only a small number of virulent and attenuated strains. Therefore, the fluorescence quantitative PCR method established using primers designed based on the UL2 gene B fragment theoretically should have specifically detected the virulent DPV strains and given negative results for the detection of the attenuated strains. However, the actual results showed that the established fluorescence quantitative PCR method also detected the attenuated DPV strains.
Therefore, we hypothesized that the UL2 gene-specific B fragment in the virulent strains was also present in the attenuated DPV strains but was separate from the UL2 gene. To validate this hypothesis, we established a PCR method for the specific amplification of the B fragment. This PCR method amplified the B fragment from five attenuated DPV strains in which the B fragment had been confirmed by sequencing. The results were consistent with the fluorescence quantitative PCR results. We did not discover the insertion of the B fragment into other locations in the DPV genome through analysis of whole gene sequences from the attenuated DPV strains in GenBank. Therefore, we speculated that either the B fragment has been separated from the DPV genome to become a free fragment or that the whole genome sequences of the existing attenuated DPV strains were not comprehensively sequenced. This issue requires further study for clarification.
The results of this study indicated that methods for the specific detection of virulent DPV strains could not be established using primers designed for the UL2 gene B fragment. To establish detection methods for virulent DPV strains, the upstream primer can be designed such that it is based on the UL2 gene A fragment, whereas the downstream primer can be designed such that it is based on the C fragment. Virulent and attenuated DPV strains can be distinguished based on the size of the amplified UL2 gene, because the size of the virulent strains is 528 bp larger than that of the attenuated strains. The results of this study provide very meaningful results to assist others and avoid repeating work that is not feasible.
The five attenuated DPV strains used for fluorescence quantitative PCR and conventional PCR detection included one attenuated strain purchased from the China Institute of Veterinary Drug Control and four commercially available attenuated vaccine strains. The same batch of the attenuated vaccine strains used in the present study was also used for the vaccination of ducks on a farm to confirm the safety of the vaccines and thereby to confirm the absence of contamination by virulent DPV strains. The ducks did not die after immunization with the DPV vaccine, thereby showing that the strains had not been contaminated with virulent DPV. Additionally, our group performed PCR amplification, cloning and sequencing of the UL2 genes from the five attenuated strains used in the present study. The PCR amplification results showed only a 474 bp attenuated strain band (the correctness of which was also confirmed by sequencing), whereas a 1002 bp virulent strain band was not observed. These results verified that the attenuated strains used in the present study were not contaminated by virulent strains, which ensured the reliability of our results.
Moreover, to avoid laboratory contamination, we asked the Fangchengguang Center for Animal Disease Control and Prevention (FCADC) to replicate our research. All materials were prepared by the FCADC, including four attenuated vaccine strains purchased from four different companies by the FCADC. The primer and all other regents were also purchased by the FCADC. The FCADC’s detected results were the same as ours; therefore their results corroborated the reliability of ours.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Campagnolo, E.R., Banerjee, M., Panigrahy, B. & Jones, R.L. (2001). An outbreak of duck viralenteritis (duck plague) in domestic muscovy ducks (Cairina moschata domesticus) in Illinois. Avian Diseases, 45, 522–528. doi: 10.2307/1592999
- Diao, Y., Lu, G., Zheng, Y., Yang, J.H., Chen, Q.P. & Liu, Y.Z. (2006). Isolation and identification and characteristics of the pathogen of a new duck plague. Chinese Journal of Veterinary Science, 26, 136–139.
- Islam, M.A.I., Samad, M.A.S., Rahman, M.B.R., Hossain, M.H. & Akter, S.A. (2005). Assessment of immunologic responses in Khaki Cambell Ducks vaccinated against duck plague. International Journal of Poultry Science, 4, 36–38. doi: 10.3923/ijps.2005.36.38
- Ji, J., Du, L.Q., Xie, Q.M., Cao, Y.C., Zuo, K.J., Xue, C.Y., Ma, J.Y., Chen, F. & Be, Y.Z. (2009). Rapid diagnosis of duck plagues virus infection by loop-mediated isothermal amplification. Research in Veterinary Science, 87, 53–58. doi: 10.1016/j.rvsc.2008.11.003
- Kumar, N.V., Reddy, Y.N. & Rao, M.V.S. (2004). Enzyme linked immunosorbent assay for detection of duck plague virus. Indian Veterinary Journal, 5, 481–483.
- Li, H., Liu, S., Han, Z., Shao, Y., Chen, S. & Kong, X. (2009a). Comparative analysis of the genes UL1 through UL7 of the duck enteritis virus and other herpesviruses of the subfamily Alphaherpesvirinae. Genetics and Molecular Biology, 32, 121–128. doi: 10.1590/S1415-47572009005000003
- Li, Y., Huang, B., Ma, X., Wu, J., Li, F., Ai, W., Song, M. & Yang, H. (2009b). Molecular characterization of the genome of duck enteritis virus. Virology, 391, 151–161. doi: 10.1016/j.virol.2009.06.018
- Lian, B., Xu, C., Cheng, A., Wang, M., Zhu, D., Luo, Q., Jia, R., Bi, F., Chen, Z., Zhou, Y., Yang, Z. & Chen, X. (2010). Identification and characterization of duck plague virus glycoprotein C gene and gene product. Virology Journal, 7, 349. doi: 10.1186/1743-422X-7-349
- Liu, S., Chen, S., Li, H. & Kong, X. (2007). Molecular characterization of the herpes simplex virus 1 (HSV-1) homologues, UL25 to UL30, in duck enteritis virus (DEV). Gene Therapy, 401, 88–96.
- Plummer, P.J., Alefantis, T., Kaplan, S., O’Connell, P., Shawky, S. & Schat, K.A. (1998). Detection of duck enteritis virus by polymerase chain reaction. Avian Diseases, 42, 554–564. doi: 10.2307/1592682
- Sandhu, T. & Shawky, S. (2003). Duck virus enteritis (duck plague). In Y.M. Saif, H.J. Barnes, J.R. Glisson, A.M. Fadly, L.R. McDougald, & D.E. Swayne (Eds.), Diseases of Poultry 11th edn (pp. 354–363). Ames: Iowa State Press.
- Walker, J.W., Pfow, C.J., Newcomb, S.S., Urban, W.D., Nadler, H.E. & Locke, L.N. (1969). Status of duck virus enteritis (duck plague) in the United States. Proceedings Annual Meeting of the United States Animal Health Association, 73, 254–279.
- Wang, J., Höper, D., Beer, M. & Osterrieder, N. (2011). Complete genome sequence of virulent duck enteritis virus (DEV) strain 2085 and comparison with genome sequences of virulent and attenuated DEV strains. Virus Research, 160, 316–325. doi: 10.1016/j.virusres.2011.07.004
- Willemse, M.J., Chalmers, W.S., Cronenberg, A.M., Pfundt, R., Strijdveen, I.G. & Sondermeijer, P.J. (1994). The gene downstream of the gC homologue in feline herpes virus type 1 is involved in the expression of virulence. Journal of General Virology, 75, 3107–3116. doi: 10.1099/0022-1317-75-11-3107
- Yang, C., Li, Q., Li, J., Zhang, G., Li, H., Xia, Y., Yang, H. & Yu, K. (2014). Comparative genomic sequence analysis between a standard challenge strain and a vaccine strain of duck enteritis virus in China. Virus Genes, 48, 296–303. doi: 10.1007/s11262-013-1009-9
- Yang, F.L., Jia, W.X., Yue, H., Luo, W., Chen, X., Xie, Y., Zen, W. & Yang, W.Q. (2005). Development of quantitative real-time polymerase chain reaction for duck enteritis virus DNA. Avian Diseases, 49, 397–400. doi: 10.1637/7338-020305R.1