ABSTRACT
Newcastle disease (ND), caused by virulent Avian avulavirus 1 (AAvV 1), affects a wide range of avian species worldwide. Recently, several AAvVs of diverse genotypes have emerged with varying genomic and residue substitutions, and subsequent clinical impact on susceptible avian species. We assessed the clinico-pathological influence of two different AAvV 1 pathotypes [wild bird originated-velogenic strain (sub-genotype VIIi, MF437287) and feral pigeon originated-mesogenic strain (sub-genotype VIm, KU885949)] in commercial broiler chickens and pigeons. The velogenic strain caused 100% mortality in both avian species while the mesogenic strain caused 0% and 30% mortality in chickens and pigeons, respectively. Both strains showed tissue tropism for multiple tissues including visceral organs; however, minor variances were observed according to host and pathotype. The observed gross and microscopic lesions were typical of AAvV 1 infection. Utilizing oropharyngeal and cloacal swabs, a comparable pattern of viral shedding was observed for both strains from each of the infected individuals of both avian species. The study concludes a varying susceptibility of chickens and pigeons to different wild bird-originated AAvV 1 pathotypes and, therefore, suggests continuous monitoring and surveillance of currently prevailing strains for effective control of the disease worldwide, particularly in disease-endemic countries.
Introduction
Viral infectious diseases lead to serious economic losses to the poultry trade worldwide (Haji-Abdolvahab et al., Citation2019). Among these, Newcastle disease (ND), caused by Newcastle disease virus (NDV) or Avian avulavirus 1 (AAvV 1), is a highly contagious disease affecting multiple avian species globally (Alexander & Senne, Citation2008). The virus belongs to the genus Orthoavulavirus within the family Paramyxoviridae (Amarasinghe et al., Citation2019). It has a mono-partite, negative-sense single-stranded RNA genome which is either 15186, 15192 or 15198 nucleotides in length (Kolakofsky et al., Citation2005). The complete genome contains six coding genes; nucleocapsid (N), phosphoprotein (P), matrix (M), fusion (F), haemagglutinin-neuraminidase (HN) and large polymerase (L) protein in the order 3′-NP-P/V/W-M-F-HN-L-5′ (Kolakofsky et al., Citation2005). Based on pathogenicity to chickens, AAvV 1 strains are categorized into four major pathotypes: velogenic (high virulent), mesogenic (moderate virulent), lentogenic (low virulent) and asymptomatic (non-virulent) (Alexander et al., Citation2012). Subsequent to infection with velogenic and mesogenic AAvV 1 strains, with a mortality of 60-100%, infected birds exhibit clinical signs corresponding to the gastrointestinal, respiratory and nervous systems depending upon the virus pathotype, target avian species and health status of birds (Kommers et al., Citation2001; Wakamatsu et al., Citation2006).
Despite mass vaccination and strict biosecurity measures, ND outbreaks are frequently observed in both vaccinated and non-vaccinated flocks (Munir et al., Citation2012; Rehmani et al., Citation2015). AAvV 1 is a continuously evolving virus with evidence of accelerated evolution of the virulent strains (Fan et al., Citation2017). In this regard, the isolation of novel avulaviruses of varying pathogenicity and genotypes from a wide range of hosts (Ramey et al., Citation2013; Shabbir et al., Citation2016; Brown & Bevins, Citation2017; Dodovski et al., Citation2017; Qiu et al., Citation2017; El Naggar et al., Citation2018; Aziz-ul-Rahman et al., Citation2018a, Citation2018b; Citation2019a, Citation2019b, Citation2019c) suggests a continuous virus evolution with subsequent implications for disease spread (Solomon et al., Citation2012; Miller et al., Citation2015; Fan et al., Citation2017; Mayahi & Esmaelizad, Citation2017). Subsequent to evolution, the emergence of novel genotypes and sub-genotypes are now increasing over a period of time (Dimitrov et al., Citation2019a). The phylogenetic analysis revealed the circulation of 21 genotypes globally and, among these, genotype VI and VII viruses are predominantly circulating and causing several outbreaks in developing countries including Pakistan (Liang et al., Citation2002; Sabra et al., Citation2017; Abolnik et al., Citation2018; Wajid et al., Citation2018; Ferreira et al., Citation2019; Dimitrov et al., Citation2019b).
Owing to the distribution of various genotypes in a particular region (Aziz-ul-Rahman et al., Citation2019c), the on-going outbreaks have been confirmed to be sub-genotypes of VI and VII viruses, which caused enormous mortality in susceptible hosts and also suggested their panzootic potential (Miller et al., Citation2015). Therefore, it is imperative to perform a clinico-pathological evaluation of currently prevalent strains to determine how the viruses are changing. The current study is an extension of our previous investigations of genetic characterization of Anseriformes- (Aziz-ul-Rahman et al., Citation2018b) and pigeon- (Akhtar et al., Citation2016) originated AAvV 1 strains because a full characterization including genomic, genotypic and clinical pathogenesis of currently prevailing strains could also improve the diagnostic aspects of disease and implementation of the control measures. Although the susceptibility of chickens and pigeons to AAvV 1 infection has been reported (Wakamatsu et al., Citation2006; Susta et al., Citation2011; Isidoro-Ayza et al., Citation2017), information related to comparative clinico-pathological assessment of currently prevailing Anseriformes- and pigeon-originated AAvV 1 viruses of varying pathogenicity and genotypes in chickens and pigeons is scarce. Besides continuous disease monitoring, surveillance and subsequent genome-based characterization, such substantial evidence calls for experiments related to infectivity potential, transmission and shedding patterns of newly characterized isolates of varying pathogenicity in their susceptible hosts so that, if required, existing disease surveillance and control strategies could be revised in disease-endemic settings. Therefore, the current study was intended to evaluate the infectious potential of field prevailing velogenic (sub-genotype VIIi) and mesogenic (sub-genotype VIm) strains in commercial chickens and pigeons under experimental and controlled conditions.
Materials and methods
Ethics statement
The study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health and Animal Research Council (https://grants.nih.gov/grants/olaw/guide-for-the-care-and-use-of-laboratory-animals.pdf). All protocols including bird management, virus challenge, tissue and swab sampling, and sacrifice of birds were approved by the Ethical Review Committee for Use of Laboratory Animals (ERCULA) of University of Veterinary and Animal Sciences (UVAS), Lahore (Permit Number: ORIC/DR-70).
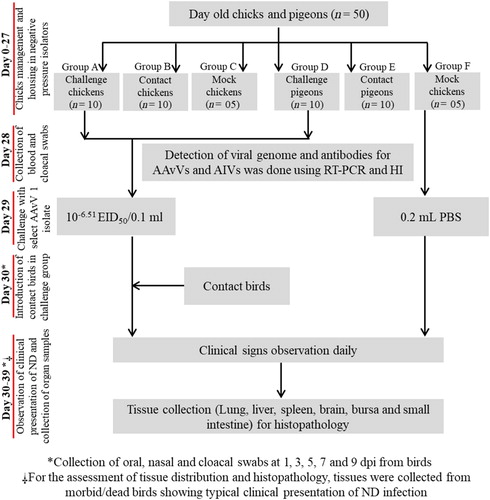
Experimental challenge of velogenic and mesogenic AAvV 1 in chickens and pigeons
Two individual challenge studies were performed separately for the assessment of the infectious potential of previously characterized velogenic (MF437287; Anas carolinensis-II-UVAS-Pak-2015) (Aziz-ul-Rahman et al., Citation2018b) and mesogenic (KU885949; Pigeon/MZS-UVAS-Pak/2014) (Akhtar et al., Citation2016) strains in commercial broiler chickens and pigeons. The velogenic strain was isolated from an asymptomatic duck (A. carolinensis) during a routine avian influenza surveillance programme, whereas; the mesogenic strain was isolated from a ND outbreak in a feral pigeon flock. Recently genomic and genotypic analysis has categorized the velogenic strain as a virus of sub-genotype VIIi (Aziz-ul-Rahman et al., Citation2018b) while the mesogenic strain was categorized as a virus of sub-genotype VIm (Akhtar et al., Citation2016). A total of 50 birds each of clinically healthy broiler chicken (25 for velogenic challenge experiment and 25 for mesogenic challenge experiment) and pigeons (25 for velogenic challenge experiment and 25 for mesogenic challenge experiment) were used. One-day-old broiler chicks were procured from an ISO certified commercial hatchery and raised in the bird experiment unit at Quality Operations Laboratory (QOL), UVAS, Lahore, Pakistan, until the beginning of experimentation. Healthy pigeons were procured from the live bird market located near Azadi Chowk (Minar-e-Pakistan), Lahore. Each challenge study was conducted for 39 days, during which chickens were housed in adequate facilities to attain maturity age till 27 days, while 2-month-old pigeons were housed for adaptation of captivity and visual inspection of any clinical sign prior to challenge. Feed and water were provided ad libitum to all birds along with general bird care as recommended by the ethical committee of the institute. All the birds were screened for the existence of viral genome in oropharyngeal and cloacal swabs, and antibodies in blood against avian influenza virus (H9N2) and AAvV 1 using reverse-transcriptase polymerase chain reaction (RT–PCR) and haemagglutination inhibition assay (HI) (Munir et al., Citation2012; Ali et al., Citation2017), respectively. Twenty-nine-day-old chickens and 62-day-old pigeons were exposed to the selected virus pathotype. The experimental birds were randomly divided into six groups. For chickens, group A (n = 10) had birds that were challenged with the velogenic strain, group B (n = 10) represented contact birds while group C (n = 05) served as mock or negative control. Similarly, for pigeons, group D (n = 10) had birds that were challenged with velogenic strain, group E (n = 10) represented contact birds while group F (n = 5) served as mock or negative control (). Regarding challenge with velogenic AAvV 1 strain, groups A and D were exposed intranasally to 10−6.51 EID50/0.1 ml of A. carolinensis-II-UVAS-Pak-2015. After 24 h of infection, the contact chickens (group B) and pigeons (group E) were mixed together with challenged birds [group B within challenged chickens (group A) and group E within challenged pigeons (group D)], accordingly. In the second challenge experiment with mesogenic AAvV 1 strain, a similar approach was employed. For chickens, group A′ (n = 10) had birds that were challenged with the mesogenic strain, group B′ (n = 10) represented contact birds while group C′ (n = 5) served as mock or negative control. Similarly, for pigeons, group D′ (n = 10) had birds that were challenged with mesogenic strain, group E′ (n = 10) represented contact birds, while group F′ (n = 5) served as mock or negative control. Regarding challenge with mesogenic AAvV 1 strain, groups A′ and D′ were exposed intranasally to 10−6.87 EID50/0.1 ml of Pigeon/MZS-UVAS-Pak/2014. Mock (groups C, C′ and F, F′) were inoculated with 0.2 ml phosphate-buffered saline (PBS). After 24 h of infection, the contact chickens (group B′) and pigeons (group E′) were mixed together with challenged birds [group B′ within challenged chickens (group A′)] and group E′ within pigeons (group D′), accordingly. All birds were monitored daily for the clinical presentation of the disease till completion of the experiment.
Viral shedding, horizontal transmission and tissue distribution of AAvV 1
Oropharyngeal and cloacal swabs were collected as aseptically as possible from challenged (groups A, A′ and D, D′) and contact birds (groups B, B′ and E, E′) on 1, 3, 5, 7 and 9 days post infection (dpi), and processed for virus isolation and identification (Munir et al., Citation2012). For this purpose, all swab samples were transferred into cryovials (2.0 ml) containing 1.5 ml brain heart infusion medium with antibiotics (penicillin 2000 IU/ml, and gentamicin 200 μg/ml) and antifungal (fungizone 1.5 μg/ml). After centrifugation at 2500 × g for 5 min, approximately 1.0 ml of each sample was filtered through a 0.22 μm syringe filter (EMD Millipore Millex™, Millipore Billerica MA, USA). A 0.2 ml aliquot of the filtrate was inoculated into 9-day-old embryonated chicken eggs. The spot haemagglutination (HA) positive harvested allantoic fluid was tested for AAvV 1 using F gene-based reverse-transcriptase polymerase chain reaction (RT–PCR) (Munir et al., Citation2012). For an assessment of tissue tropism, tissue samples (n = 19) including feather follicle, breast muscle, brain, whole eye, harderian glands, trachea, tongue, oesophagus, gizzard, proventriculus, liver, heart, lungs, kidney, spleen, bursa, small intestine, caecum and caecal tonsils were collected from recently deceased/diseased birds showing typical signs of ND. These tissues were then processed individually for RNA extraction following the manufacturer’s instructions (QIAmp Viral RNA extraction Mini Kit, Qiagen, Valenica, CA), and F gene-based molecular identification via RT–PCR (Munir et al., Citation2012).
Histopathological examination of tissues
In order to assess the severity of infection in chickens and pigeons to wild bird-originated velogenic and mesogenic AAvV 1 strains, the tissue samples were collected from birds immediately after death, and from those infected birds of groups A, A′, B, B′, D, D′, E and E′, showing severe typical clinical signs of ND infection (respiratory and/or nervous signs) regardless of timeline. Selected tissue samples (brain, liver, trachea, lungs, kidney, spleen, bursa and intestine) were kept separately in 10% neutral buffered formalin (NBF) solution for haematoxylin and eosin staining for subsequent microscopic changes under a light microscope (Carleton et al., Citation1980).
Results
Clinical presentation of velogenic AAvV 1 infection in chickens and pigeons
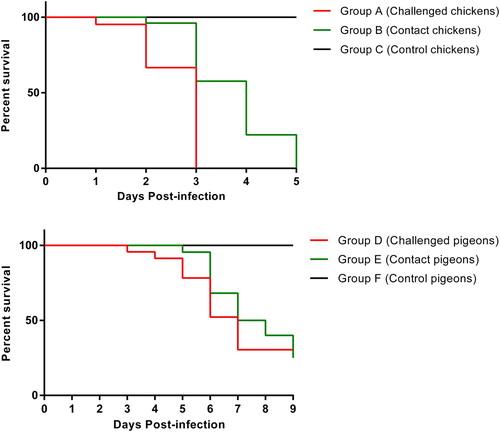
The clinical presentation showed variations in severity level and duration of clinical signs between the challenge (groups A and D) and contact birds (groups B and E) over time of post-infection. The challenged and contact chickens (groups A and B) showed 100% mortality by 6 dpi. In contrast, challenged and contact pigeons (groups D and E) showed 100% morbidity and mortality by 10 dpi. Briefly, the severity of clinical signs was more pronounced in chickens compared to pigeons. The commonly observed clinical signs were general sickness (anorexia, lethargy and depression), reluctance to move, respiratory distress (open mouth breathing, sneezing, coughing, respiratory sounds, oculonasal discharge) and diarrhoea as a gastric sign. No neurological signs were noted in chickens (groups A and B). Sudden death with no-to-negligible clinical signs was observed only in group A but no sudden death was observed in any bird of groups D and E. Moreover, neurological signs were observed in pigeons of groups D and E. Birds of group A showed clinical signs from 2 dpi with mortality of three chickens. The clinical infection became exacerbated and peaked at 3 dpi with the death of four chickens. After 3 dpi, minor clinical signs were observed in one bird of group B. At the end of 4 dpi, the remaining challenged chickens (group A) also succumbed. At 5 dpi, severe clinical presentation of ND was observed in three birds of group B (contact chickens) and all contact birds died at the end of 6 dpi (). The pigeons of group D exhibited clinical signs from 3rd dpi with mild clinical signs of respiratory distress. The clinical infection was exacerbated and peaked at 4 dpi with the death of two pigeons in group D. At 5 dpi, nervous signs were observed in a few birds of group D with the death of three pigeons. After 5 dpi, mild clinical signs were observed in few birds of group E. At the end of 6 dpi, two pigeons from group D were found dead and clinical infection was enhanced and peaked with death of three pigeons in group E. At the end of 7 dpi, two pigeons from group D and four pigeons from group E were found dead. Subsequently, three deaths were observed in birds of each group D and E at 8 dpi (). The mock-infected (groups C and F) remained normal throughout the experiment. Necropsy revealed characteristic lesions corresponding to ND infection and included haemorrhages in lungs and liver, enlarged liver, congested kidneys, mottled spleen, pinpoint haemorrhages in proventricular glands and oedematous bursa. The disease outcomes coupled with gross pathognomic lesions revealed a remarkable difference in the susceptibility of the two avian species; chickens (groups A′, A, B, B′) and pigeons (groups D, D′, E, E′).
Virus shedding, horizontal transmission and velogenic AAvV 1 tissue distribution
Throughout the whole experiment, the collected oropharyngeal and cloacal swabs showed a discrete pattern of virus shedding without any noticeable difference with respect to dpi within the same group of challenged chickens (group A) and contact chickens (group B). Virus shedding was detected in group A form 2 dpi. On the other hand, it was detected in group B from 4 dpi. Similarly, for pigeons, virus shedding was detected in pigeons (group D) from 3 dpi onwards while, in contact birds, it was detected from 5 dpi onwards. Mock-infected birds (groups C and F) remained virus-negative throughout the whole experiment. In order to assess the tissue tropism of velogenic AAvV 1, the collected tissues (n = 19) from dead or diseased birds (pigeons and chickens) showing typical clinical signs and necropsy lesions were processed for detection of the viral RNA. For chickens (groups A and B), the virus RNA was detected in all tissue samples. However, for pigeon (groups D and E), the existence of viral genome was detected in all tissues except kidney, feather follicle, heart and breast muscle.
Histopathological findings for velogenic AAvV 1 infection
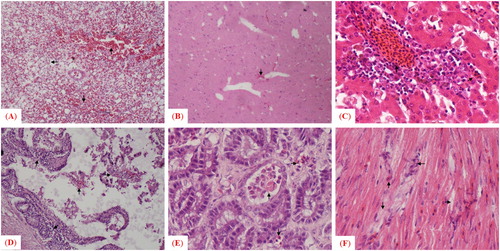
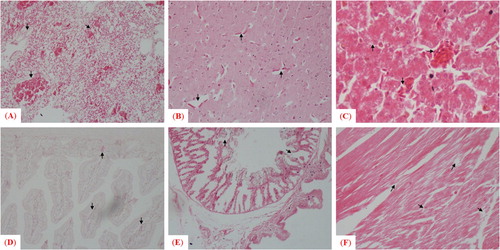
The velogenic AAvV 1 strain caused notable histopathological lesions in morbid birds of both avian species. For chickens, the microscopic findings were consistent with the afore-mentioned gross lesions such as congestion, haemorrhages with mononuclear inflammatory cell infiltration in sub-mucosa of trachea, lung, kidney and spleen, a mild congestion in brain, degeneration of hepatocytes, fatty changes (vacuoles in hepatocytes), infiltration of inflammatory cells in the portal cord of the liver, damaged basal membrane and degeneration in follicles in bursa of Fabricius along with presence of dead/necrotic tissue masses, and sloughing of epithelium in the small intestine. In contrast, the microscopic findings were comparatively less pronounced in pigeons (groups D and E) than to chickens (groups A and B). Lesions included mild congestion in brain, disruption of cardiac fibres with accumulation of immune cells in heart, congestion in parabronchial blood capillaries, haemorrhages with mononuclear inflammatory cells infiltration in lungs, fatty changes evidenced by vacuoles of varying sizes in the cytoplasm of hepatocytes, infiltration of inflammatory cells in the portal cord, engorgement of sinusoidal capillaries with red blood cells and Kupffer cells in liver, sloughing of epithelium in the small intestine, and infiltration of inflammatory cells in the gizzard ().
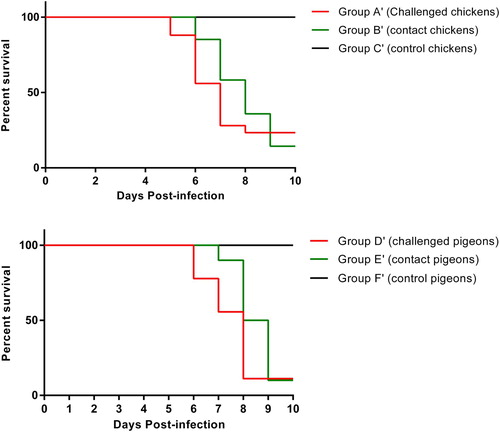
Clinical presentation of mesogenic AAvV 1 infection in chickens and pigeons
Based on severity and duration of clinical infection, variations in the clinical signs were observed among challenged birds (groups A′ and D′) and contact birds (groups B′ and E′) over a designated time period. The challenged and contact chickens (groups A′ and B′) showed 40% morbidity without any mortality by 10 dpi. Notably, neurological signs were also observed in chickens of groups A′ and B′, however, no sudden death was observed in any group. Birds of group A showed mild clinical signs from 5 dpi onward. Minor clinical signs were observed in the birds of group B′ by 6 dpi (). The challenged and contact pigeons (groups D′ and E′) showed 50% morbidity and 30% mortality by 10th dpi. Clinical signs, including respiratory and nervous signs, were also observed in pigeons of groups D′ and E′. Similar, but less severe, clinical signs were observed in all birds as reported in the previous velogenic-challenge experiment. The birds of group D′ showed clinical signs from 6 dpi. At 7 dpi, mild nervous signs were observed in a few birds of group E. After 8 dpi, mild respiratory signs were also observed in few birds of groups D′ and E′ (). The control birds (groups C′ and F′) remained normal during the whole experiment. Upon post mortem examination of infected/morbid birds, characteristic necropsy lesions, including haemorrhages and congestion in brains and lungs, enlarged liver, congested kidneys, mottled spleen, pinpoint haemorrhages in proventricular glands and oedematous bursa, were observed.
Virus shedding, horizontal transmission and mesogenic AAvV 1 tissue distribution
Virus shedding was detectable in challenged chickens of group A′ from 5 dpi onward, whereas the virus was detected in swabs of contact chickens of group B′ from 6 dpi onward. Throughout the whole experiment, the oropharyngeal and cloacal swabs showed a discrete pattern of virus shedding without any noticeable difference with respect to dpi within the same group of challenged and contact chickens (groups A′ and B′). Similarly, for pigeons, virus shedding was detectable in challenged pigeons (group D′) from 6 dpi onward while, in contact pigeons (group E′), it was detected from 7 dpi onward. Collectively, the virus was detected in cloacal swabs from 6 dpi onward, while it was detectable from 7 dpi onward in oropharyngeal swabs. Mock birds (groups C′ and F′) remained virus-negative during the whole experiment.
In order to assess the tissue tropism of mesogenic AAvV 1, the tissues (n = 19) collected from birds (pigeons and chickens) showing typical clinical signs and necropsy lesions were processed for the detection of viral RNA. For chickens (groups A′ and B′), the virus RNA was detected in most of the tissue samples except heart, kidney, feather follicle, tongue, oesophagus, caecum, caecal tonsils and liver. However, for pigeons (groups D′ and E′), the presence of viral genome was detected in all tissues except heart, kidney, breast muscle, feather follicle, tongue and bursa of Fabricius.
Histopathological findings for mesogenic AAvV 1 infection
The histopathological changes, including haemorrhages with mononuclear inflammatory cell infiltration in some areas of tracheal tissues coupled with absence of pseudostratified columnar epithelium in trachea, haemorrhages and congestion with degenerative changes in the lamina propria of lung, congestion in peritubular capillaries, cellular swelling in renal epithelial cells and coagulative necrosis in renal tubules were observed in tissues collected from morbid chickens. Some renal tubules had epithelial cells separated from the basement membrane, congestion, haemorrhages and necrotic degeneration in spleen, mild congestion in brain, venous congestion, degeneration in hepatocyte, fatty changes (vacuoles in hepatocytes), infiltration of inflammatory cells in the portal cord in liver, damaged basal membrane and degeneration in follicles in bursa of Fabricius and presence of dead/necrotic tissue mass, sloughing of epithelial cells in the lumen, dropout of intestinal villi in the small intestine ().
Figure 5. Microscopic examination of histopathologic changes in different tissues collected from chickens infected with the mesogenic strain. Arrows indicate histologic and pathologic lesions in (A) trachea, (B) lung, (C) kidney, (D) spleen, (E) brain, (F) liver, (G) bursa of Fabricius and (H) small intestine.
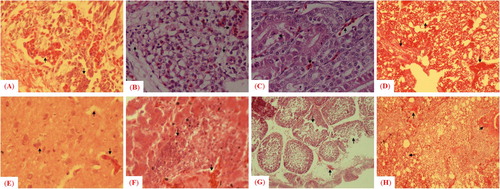
In contrast to chickens, the histopathological alterations were comparatively less pronounced in pigeons (groups D′ and E′). Microscopic lesions included congestion, haemorrhages with mononuclear inflammatory cells infiltration in lung, mild congestion in brain, fatty changes evidenced, infiltration of inflammatory cells in portal cord of liver, presence of dead/necrotic tissue masses, dropout of epithelium in small intestine, infiltration of inflammatory cells in gizzard and disruption of cardiac fibres in heart. None of the tissues collected from birds of the control group had any apparent histological and pathological changes ().
Discussion
The experimental studies under controlled conditions enable us to properly evaluate the infectivity and transmissibility of field prevailing AAvV 1 strains, including onset of clinical signs, mortality, virus shedding and transmission to healthy birds (Panshin et al., Citation2002; Kapczynski et al., Citation2006; Wakamatsu et al., Citation2006; Carrasco et al., Citation2008; Alexander et al., Citation2012; Guo et al., Citation2014; Desingu et al., Citation2017). Therefore, owing to the greater prevalence of genetically diverged isolates than previously known, the current study was designed to evaluate the comparative pathogenicity of currently circulating AAvV 1 strains of varying pathogenicity (velogenic and mesogenic strains) in commercial chickens and pigeons. The oculonasal route was used to induce the infection in both avian species simply because it is considered a natural route of infection in field conditions (Alexander et al., Citation2012). In fact, with the same route of infection, true clinical presentation of ND has been evidenced in various susceptible avian species in captivity (Otim et al., Citation2006; Piacenti et al., Citation2006; Carrasco et al., Citation2008; Susta et al., Citation2010; Guo et al., Citation2014; Kang et al., Citation2016: Desingu et al., Citation2017). We found a higher rate of morbidity and mortality in both avian species upon exposure to a wild bird-originated velogenic AAvV 1 strain than upon exposure to a pigeon-originated mesogenic AAvV 1 strain. Similarly, velogenic strains have been frequently reported from chickens and pigeons with high morbidity and mortality (Aldous et al., Citation2010; Alexander et al., Citation2012), whereas mesogenic strains showed relatively less mortality in both commercial chickens and pigeons (Kommers et al., Citation2001; Kim et al., Citation2008). In fact, velogenic strains caused a high degree of infection in chickens, while; mesogenic strains show a high degree of infection in pigeons (Kommers et al., Citation2001; Kim et al., Citation2008). This is because; the course of infection with a pigeon-originated AAvV 1 variant in chickens can be very mild even though the strain is mesogenic due to its classification into pathotypes, which is actually dependent on virulence assessment via intracerebral inoculation in one-day-old chickens but not in pigeons (Meulemans et al., Citation2002). Along with intracerebral pathogenicity index in one-day-old chickens, disease severity in susceptible hosts could serve as a reference index for AAvV pathotyping. However, the accuracy and precision of these indexes are questioned to some extent, especially when used to evaluate viruses isolated from wild birds (Guo et al., Citation2014).
There is still controversy over the susceptibility of chickens and pigeons to viruses of varying pathogenicity. For instance, a few studies demonstrated a disease of less severity in chickens upon challenge with viruses of velogenic and mesogenic strains (Panshin et al., Citation2002; Guo et al., Citation2014). A severe form of infection was observed in chickens upon challenge with viruses of velogenic and mesogenic strains (Ezema et al., Citation2016; Ren et al., Citation2017; Xue et al., Citation2017). Likewise, a variable degree of infection was also observed in pigeons inoculated with velogenic and mesogenic strains (Wakamatsu et al., Citation2006; Guo et al., Citation2014; Carrasco et al., Citation2016). However, some of the studies argued that wild bird- (including pigeon) originated avulaviruses need further passages in embryonated chicken eggs in order to adapt and subsequently infect chickens (Kommers et al., Citation2003). Mesogenic strains of pigeon origin may evolve into virulent viruses and, therefore, lead to major outbreaks by causing infection in chickens (Dortmans et al., Citation2011a). Differences in the susceptibility of chickens to wild bird-originated AAvV 1 strains were observed because wild bird originated strains may gain virulence after a variable number of passages which was not performed in the current study. Therefore, host specificity is not only a typical characteristic of wild bird-originated AAvV 1 strains because a vast range of hosts is susceptible to different strains. Such findings, coupled with the observation reported in this study, highlight the potential of wild bird-originated strains to cause infection in commercial chickens (Karamendin et al., Citation2016).
The clinico-pathological findings of the current study revealed nervous signs in both avian species upon challenge with mesogenic strains, whereas nervous signs were observed exclusively in pigeons when challenged with the velogenic strain. Similar to findings reported previously (Kapczynski et al., Citation2006; Wakamatsu et al., Citation2006; Carrasco et al., Citation2008), this indicates a varying pattern of tissue tropism of both species for each of the avulaviruses and the fact that chickens and pigeons are not equally susceptible to infection by the same strain (Wakamatsu et al., Citation2006). Indeed, variations in disease severity involving varying tissues have been well documented among multiple avian species exposed with the same strain of avulavirus (Piacenti et al., Citation2006; Kang et al., Citation2016). A development of viscerotropic and neurotropic form of infection has been reported previously when chickens were inoculated with the velogenic strain (Oladele et al., Citation2005; Bobbo et al., Citation2013; Igwe et al., Citation2014), whereas only the neurotropic form of infection was evidenced in pigeons upon exposure to the velogenic strain (Kapczynski et al., Citation2006). The velogenic strain, isolated from an epidemic in a flock showing both respiratory and nervous signs, exhibited only a neurotropic form of infection following experimental infection in chickens (Igwe et al., Citation2014). Involving the central nervous system and respiratory tract, mild to moderate lesions in chickens have previously been reported upon exposure to a mesogenic strain (Wakamatsu et al., Citation2006; Susta et al., Citation2010). A possible reason for such variations in tissue tropism, and a subsequent variable degree of infection in multiple avian species, could be linked to the presence of proteases or furin-like enzymes in different tissues of susceptible/infected hosts (Seal et al., Citation2005).
Consistent with previous observations (Panshin et al., Citation2002; Desingu et al., Citation2017), when challenged with velogenic and mesogenic strains, we evidenced a variable pattern of viral shedding from both avian species. Viral shedding was detected in both oropharyngeal and cloacal swabs in a pattern comparable to previous studies (Kapczynski et al., Citation2006; Wakamatsu et al., Citation2006) where the virus was detected from oral and tracheal swabs starting from 2 dpi. We found detection of virus in cloacal swabs earlier than oropharyngeal swabs. Some minor variations, in this regard, have previously been documented. For instance, similar to our study observations, the onset of virus shedding was detected earlier in cloacal swabs starting from 2 to 3 dpi (Kapczynski et al., Citation2006; Wakamatsu et al., Citation2006; Carrasco et al., Citation2009). Likewise, in a previous study, virus shedding through the cloaca was also observed from 5 dpi onward (Carrasco et al., Citation2008). In contrast, the onset of virus shedding was detected in oropharyngeal swabs starting from 5 to 9 dpi (Dortmans et al., Citation2011b; Dai et al., Citation2014). The NDV infection was also evident in contact chickens and pigeons where virus distribution among different organs or tissues was identified. Such type of transmission is not uncommon for velogenic or mesogenic AAvV 1 strains (Shabbir et al., Citation2016; Desingu et al., Citation2017). Similar to previous studies, a variable pattern of tissue tropism was evidenced depending on the pathotypes used for challenge and the avian species involved (Pandarangga et al., Citation2016; Shabbir et al., Citation2016; Igwe et al., Citation2018). Though comparable, in contrast to chickens, the distribution of viral RNA was not detected in all of the tissue samples collected from pigeon. Such a pattern of varying tissue tropism has also been evidenced previously (Guo et al., Citation2014). Taken together, the current study revealed that both types of AAvV 1 strains have the potential to target multiple tissues which, therefore, could be utilized for either detection and/or isolation of the virus. In this regard, the potential of molecular assays such as RT–PCR has previously been validated for accurate detection of the viral genome using a range of tissues from the diseased/deceased host (Barbezange & Jestin, Citation2002).
Conclusion
Although there are variations in disease severity, transmission, virus shedding, and tissue tropism, wild bird-originated velogenic and mesogenic AAvV 1 strains have the potential to induce infection in both chickens and pigeons. These wild bird-originated AAvV 1 strains are circulating in the environment, may cause infection when coming into contact with commercial birds, and outbreaks may occur, with considerable economic losses to poultry industries particularly in disease-endemic countries. A lack of appropriate biosecurity measures is not uncommon in developing countries where disease is endemic and disease occurrence is frequent. Therefore, integrated continuous disease surveillance, coupled with biosecurity measures including vaccination regimen of available live and killed ND vaccines, are crucial for an efficient disease combat in a disease-endemic setting.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Aziz-ul-Rahman http://orcid.org/0000-0002-3342-4462
Muhammad Zubair Shabbir http://orcid.org/0000-0002-3562-007X
References
- Abolnik, C., Mubamba, C., Wandrag, D.B., Horner, R., Gummow, B., Dautu, G. & Bisschop, S.P. (2018). Tracing the origins of genotype VII h Newcastle disease in Southern Africa. Transboundary and Emerging Diseases, 65, e393–e403. doi: 10.1111/tbed.12771
- Akhtar, S., Muneer, M.A., Muhammad, K., Tipu, M.Y., Rabbani, M. & Shabbir, M.Z. (2016). Genetic characterization and phylogeny of pigeon paramyxovirus isolate (PPMV-1) from Pakistan. SpringerPlus, 5, 1295. doi: 10.1186/s40064-016-2939-1
- Aldous, E.W., Seekings, J.M., McNally, A., Nili, H., Fuller, C.M., Irvine, R.M., Alexander, D.J. & Brown, I.H. (2010). Infection dynamics of highly pathogenic avian influenza and virulent avian paramyxovirus type 1 viruses in chickens, turkeys and ducks. Avian Pathology, 39, 265–273. doi: 10.1080/03079457.2010.492825
- Alexander, D.J., Aldous, E.W. & Fuller, C.M. (2012). The long view: a selective review of 40 years of Newcastle disease research. Avian Pathology, 41, 329–335. doi: 10.1080/03079457.2012.697991
- Alexander, D.J. & Senne, D.A. (2008). Newcastle disease. In Y.M. Saif, A.M. Fadly, J.R. Glisson, L.R. Mcdougald, L.K. Nolan & D.E. Swayne (Eds.), Diseases of poultry (12th edn.) (pp. 75–100). Blackwell publishing, Ames, Iowa, USA.
- Ali, M., Yaqub, T., Mukhtar, N., Imran, M., Ghafoor, A., Shahid, M.F., Yaqub, S., Smith, G.J., Su, Y.C. & Naeem, M. (2017). Prevalence and phylogenetics of H9N2 in backyard and commercial poultry in Pakistan. Avian Diseases, 62, 416–424. doi: 10.1637/11690-062117-ResNote.1
- Amarasinghe, G.K., Ayllón, M.A., Bào, Y., Basler, C.F., Bavari, S., Blasdell, K.R., Briese, T., Brown, P.A., Bukreyev, A., Balkema-Buschmann, A. & Buchholz, U.J. (2019). Taxonomy of the order Mononegavirales: update 2019. Archives of Virology, 164, 1967–1980. doi: 10.1007/s00705-019-04247-4
- Aziz-ul-Rahman, Munir, M. & Shabbir, M.Z. (2018a). Comparative evolutionary and phylogenomic analysis of avian avulaviruses 1–20. Molecular Phylogenetics and Evolution, 127, 931–951. doi: 10.1016/j.ympev.2018.06.040
- Aziz-ul-Rahman, Munir, M. & Shabbir, M.Z. (2019c). A comparative genomic and evolutionary analysis of circulating strains of avian avulavirus 1 in Pakistan. Molecular Genetics and Genomics, 1–21.
- Aziz-ul-Rahman, & Shabbir, M.Z. (2019a). A comparative phylogenomic analysis of avian avulavirus 1 isolated from non-avian hosts: conquering new frontiers of zoonotic potential among species. Archives of Virology, 164, 1771–1780. doi: 10.1007/s00705-019-04276-z
- Aziz-ul-Rahman, Yaqub, T., Imran, M., Habib, M., Sohail, T., Furqan Shahid, M., Munir, M. & Shabbir, M.Z. (2018b). Phylogenomics and infectious potential of avian avulaviruses species-Type 1 isolated from healthy green-winged teal (Anas carolinensis) from a wetland sanctuary of Indus river. Avian Diseases, 62, 404–415.
- Aziz-ul-Rahman, Yaqub, T., Imran, M., Habib, M., Sohail, T., Mukhtar, N., Shahid, M.F., Munir, M. & Shabbir, M.Z. (2019b). Sequence analysis and biological characterization of virulent avian avulavirus 1 isolated from asymptomatic migratory fowl. Acta Virologica, 63, 223–228. doi: 10.4149/av_2019_208
- Barbezange, C. & Jestin, V. (2002). Development of a RT-nested PCR test detecting pigeon paramyxovirus-1 directly from organs of infected animals. Journal of Virological Methods, 106, 197–207. doi: 10.1016/S0166-0934(02)00148-9
- Bobbo, A.G., Baba, S.S., Yahaya, M.S. & El-Yuguda, A.D. (2013). Susceptibility of three phenotypes of village chickens to Newcastle disease in Adamawa State. Alexandria Journal of Veterinary Sciences, 39, 133–140.
- Brown, V.R. & Bevins, S.N. (2017). A review of virulent Newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Veterinary Research, 48, 68. doi: 10.1186/s13567-017-0475-9
- Carleton, H.M., Drury, R.A. & Wallington, E.A. (1980). Carleton’s histological technique. Oxford: Oxford University Press.
- Carrasco, A.D.O.T., Seki, M.C., Benevenute, J.L., Ikeda, P. & Pinto, A.A. (2016). Experimental infection with Brazilian Newcastle disease virus strain in pigeons and chickens. Brazilian Journal of Microbiology, 47, 231–242. doi: 10.1016/j.bjm.2015.07.001
- Carrasco, A.D.O.T., Seki, M.C., de Freitas Raso, T., Paulillo, A.C. & Pinto, A.A. (2008). Experimental infection of Newcastle disease virus in pigeons (Columba livia): humoral antibody response, contact transmission and viral genome shedding. Veterinary Microbiology, 129, 89–96. doi: 10.1016/j.vetmic.2007.11.012
- Carrasco, A.O.T., Seki, M.C., de Sousa, R.L., Raso, T.F. & Pinto, A.A. (2009). Protection levels of vaccinated pigeons (Columba livia) against a highly pathogenic Newcastle disease virus strain. Tropical Animal Health and Production, 41, 1325–1333. doi: 10.1007/s11250-009-9318-7
- Dai, Y., Cheng, X., Liu, M., Shen, X., Li, J., Yu, S., Zou, J. & Ding, C. (2014). Experimental infection of duck origin virulent Newcastle disease virus strain in ducks. BMC Veterinary Research, 10, 164. doi: 10.1186/1746-6148-10-164
- Desingu, P.A., Singh, S.D., Dhama, K., Kumar, O.V., Malik, Y.S. & Singh, R. (2017). Clinicopathological characterization of experimental infection in chickens with sub-genotype VIIi Newcastle disease virus isolated from peafowl. Microbial Pathogenesis, 105, 8–12. doi: 10.1016/j.micpath.2017.01.057
- Dimitrov, K.M., Abolnik, C., Afonso, C.L., Albina, E., Bahl, J., Berg, M., Briand, F.X., Brown, I.H., Choi, K.S., Chvala, I. & Diel, D.G. (2019a). Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infection, Genetics and Evolution, 74: 103917. doi: 10.1016/j.meegid.2019.103917
- Dimitrov, K.M., Ferreira, H.L., Pantin-Jackwood, M.J., Taylor, T.L., Goraichuk, I.V., Crossley, B.M., Killian, M.L., Bergeson, N.H., Torchetti, M.K., Afonso, C.L. & Suarez, D.L. (2019b). Pathogenicity and transmission of virulent Newcastle disease virus from the 2018–2019 California outbreak and related viruses in young and adult chickens. Virology, 531, 203–218. doi: 10.1016/j.virol.2019.03.010
- Dodovski, A., Cvetkovikj, I., Krstevski, K., Naletoski, I. & Savić, V. (2017). Characterization and epidemiology of pigeon paramyxovirus type-1 viruses (PPMV-1) isolated in Macedonia. Avian Diseases, 61, 146–152. doi: 10.1637/11517-101816-Reg.1
- Dortmans, J.C.F.M., Koch, G., Rottier, P.J.M. & Peeters, B.P.H. (2011b). A comparative infection study of pigeon and avian paramyxovirus type 1 viruses in pigeons: evaluation of clinical signs, virus shedding and seroconversion. Avian Pathology, 40, 125–130. doi: 10.1080/03079457.2010.542131
- Dortmans, J.C., Rottier, P.J., Koch, G. & Peeters, B.P. (2011a). Passaging of a Newcastle disease virus pigeon variant in chickens results in selection of viruses with mutations in the polymerase complex enhancing virus replication and virulence. Journal of General Virology, 92, 336–345. doi: 10.1099/vir.0.026344-0
- El Naggar, R.F., Rohaim, M.A., Bazid, A.H., Ahmed, K.A., Hussein, H.A. & Munir, M. (2018). Biological characterization of wild-bird-origin avian avulavirus 1 and efficacy of currently applied vaccines against potential infection in commercial poultry. Archives of Virology, 163, 2743–2755. doi: 10.1007/s00705-018-3916-5
- Ezema, W.S., Eze, D.C., Shoyinka, S.V.O. & Okoye, J.O.A. (2016). Atrophy of the lymphoid organs and suppression of antibody response caused by velogenic Newcastle disease virus infection in chickens. Tropical Animal Health and Production, 48, 1703–1709. doi: 10.1007/s11250-016-1147-x
- Fan, W., Wang, Y., Wang, S., Cheng, Z., Guo, H., Zhao, X. & Liu, J. (2017). Virulence in Newcastle disease virus: a genotyping and molecular evolution spectrum perspective. Research in Veterinary Science, 111, 49–54. doi: 10.1016/j.rvsc.2016.12.001
- Ferreira, H.L., Taylor, T.L., Absalon, A.E., Dimitrov, K.M., Cortés-Espinosa, D.V., Butt, S.L., Marín-Cruz, J.L., Goraichuk, I.V., Volkening, J.D., Suarez, D.L. & Afonso, C.L. (2019). Presence of Newcastle disease viruses of sub-genotypes Vc and VIn in backyard chickens and in apparently healthy wild birds from Mexico in 2017. Virus Genes, 12, 1–1.
- Guo, H., Liu, X., Xu, Y., Han, Z., Shao, Y., Kong, X. & Liu, S. (2014). A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Veterinary Microbiology, 168, 88–97. doi: 10.1016/j.vetmic.2013.11.002
- Haji-Abdolvahab, H., Ghalyanchilangeroudi, A., Bahonar, A., Ghafouri, S.A., Marandi, M.V., Mehrabadi, M.H.F. & Tehrani, F. (2019). Prevalence of avian influenza, Newcastle disease, and infectious bronchitis viruses in broiler flocks infected with multifactorial respiratory diseases in Iran, 2015–2016. Tropical Animal Health and Production, 51, 689–695. doi: 10.1007/s11250-018-1743-z
- Igwe, A.O., Afonso, C.L., Ezema, W.S., Brown, C.C. & Okoye, J.O.A. (2018). Pathology and distribution of velogenic viscerotropic Newcastle disease virus in the reproductive system of vaccinated and unvaccinated laying hens (Gallus gallus domesticus) by immunohistochemical labelling. Journal of Comparative Pathology, 159, 36–48. doi: 10.1016/j.jcpa.2017.12.009
- Igwe, O.A., Ezema, S.W., Eze, C.D. & Okoye, O.A. (2014). Experimental velogenic Newcastle disease can be very severe and viscerotropic in chickens but moderate and neurotropic in Guinea fowls. International Journal of Poultry Science, 13, 582–590. doi: 10.3923/ijps.2014.582.590
- Isidoro-Ayza, M., Afonso, C.L., Stanton, J.B., Knowles, S., Ip, H.S., White, C.L., Fenton, H., Ruder, M.G., Dolinski, A.C. & Lankton, J. (2017). Natural infections with pigeon paramyxovirus serotype 1: pathologic changes in Eurasian collared-doves (Streptopelia decaocto) and rock pigeons (Columba livia) in the United States. Veterinary Pathology, 54, 695–703. doi: 10.1177/0300985817695782
- Kang, Y., Xiang, B., Yuan, R., Zhao, X., Feng, M., Gao, P., Li, Y., Li, Y., Ning, Z. & Ren, T. (2016). Phylogenetic and pathotypic characterization of Newcastle disease viruses circulating in South China and transmission in different birds. Frontiers in Microbiology, 7, 119. doi: 10.3389/fmicb.2016.00119
- Kapczynski, D.R., Wise, M.G. & King, D.J. (2006). Susceptibility and protection of naïve and vaccinated racing pigeons (Columbia livia) against exotic Newcastle disease virus from the California 2002–2003 outbreak. Avian Diseases, 50, 336–341. doi: 10.1637/7479-112905R.1
- Karamendin, K., Kydyrmanov, A., Seidalina, A., Asanova, S., Daulbayeva, K., Kasymbekov, Y., Khan, E., Fereidouni, S., Starick, E., Zhumatov, K. & Sayatov, M. (2016). Circulation of avian paramyxoviruses in wild birds of Kazakhstan in 2002–2013. Virology Journal, 13, 23. doi: 10.1186/s12985-016-0476-8
- Kim, L.M., King, D.J., Guzman, H., Tesh, R.B., da Rosa, A.P.T., Bueno, R., Dennett, J.A. & Afonso, C.L. (2008). Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. Journal of Clinical Microbiology, 46, 3303–3310. doi: 10.1128/JCM.00644-08
- Kolakofsky, D., Roux, L., Garcin, D. & Ruigrok, R.W. (2005). Paramyxovirus mRNA editing, the ‘rule of six’ and error catastrophe: a hypothesis. Journal of General Virology, 86, 1869–1877. doi: 10.1099/vir.0.80986-0
- Kommers, G.D., King, D.J., Seal, B.C. & Brown, C.C. (2001). Virulence of pigeon-origin Newcastle disease virus isolates for domestic chickens. Avian Diseases, 45, 906–921. doi: 10.2307/1592870
- Kommers, G.D., King, D.J., Seal, B.S. & Brown, C.C. (2003). Virulence of six heterogeneous-origin Newcastle disease virus isolates before and after sequential passages in domestic chickens. Avian Pathology, 32, 81–93. doi: 10.1080/0307945021000070750
- Liang, R., Cao, D.J., Li, J.Q., Chen, J., Guo, X., Zhuang, F.F. & Duan, M.X. (2002). Newcastle disease outbreaks in western China were caused by the genotypes VIIa and VIII. Veterinary Microbiology, 87, 193–203. doi: 10.1016/S0378-1135(02)00050-0
- Mayahi, V. & Esmaelizad, M. (2017). Molecular evolution and epidemiological links study of Newcastle disease virus isolates from 1995 to 2016 in Iran. Archives of Virology, 162, 3727–3743. doi: 10.1007/s00705-017-3536-5
- Meulemans, G., Berg, T.V., Decaesstecker, M. & Boschmans, M. (2002 Oct 1). Evolution of pigeon Newcastle disease virus strains. Avian Pathology, 31, 515–519. doi: 10.1080/0307945021000005897
- Miller, P.J., Haddas, R., Simanov, L., Lublin, A., Rehmani, S.F., Wajid, A., Bibi, T., Khan, T.A., Yaqub, T., Setiyaningsih, S. & Afonso, C.L. (2015). Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infection, Genetics and Evolution, 29, 216–229. doi: 10.1016/j.meegid.2014.10.032
- Munir, M., Cortey, M., Abbas, M., Afzal, F., Shabbir, M.Z., Khan, M.T., Ahmed, S., Ahmad, S., Baule, C., Ståhl, K. & Zohari, S. (2012). Biological characterization and phylogenetic analysis of a novel genetic group of Newcastle disease virus isolated from outbreaks in commercial poultry and from backyard poultry flocks in Pakistan. Infection, Genetics and Evolution, 12, 1010–1019. doi: 10.1016/j.meegid.2012.02.015
- Oladele, S.B., Nok, A.J., Esievo, K.A.N., Abdu, P.A. & Useh, N.M. (2005). Haemagglutination inhibition antibodies, rectal temperatures and total protein of chickens infected with a local Nigerian isolate of velogenic Newcastle disease virus. Veterinary Research Communications, 29, 171–179. doi: 10.1023/B:VERC.0000047495.03341.2b
- Otim, O.M., Christensen, H., Mukiibi, G.M. & Bisgaard, M. (2006). A preliminary study of the role of ducks in the transmission of Newcastle disease virus to in-contact rural free-range chickens. Tropical Animal Health & Production, 38, 285–289. doi: 10.1007/s11250-006-4309-4
- Pandarangga, P., Brown, C.C., Miller, P.J., Haddas, R., Rehmani, S.F., Afonso, C.L. & Susta, L. (2016). Pathogenesis of new strains of Newcastle disease virus from Israel and Pakistan. Veterinary Pathology, 53, 792–796. doi: 10.1177/0300985815622972
- Panshin, A., Shihmanter, E., Weisman, Y., Örvell, C. & Lipkind, M. (2002). Antigenic heterogeneity among the field isolates of Newcastle disease virus (NDV) in relation to the vaccine strain: 1. Studies on viruses isolated from wild birds in Israel. Comparative Immunology, Microbiology and Infectious Diseases, 25, 95–108. doi: 10.1016/S0147-9571(01)00026-1
- Piacenti, A.M., King, D.J., Seal, B.S., Zhang, J. & Brown, C.C. (2006). Pathogenesis of Newcastle disease in commercial and specific pathogen-free turkeys experimentally infected with isolates of different virulence. Veterinary Pathology, 43, 168–178. doi: 10.1354/vp.43-2-168
- Qiu, X., Meng, C., Zhan, Y., Yu, S., Li, S., Ren, T., Yuan, W., Xu, S., Sun, Y., Tan, L. & Song, C. (2017). Phylogenetic, antigenic and biological characterization of pigeon paramyxovirus type 1 circulating in China. Virology Journal, 14, 186. doi: 10.1186/s12985-017-0857-7
- Ramey, A.M., Reeves, A.B., Ogawa, H., Ip, H.S., Imai, K., Bui, V.N., Yamaguchi, E., Silko, N.Y. & Afonso, C.L. (2013). Genetic diversity and mutation of avian paramyxovirus serotype 1 (Newcastle disease virus) in wild birds and evidence for intercontinental spread. Archives of Virology, 158, 2495–2503. doi: 10.1007/s00705-013-1761-0
- Rehmani, S.F., Wajid, A., Bibi, T., Nazir, B., Mukhtar, N., Hussain, A., Lone, N.A., Yaqub, T. & Afonso, C.L. (2015). Presence of virulent Newcastle disease virus in vaccinated chickens in farms in Pakistan. Journal of Clinical Microbiology, 53, 1715–1718. doi: 10.1128/JCM.02818-14
- Ren, S., Wang, C., Zhang, X., Zhao, L., Wang, X., Yao, W., Han, Q., Wang, Y., Fan, M., Gao, X. & Xiao, S. (2017). Phylogenetic and pathogenic characterization of a pigeon paramyxovirus type 1 isolate reveals cross-species transmission and potential outbreak risks in the northwest region of China. Archives of Virology, 162, 2755–2767. doi: 10.1007/s00705-017-3422-1
- Sabra, M., Dimitrov, K.M., Goraichuk, I.V., Wajid, A., Sharma, P., Williams-Coplin, D., Basharat, A., Rehmani, S.F., Muzyka, D.V., Miller, P.J. & Afonso, C.L. (2017). Phylogenetic assessment reveals continuous evolution and circulation of pigeon-derived virulent avian avulaviruses 1 in Eastern Europe, Asia, and Africa. BMC Veterinary Research, 13, 291. doi: 10.1186/s12917-017-1211-4
- Seal, B.S., Wise, M.G., Pedersen, J.C., Senne, D.A., Alvarez, R., Scott, M.S., King, D.J., Yu, Q. & Kapczynski, D.R. (2005). Genomic sequences of low-virulence avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from live-bird markets in North America not related to commonly utilized commercial vaccine strains. Veterinary Microbiology, 106, 7–16. doi: 10.1016/j.vetmic.2004.11.013
- Shabbir, M.Z., Akhtar, S., Tang, Y., Yaqub, T., Ahmad, A., Mustafa, G., Alam, M.A., Santhakumar, D. & Nair, V. (2016). Infectivity of wild bird origin avian Paramyxovirus serotype 1 and vaccine effectiveness in chickens. Journal of General Virology, 97, 3161–3173. doi: 10.1099/jgv.0.000618
- Solomon, P., Abolnik, C., Joannis, T.M. & Bisschop, S. (2012). Virulent Newcastle disease virus in Nigeria: identification of a new clade of sub-lineage 5f from livebird markets. Virus Genes, 44, 98–103. doi: 10.1007/s11262-011-0678-5
- Susta, L., Miller, P.J., Afonso, C.L. & Brown, C.C. (2011). Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Veterinary Pathology, 48, 349–360. doi: 10.1177/0300985810375806
- Susta, L., Miller, P.J., Afonso, C.L., Estevez, C., Yu, Q., Zhang, J. & Brown, C.C. (2010). Pathogenicity evaluation of different Newcastle disease virus chimeras in 4-week-old chickens. Tropical Animal Health and Production, 42, 1785–1795. doi: 10.1007/s11250-010-9638-7
- Wajid, A., Dundon, W.G., Hussain, T. & Babar, M.E. (2018). Pathotyping and genetic characterization of avian avulavirus-1 from domestic and wild waterfowl, geese and black swans in Pakistan, 2014 to 2017. Archives of Virology, 163, 2513–2518. doi: 10.1007/s00705-018-3902-y
- Wakamatsu, N., King, D.J., Kapczynski, D.R., Seal, B.S. & Brown, C.C. (2006). Experimental pathogenesis for chickens, turkeys, and pigeons of exotic Newcastle disease virus from an outbreak in California during 2002-2003. Veterinary Pathology, 43, 925–933. doi: 10.1354/vp.43-6-925
- Xue, C., Xu, X., Yin, R., Qian, J., Sun, Y., Wang, C., Ding, C., Yu, S., Hu, S., Liu, X. & Cong, Y. (2017). Identification and pathotypical analysis of a novel VIk sub-genotype Newcastle disease virus obtained from pigeon in China. Virus Research, 238, 1–7. doi: 10.1016/j.virusres.2017.05.011