ABSTRACT
Numerous studies have shown that viruses can utilize or manipulate ribosomal proteins to achieve viral protein biosynthesis and replication. In our recent studies using proteomics analysis of virus-infected cells, we found that ribosomal protein L18 (RPL18) was the highest up-regulated differentially expressed protein, along with the increasingly expressed viral proteins later in Newcastle disease virus (NDV) infection. However, the association of RPL18 with viral protein biosynthesis and NDV replication remains unclear. In this study, we found that the expression and transcription levels of RPL18 was reduced early in NDV infection but increased later in NDV infection. In addition, the presence of cytoplasmic NDV matrix (M) protein was responsible for the increased expression of RPL18 in both virus-infected cells and plasmid-transfected cells. Moreover, cytoplasmic M protein increased RPL18 expression in a dose-dependent manner, even though they did not interact with each other. Furthermore, siRNA-mediated knockdown of RPL18 or overexpression of RPL18 dramatically reduced or enhanced NDV replication by decreasing or increasing viral protein translation rather than viral RNA synthesis and transcription. Taken together, these results suggested that the increased expression of RPL18 might be associated with the physical clumping together of the M protein, which in turn promoted viral protein biosynthesis and NDV replication.
The increased expression of RPL18 is associated with the presence of cytoplasmic M protein.
Cytoplasmic M protein increases RPL18 expression in a dose-dependent manner.
Knockdown of RPL18 reduces NDV replication by decreasing viral protein translation.
Overexpression of RPL18 enhances NDV replication by increasing viral protein translation.
RESEARCH HIGHLIGHTS
Introduction
Newcastle disease virus (NDV) is one of the important avian pathogens that usually cause considerable economic losses to the poultry industry worldwide (Aldous & Alexander, Citation2001). NDV has a non-segmented, negative-sense, and single-stranded RNA genome, which encodes six structural proteins in the order 3´-NP-P-M-F-HN-L-5´ (Peeters et al., Citation2000). Among these viral proteins, the matrix (M) protein is located at the inner surface of viral envelope and forms an outer protein shell around the nucleocapsid, constituting a bridge between the viral envelope and nucleocapsid (Battisti et al., Citation2012). It has been demonstrated that the NDV M protein can shuttle between the nucleus and cytoplasm via its intrinsic nuclear localization signal (NLS) (Peeples et al., Citation1992; Coleman & Peeples, Citation1993) and nuclear export signal (NES) (Duan et al., Citation2013). Early during NDV infection, the nuclear localization of M protein is considered to regulate viral RNA synthesis and transcription and inhibit host cell transcription (Duan et al., Citation2019), but later during infection the M protein is localized mainly in the cytoplasm as well as associated with cell membranes, where it participates in the assembly and budding of progeny virions (Pantua et al., Citation2006). In recent years, an increasing number of studies have investigated the relationship between M protein and the replication and pathogenicity of NDV. For example, the N-terminal motif 23FPIV26 (Duan, Hu et al., Citation2014), amino acid R42 (Duan, Li et al., Citation2014), and the C-terminal motifs 247KKGKKVIFDKIEEKIRR263 (Duan et al., Citation2018) and 275GPL277 (Xu et al., Citation2016) in the M protein are confirmed to affect the virulence and replication of NDV. Moreover, the interactions of M protein with host proteins, such as host charged multivesicular body protein 4B (Li et al., Citation2013), nucleophosmin (Duan, Chen et al., Citation2014), importin β1 (Duan et al., Citation2018), and bromodomain-containing protein 2 (Duan, Han et al., Citation2020), are also essential for the replication and pathogenicity of NDV. Therefore, these findings demonstrate that the M protein is a multifunctional nucleocytoplasmic trafficking protein and plays a crucial role in the life cycle of NDV.
In recent years, an increasing number of studies have focused on the association of ribosomal proteins (RPs) with virus replication. For example, ribosomal protein L4 (RPL4) is found to interact with infectious bursal disease virus VP3 protein, and knockdown of RPL4 results in the reduction of viral protein expression and virus replication (Chen et al., Citation2016). In addition, foot-and-mouth disease virus (FMDV) VP1 protein interacting with ribosomal protein SA (RPSA) is beneficial to abrogate the RPSA-mediated suppressive role in MAPK pathway activation and promote FMDV replication (Zhu et al., Citation2020). Meanwhile, a recent study has found that the interaction of FMDV 3Cpro protease with ribosomal protein L13 (RPL13) plays an important role in antagonizing the RPL13-mediated antiviral activity (Guan et al., Citation2021). These results indicate that viral proteins can interact with RPs to promote virus replication in a variety of pathways. In recent years, several studies using proteomics analysis of virus-infected cells also revealed that the expression of most RPs shows a marked difference during the early and late stages of virus infection (Lu et al., Citation2004; Hu et al., Citation2017; Becker et al., Citation2018; Li et al., Citation2020). In our recent studies, we similarly found that most of the RPs showed significantly down-regulated expression early in NDV infection but exhibited remarkably up-regulated expression later in NDV infection (Duan, Yuan et al., Citation2020). Of these RPs, ribosomal protein L18 (RPL18) was the most up-regulated protein, along with the increasing expression of viral proteins later in NDV infection. It is noteworthy that RPL18 has been reported to participate in viral protein biosynthesis and enhance virus replication (Li, Citation2019; Dong et al., Citation2021; Miller et al., Citation2021). Therefore, we hypothesized that the increased expression of RPL18 might be a benefit for viral protein translation and NDV replication. To this end, the experiments including virus infection, plasmid transfection, small interfering RNA (siRNA)-mediated knockdown of RPL18 and overexpression of RPL18 were performed. We found that the presence of cytoplasmic M protein was responsible for the increased expression of RPL18 in a dose-dependent manner, but there was no interaction between them. In addition, knockdown of RPL18 or overexpression of RPL18 dramatically reduced or enhanced NDV replication by decreasing or increasing viral protein translation rather than viral RNA synthesis and transcription. This finding revealed for the first time that the association of RPL18 with NDV M protein represents an important mechanism for NDV replication.
Materials and methods
Cells, viruses, and antibodies
BSR-T7/5 cells (a baby hamster kidney cell line (BHK-21) stably expressing T7 RNA polymerase) were a kind gift from Prof. Xiufan Liu (Key Laboratory of Animal Infectious Diseases, Yangzhou University, Yangzhou, China). The virulent NDV strain SS1 (GenBank no. KP742770.1) and the recombinant SS1 stably expressing green fluorescent protein (rSS1GFP) were reported in our previous studies (Duan et al., Citation2015; Duan et al., Citation2018). The mouse anti-M and rabbit anti-NP polyclonal antibodies were prepared by Wuhan GeneCreate Biological Engineering Co., Ltd (Wuhan, China). The rabbit anti-RPL18 (DF3700), histone H3 (DF6932), and GAPDH (T0004) polyclonal antibodies were purchased from Affinity Biosciences (Cincinnati, OH, USA). The mouse monoclonal antibodies against HA tag (66006-2-Ig), Myc tag (60003-2-Ig), and GFP tag (66002-1-Ig) were purchased from Proteintech Group, Inc (Chicago, IL, USA).
Plasmid constructions
The open reading frame (ORF) of NDV M, NP, P, F, HN and L genes (GenBank no. KP742770.1) was amplified from the full-length cDNA plasmid pNDV/SS1GFP or pNDV/SS1GFP-M/NLSm (Duan et al., Citation2018) and then subcloned into the plasmids pCMV-HA and pCMV-Myc (Thermo Fisher, Waltham, MA, USA) to generate pCMV-HA-M, pCMV-HA-M/NLSm, pCMV-Myc-NP, pCMV-Myc-P, pCMV-Myc-F, pCMV-Myc-HN, and pCMV-Myc-L, respectively. In addition, the ORF of hamster RPL18 gene (EMBL no. EGW03967.1) was amplified from cDNA derived from BSR-T7/5 cells and then used to construct the recombinant plasmid pCMV-Myc-RPL18. Primers used for the construction of the above recombinant plasmids are shown in , and all enzymes used for cloning procedures were purchased from Thermo Fisher (Waltham, MA, USA). All the constructed recombinant plasmids were confirmed by PCR, restriction digestion and DNA sequencing.
Table 1. Details of primers and siRNAs used in this study.
Cell culture and virus infection
BSR-T7/5 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NE, USA) supplemented with 10% foetal bovine serum (FBS) (Gibco, Grand Island, NE, USA) and cultured at 37°C under 5% CO2. For virus infection experiment, 5 × 105 BSR-T7/5 cells grown to 80% confluence in 6-well plates were infected with rSS1GFP at a mutiplicity of infection (MOI) of 1. At 6, 12, 18, and 24 h post-infection (hpi), nuclear and cytoplasmic fraction proteins were extracted using Nuclear and Cytoplasmic Protein Extraction kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. All protein concentrations were detected using the Bicinchoninic Acid (BCA) Protein Assay kit (Beyotime, Shanghai, China). Then equal amounts of different proteins from nuclear and cytoplasmic extracts were immunoblotted with anti-M or anti-RPL18 antibody together with anti-histone H3 and anti-GAPDH antibodies, respectively. The relative expression levels of the M and RPL18 proteins to control histone H3 or GAPDH expression were determined by densitometry using ImageJ software version 1.8.0. In addition, quantitative real-time PCR (qRT-PCR) was used to detect the transcription level of endogenous RPL18 gene in rSS1GFP-infected cells as previously described (Duan, Yuan et al., Citation2020).
Plasmid transfection
5 × 105 BSR-T7/5 cells grown to 80% confluence in 6-well plates were transfected with a total of 2 μg of each plasmid (pCMV-HA, pCMV-HA-M, pCMV-HA-M/NLSm, pCMV-Myc, pCMV-Myc-NP, pCMV-Myc-P, pCMV-Myc-L, pCMV-Myc-F, pCMV-Myc-HN) using TurboFect Transfection Reagent (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Thirty-six hours after transfection, BSR-T7/5 cells expressing the recombinant proteins were rinsed with phosphate-buffered saline (PBS), fixed with pre-cooled 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, and then used for indirect immunofluorescence assay as described previously (Duan et al., Citation2018). Meanwhile, the expression of these recombinant proteins in nuclear and cytoplasmic extracts, and the effect of the recombinant proteins on the expression of endogenous RPL18 protein were examined by western blotting. In addition, BSR-T7/5 cells were transfected with plasmid pCMV-HA or plasmid pCMV-HA-M/NLSm at a dose of 1.0, 2.0 or 4.0 μg. The expression levels of HA, HA-M/NLSm, or RPL18 in cells transfected with different doses of plasmid were examined at 36 h post-transfection (hpt). The relative expression level of RPL18 compared to control GAPDH was determined by densitometry using ImageJ software version 1.8.0.
Co-immuprecipitation (Co-IP) assay
5 × 105 BSR-T7/5 cells cultured in 6-well plates were infected with NDV strain SS1 or co-transfected with the indicated plasmids co-expressing the Myc tag and HA-M or HA-M/NLSm, Myc-RPL18 and HA-M or HA-M/NLSm. Twenty-four hours after infection or 36 h after transfection, cells were washed three times with PBS and then lysed with immunoprecipitation lysis buffer (Pierce, Illinois, USA). The supernatants were collected after centrifugation and then incubated with an anti-M, anti-RPL18, or anti-Myc antibody overnight at 4°C. The immune complexes were recovered by adsorption to protein A + G-Sepharose (Millipore Sigma, Burlington, MA, USA) for 3 h at 4°C. After three washes with immunoprecipitation lysis buffer, the immunoprecipitates were detected by western blotting using anti-RPL18, anti-M, or anti-HA antibody. Because the M protein and NP protein of NDV are known to interact with each other (Pantua et al., Citation2006), cells co-transfected with the plasmids expressing the Myc tag and HA-M or HA-M/NLSm, Myc-NP and HA-M or HA-M/NLSm were used as positive controls to verify the accuracy of Co-IP assay.
siRNA treatment and virus infection
The information of siRNAs designed to reduce the expression of RPL18 gene in BSR-T7/5 cells are shown in . During the siRNA transfection experiment, 5 × 105 BSR-T7/5 cells cultured in 6-well plates were transfected with the RPL18 siRNAs or negative control siRNA using siRNA transfection reagent LipofectamineTM RNAiMAX (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions. Forty-eight hours after transfection, the knockdown efficiency of RPL18 siRNAs was examined by western blotting. In addition, RPL18 siRNA#2- or control siRNA-treated cells or normal cells were infected with rSS1GFP at an MOI of 1. The cell culture supernatants were then collected at 6, 12, 24, 36, 48, and 72 hpi, and the virus titres were titrated using 50% tissue culture infective dose (TCID50) assay in BSR-T7/5 cells. Meanwhile, the effect of RPL18 knockdown on the RNA synthesis, transcription, and translation of NDV was detected at 12 and 24 hpi, respectively, as previously described (Duan et al., Citation2019). Moreover, 100 μg/ml cycloheximide (CHX) (Millipore Sigma, Burlington, MA, USA) was added to RPL18 siRNA#2- or control siRNA-treated cells at 24 hpi, and the half-life of M, NP and GFP proteins were determined at 0, 2, 4, 6 and 8 h, as described elsewhere (Gupta & Ono, Citation1997).
RPL18 overexpression and virus infection
To further investigate the effect of RPL18 overexpression on the replication of NDV, low-passage BSR-T7/5 cells at 80% confluence in 6-well plates were transfected with 2.0 μg of pCMV-Myc-RPL18 or pCMV-Myc, respectively. Thirty-six hours after transfection, the expression of Myc-RPL18 and Myc was verified by fluorescence observation, and the effect of Myc-RPL18 or Myc overexpression on the expression of RPL18 protein was examined by qRT-PCR and western blotting, respectively. In addition, the plasmid-transfected BSR-T7/5 cells or normal cells at 36 hpt were used to infect rSS1GFGP at an MOI of 1. The cell culture supernatants were collected at the indicated time-points (6, 12, 24, 36, 48, and 72 hpi), and the virus titres were determined using TCID50 assay in BSR-T7/5 cells. Moreover, the effect of RPL18 overexpression on the RNA synthesis, transcription, and translation of NDV was detected at 12 and 24 hpi, as previously described (Duan et al., Citation2019).
Data analysis and statistics
Kolmogorov–Smirnov test was performed to confirm the normality of the data, which were then analyzed by ANOVA test using the statistical programme Statistica (Statsoft, Tulsa, OK, USA). All experiments were repeated at least three times, and the results are presented as the mean ± standard deviation (SD). A P-value < 0.05 was considered statistically significant. P-values are indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
The increased expression of RPL18 is associated with the presence of cytoplasmic NDV M protein
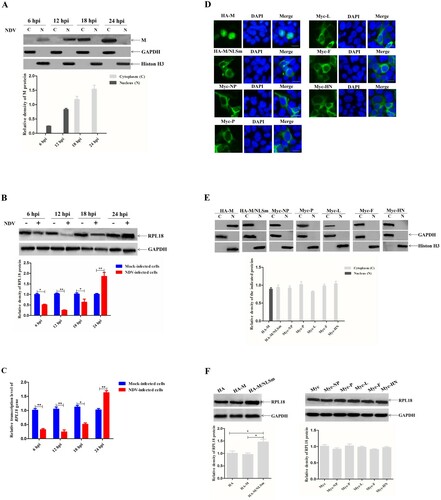
To investigate the association of RPL18 with NDV M protein, the effect of M protein on the expression of RPL18 in NDV-infected or plasmid-transfected cells was first examined. The results showed that the NDV M protein was detected in the nucleus early in infection (at 6 and 12 hpi), but in the cytoplasm later in infection (at 18 and 24 hpi), and the expression of M protein exhibited an increased pattern from 6 to 24 hpi ((A)), which was consistent with the previous findings (Peeples et al., Citation1992; Duan, Yuan et al., Citation2020). In addition, the expression of RPL18 protein was reduced from 6 to 12 hpi, while it was markedly increased from 18 to 24 hpi ((B)). Meanwhile, the transcription level of RPL18 gene in NDV-infected cells was consistent with the expression pattern of RPL18 ((C)). Next, the effect of viral structural proteins on RPL18 expression in plasmid-transfected cells was investigated. We found showed that, in addition to the predominantly nuclear localization of HA-M, the other recombinant proteins, including HA-M/NLSm harbouring NLS mutation, Myc-NP, Myc-P, Myc-L, Myc-F, and Myc-HN, were mainly localized in the cytoplasm ((D)). Similarly, the distribution of these recombinant viral proteins detected in nuclear and cytoplasmic extracts was consistent with the results of fluorescence observation, and there was no significant difference in expression level among them ((E)). However, it was strange that only the recombinant protein HA-M/NLSm localized in the cytoplasm increased the expression level of RPL18 ((F)). Therefore, these results suggested that the presence of cytoplasmic NDV M protein was associated with the increased expression of RPL18.
Figure 1. The increased expression of RPL18 is associated with the presence of cytoplasmic NDV M protein. The expression level of M protein (A) and RPL18 (B) was examined by western blotting. BSR-T7/5 cells were infected with NDV strain rSS1GFP at an MOI of 1, and the expression of M protein and RPL18 was detected at 6, 12, 18 and 24 hpi, respectively. The relative expression levels of the M protein and RPL18 compared to control GAPDH or histone H3 expression levels were determined by densitometry using ImageJ software version 1.8.0. (C) The effect of rSS1GFP infection on the transcription level of RPL18 gene at 6, 12, 18 and 24 hpi. (D) The subcellular localization of HA- or Myc-tagged viral proteins. The recombinant plasmids were transfected into BSR-T7/5 cells, and indirect immunofluorescence assay was used to observe the fluorescence of HA- and Myc-tagged viral proteins at 36 hpt. DAPI was used to detect nuclei. The original magnification was 1 × 200. Scale bars represent 10 μm. (E) The expression of HA- or Myc-tagged viral proteins was examined by western blotting. The recombinant plasmids were transfected into BSR-T7/5 cells, and the recombinant proteins in nuclear (N) and cytoplasmic (C) extracts were detected at 36 hpt. The relative expression levels of the recombinant proteins compared to control GAPDH or histone H3 expression levels were determined by densitometry using ImageJ software version 1.8.0. (F) The effect of HA- or Myc-tagged viral proteins on the expression of RPL18. BSR-T7/5 cells were transfected with the recombinant plasmids and the expression of RPL18 was examined at 36 hpt. The relative expression level of RPL18 compared to control GAPDH expression level was determined by densitometry using ImageJ software version 1.8.0.
Cytoplasmic NDV M protein increases RPL18 expression in a dose-dependent manner
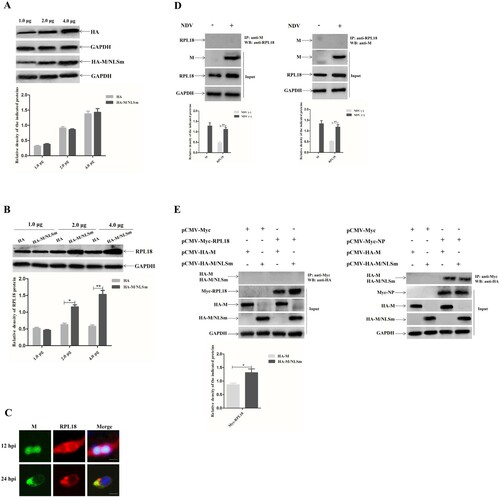
The relationship between the quantity of cytoplasmically expressed M protein and RPL18 expression and the interaction of M protein with RPL18 were further investigated. The results of different amounts (1.0, 2.0, or 4.0 μg) of plasmid-transfected cells showed that the recombinant protein HA-M/NLSm and the HA tag were more highly expressed in cells transfected with 4.0 μg of plasmid at 36 hpt ((A)). Meanwhile, the expression level of RPL18 was relatively higher in cells transfected with 2.0 or 4.0 μg of pCMV-HA-M/NLSm (upregulated 1.64 and 2.85 times, respectively) than that of the pCMV-HA group ((B)). In addition, the fluorescence co-localization experiment revealed that the M protein and RPL18 were distributed in the nucleus and cytoplasm at 12 hpi, and then they were co-localized in the cytoplasm at 24 hpi ((C)). However, neither the M protein nor the M/NLSm protein interacted with RPL18 in either NDV-infected cells ((D)) or in plasmid co-transfected cells ((E), left) using Co-IP assay. As a positive control, the M protein and M/NLSm protein could be detected to interact with viral NP protein ((E), right), demonstrating that it was a true lack of M and RPL18 interaction rather than a technical failure of the Co-IP assay. Together, these results indicated that cytoplasmic NDV M protein could increase RPL18 expression in a dose-dependent manner, even though there was no interaction between them.
Figure 2. Cytoplasmic NDV M protein increases RPL18 expression in a dose-dependent manner. (A) The expression levels of HA and HA-M/NLSm in cells transfected with different doses of pCMV-HA or pCMV-HA-M/NLSm. BSR-T7/5 cells were transfected with the plasmid pCMV-HA or pCMV-HA-M/NLSm at a dose of 1.0, 2.0 or 4.0 μg. The expression levels of HA and HA-M/NLSm in plasmid-transfected cells were examined at 36 hpt. The relative expression level of the indicated proteins to control GAPDH expression level was determined by densitometry using ImageJ software version 1.8.0. (B) The expression level of RPL18 in cells transfected with different doses (1.0, 2.0 or 4.0 μg) of pCMV-HA or pCMV-HA-M/NLSm. The relative expression level of RPL18 to control GAPDH expression level was determined by densitometry using ImageJ software version 1.8.0. (C) Fluorescence co-localization of M protein and RPL18 in NDV-infected cells. BSR-T7/5 cells were infected with NDV strain SS1 at an MOI of 1. The subcellular localization of M protein and RPL18 was examined by indirect immunofluorescence assay at 12 and 24 hpi, respectively. DAPI was used to detect nuclei. The original magnification was 1 × 200. Scale bars represent 10 μm. Verification of the interaction between M protein and RPL18 in virus-infected cells (D) and plasmid co-transfected cells (E) by Co-IP assay. The cell supernatants in rSS1GFP-infected cells or plasmid co-transfected cells were collected at 24 hpi and 36 hpt, respectively. The co-immunoprecipitation assay was carried out to detect the interaction between M protein and RPL18 using the corresponding antibodies. The interaction between M protein and NP protein verified by Co-IP assay was used as a positive control. The relative expression level of M protein and RPL18 to control GAPDH expression level was determined by densitometry using ImageJ software version 1.8.0.
Knockdown of RPL18 reduces NDV replication by decreasing viral protein translation
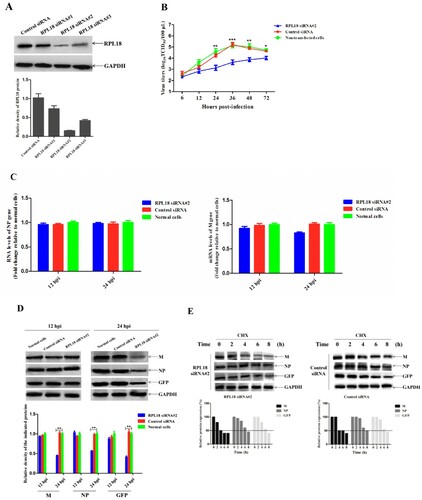
To further explore the role of RPL18 in NDV replication, siRNA-mediated knockdown of RPL18 was performed. The results of RPL18 siRNA interference effect showed that RPL18 siRNA#2 could more effectively reduce the expression level of RPL18 than other RPL18 siRNAs and control siRNA ((A)), with no marked toxic or nonspecific effect on the viability of those cells shown using trypan blue exclusion and MTT assays (data not shown). The replication ability of NDV in RPL18 siRNA#2- or control siRNA-treated cells or normal cells was then evaluated. The multicycle growth kinetic analysis revealed that the virus titres in RPL18 siRNA#2-treated cells were relatively lower (decreased from 1.2 to 1.8 titre) than those in control siRNA-treated cells or normal cells from 24 to 48 hpi (P < 0.01) ((B)). Then the effect of RPL18 knockdown on viral RNA synthesis, transcription, and translation was examined. We found that the relative viral RNA level (corresponding to the NP gene) and transcription level (corresponding to the M gene) showed no significant difference between RPL18 siRNA#2- and control siRNA-treated cells or normal cells at 12 and 24 hpi ((C)). However, although the expression levels of viral M and NP proteins and exogenous GFP were not changed at 12 hpi, they were obviously decreased in RPL18 siRNA#2-treated cells compared with control siRNA-treated cells or normal cells at 24 hpi (P < 0.01) ((D)). Interestingly, the half-life of M, NP, and GFP proteins was not changed after RPL18 knockdown ((E)). Overall, these results indicated that siRNA-mediated knockdown of RPL18 reduced NDV replication by decreasing viral protein translation.
Figure 3. siRNA-mediated knockdown of RPL18 reduces NDV replication by decreasing viral protein translation. (A) The effect of RPL18 siRNAs or control siRNA on the expression of RPL18. BSR-T7/5 cells grown in 6-well plates were transfected with the indicated siRNAs (10 μM) at a dose of 30 pmol, and the knockdown efficiency of RPL18 was detected by Western blotting at 48 hpt. The expression level of RPL18 relative to control GAPDH expression level was determined by densitometry using ImageJ software version 1.8.0. (B) The growth kinetics of rSS1GFP in RPL18 siRNA#2- or control siRNA-treated cells or normal cells. The cell culture supernatants were collected at the indicated time-points (6, 12, 24, 36, 48 and 72 hpi), and the viral titres were determined using TCID50 assay in BSR-T7/5 cells. The effect of RPL18 knockdown on viral RNA synthesis and transcription (C), and viral protein translation (D) at 12 and 24 hpi, respectively. The relative expression levels of the M, NP, and GFP proteins compared to control GAPDH expression levels were determined by densitometry using ImageJ software version 1.8.0. (E) The effect of RPL18 knockdown on the half-life of M, NP and GFP proteins. 100 μg/ml CHX was added to RPL18 siRNA#2- or control siRNA-treated cells at 24 hpi, and the half-life of M, NP and GFP proteins were detected at 0, 2, 4, 6 and 8 h. The relative expression levels of the M, NP, and GFP proteins compared to control GAPDH expression levels were determined by densitometry using ImageJ software version 1.8.0.
Overexpression of RPL18 enhances NDV replication by increasing viral protein translation
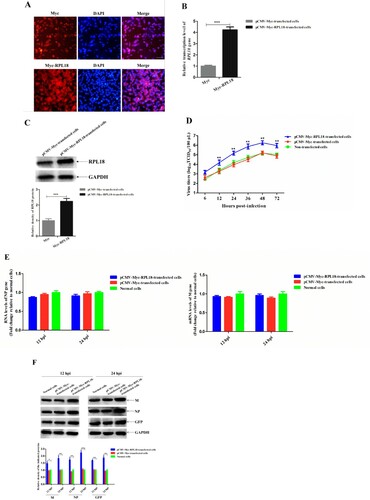
Because knockdown of RPL18 reduced NDV replication, it was intriguing to further investigate whether RPL18 overexpression could also affect NDV replication. Therefore, the effect of overexpression of Myc-RPL18 or Myc on the replication of NDV was examined. As shown in (A), the expression of Myc-RPL18 or Myc in plasmid-transfected cells was clearly observed by fluorescence observation. The results of qRT-PCR and western blotting analysis showed that the transcription level and expression level of RPL18 in pCMV-Myc-RPL18-transfected cells were much higher than that in pCMV-Myc-transfected cells (P < 0.001) ((B,C)). In addition, overexpression of Myc-RPL18 increased the virus titres (from 1.1 to 1.4 titre) of rSS1GFP from 12 to 72 hpi (P < 0.01) ((D)). However, the relative viral RNA level (corresponding to the NP gene) and transcription level (corresponding to the M gene) between pCMV-Myc-RPL18- and pCMV-Myc-transfected cells or normal cells had no significant difference at 12 and 24 hpi ((E)), which was similar to the RPL18 siRNA treatment experiment. By contrast, the expression levels of viral M and NP proteins and exogenous GFP protein were significantly decreased in pCMV-Myc-RPL18-transfected cells compared with pCMV-Myc-transfected cells or normal cells at 12 and 24 hpi (P < 0.01) ((F)). Therefore, these results suggested that overexpression of RPL18 enhanced NDV replication by increasing viral protein translation.
Figure 4. Overexpression of RPL18 enhances NDV replication by increasing viral protein translation. (A) Fluorescence observation of Myc and Myc-RPL18 in plasmid-transfected cells. BSR-T7/5 cells were transfected with 2.0 μg of pCMV-Myc or pCMV-Myc-RPL18 to overexpress Myc or Myc-RPL18. The fluorescence of Myc and Myc-RPL18 was observed under an inverted fluorescence microscope at 36 hpt. DAPI was used to detect nuclei. The original magnification was 1 × 100. Scale bars represent 10 μm. The effect of Myc or Myc-RPL18 overexpression on the transcription level (B) and expression level (C) of endogenous RPL18 in BSR-T7/5 cells. The transcription level and expression level of the endogenous RPL18 in pCMV-Myc- or pCMV-Myc-RPL18-transfected cells were detected by qRT-PCR and western blotting at 36 hpt, respectively. (D) The growth kinetics of rSS1GFP in pCMV-Myc- or pCMV-Myc-RPL18-transfected cells or non-transfected cells. The cell culture supernatants were collected at the indicated time-points (6, 12, 24, 36, 48, and 72 hpi), and the viral titres were determined using TCID50 assay in BSR-T7/5 cells. The effect of RPL18 overexpression on viral RNA synthesis and transcription (E), and viral protein translation (F) at 12 and 24 hpi, respectively. The relative expression levels of the M, NP and GFP proteins compared to control GAPDH expression levels were determined by densitometry using ImageJ software version 1.8.0.
Discussion
Currently, the more familiar functions of NDV M protein are: (i) the core role of M protein in the assembly and budding of NDV progeny virions (Pantua et al., Citation2006; Duan, HU et al., Citation2014), and (ii) the regulation of viral RNA synthesis and transcription and host cell transcription inhibition of M protein in the nucleus (Duan et al., Citation2019; Duan, Yuan et al., Citation2020), which are analogous to the M proteins of human respiratory syncytial virus (Ghildyal et al., Citation2003; Forster et al., Citation2015), vesicular stomatitis virus (Raux et al., Citation2010; Rajani et al., Citation2012), and measles virus (Yu et al., Citation2016; Ke et al., Citation2018). In recent years, studying virus-host interactions has remained an important strategy to understand the functions of M protein and the replication of NDV. In addition to the reported interactions between NDV M protein and host proteins (Molouki et al., Citation2011; Li et al., Citation2013; Duan, Chen et al., Citation2014; Duan et al., Citation2018; Shah et al., Citation2019; Duan, Han et al., Citation2020), our recent studies also found that M protein can indirectly affect the expression of host proteins to regulate NDV replication. For example, the increased expression of prospero homeobox 1 and decreased expression of aryl hydrocarbon receptor caused by nuclear localization of M protein enhanced the viral RNA synthesis and NDV replication (Duan et al., Citation2019). Moreover, the cytoplasmic M protein could inhibit TIFA expression in a dose-dependent manner and promote NDV replication by down-regulating TIFA/TRAF6/NF-κB-mediated production of cytokines (Duan, Yuan et al., Citation2020), thus demonstrating the involvement of M protein in NDV immune evasion. Together, all these findings accelerate our understanding of the potential functions of M protein and the underlying replication mechanism of NDV.
In recent years, numerous reviews are now describing the interactions of various types of viral proteins with RPs to regulate virus replication (Li, Citation2019; Rofeal & Abd El-Malek, Citation2020; Dong et al., Citation2021; Miller et al., Citation2021). However, there is no available information about the involvement of RPs in the replication of NDV and other paramyxoviruses. In our recent studies using quantitative proteomics analysis of NDV-infected cells, we found that most of the RPs showed the decreased expression patterns in parental NDV-infected BSR-T7/5 cells at 12 hpi, and then exhibited increased expression levels at 24 hpi when compared to the mutant NDV harbouring a NLS mutation in the M protein (Duan, Yuan et al., Citation2020). Because NDV is the most effective paramyxovirus at inhibiting the production of host proteins (Hightower & Bratt, Citation1974), the down-regulated expression of RPs early in NDV infection might be related to the shutoff of global cellular protein synthesis due to the competition of NDV with the host mRNAs for translation factors and ribosomes (Zhan et al., Citation2020). Interestingly, among these differentially expressed proteins (DEPs), RPL18 was the highest up-regulated DEP during NDV infection at 24 hpi, and the viral proteins were also increasingly expressed at this time point (Duan, Yuan et al., Citation2020). This finding suggested that the up-regulated expression of RPL18 might be associated with the presence of M protein in the cytoplasm. Consistent with this hypothesis, in this study, we demonstrated that the existence of cytoplasmic M protein was responsible for the increased expression of RPL18 in both NDV-infected cells and pCMV-HA-M/NLSm-transfected cells. More importantly, cytoplasmic M protein increased RPL18 expression in a dose-dependent manner. Therefore, the association of RPL18 with M protein in regulating NDV replication is worthy of further study.
Recently, several studies have reported the functional relationship between RPL18 and virus replication; for example, the interactions of RPL18 with cauliflower mosaic virus P6 protein and rice stripe tenuivirus nucleocapsid protein promote viral translation and replication (Leh et al., Citation2000; Li et al., Citation2018). A recent study has shown that infectious bursal disease virus VP3 protein interacts with chicken RPL18, and knockdown of RPL18 remarkably suppresses infectious bursal disease virus replication by enhancing type I interferon expression (Wang et al., Citation2018). In addition, RPL18 is found to be incorporated into Ebola and Marburg virions, and the reduced expression of RPL18 effectively interferes with viral infection (Spurgers et al., Citation2010). However, unlike what has been reported in these findings, the RPL18 was neither incorporated into NDV virions (Ren et al., Citation2012) nor found to interact with NDV M protein. Even so, knockdown of RPL18 or overexpression of RPL18 reduced or enhanced NDV replication by decreasing or increasing viral protein translation rather than viral RNA synthesis and transcription, indicating that the increased expression of RPL18 might be associated with the physical clumping together of the M protein, which in turn promoted viral protein biosynthesis and NDV replication. To date, none of the host proteins interacting with M protein are known to be related to NDV viral translation. Therefore, our findings demonstrate for the first time that the association of RPL18 with NDV M protein is important for viral translation and replication, which provides useful information about the replication mechanism of NDV and other paramyxoviruses. However, how M protein increases RPL18 expression to regulate NDV replication remains to be further elucidated.
Ethical statement
This study does not contain any studies with human participants or animals performed by any of the authors.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aldous, E.W. & Alexander, D.J. (2001). Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathology, 30, 117–128.
- Battisti, A.J., Meng, G., Winkler, D.C., McGinnes, L.W., Plevka, P., Steven, A.C., Morrison, T.G. & Rossmann, M.G. (2012). Structure and assembly of a paramyxovirus matrix protein. Proceedings of the National Academy of Sciences of the United States of America, 109, 13996–14000.
- Becker, A.C., Gannagé, M., Giese, S., Hu, Z., Abou-Eid, S., Roubaty, C., Paul, P., Bühler, L., Gretzmeier, C., Dumit, V.I., Kaeser-Pebernard, S., Schwemmle, M., Münz, C. & Dengjel, J. (2018). Influenza A virus induces autophagosomal targeting of ribosomal proteins. Molecular & Cellular Proteomics, 17, 1909–1921.
- Chen, Y., Lu, Z., Zhang, L., Gao, L., Wang, N., Gao, X., Wang, Y., Li, K., Gao, Y., Cui, H., Gao, H., Liu, C., Zhang, Y., Qi, X. & Wang, X. (2016). Ribosomal protein L4 interacts with viral protein VP3 and regulates the replication of infectious bursal disease virus. Virus Research, 211, 73–78.
- Coleman, N.A. & Peeples, M.E. (1993). The matrix protein of Newcastle disease virus localizes to the nucleus via a bipartite nuclear localization signal. Virology, 195, 596–607.
- Dong, H.J., Zhang, R., Kuang, Y. & Wang, X.J. (2021). Selective regulation in ribosome biogenesis and protein production for efficient viral translation. Archives of Microbiology, 203, 1021–1032.
- Duan, Z., Chen, J., Xu, H., Zhu, J., Li, Q., He, L., Liu, H., Hu, S. & Liu, X. (2014). The nucleolar phosphoprotein B23 targets Newcastle disease virus matrix protein to the nucleoli and facilitates viral replication. Virology, 452–453, 212–222.
- Duan, Z., Deng, S., Ji, X., Zhao, J., Yuan, C. & Gao, H. (2019). Nuclear localization of Newcastle disease virus matrix protein promotes virus replication by affecting viral RNA synthesis and transcription and inhibiting host cell transcription. Veterinary Research, 50, 22.
- Duan, Z., Han, Y., Zhou, L., Yuan, C., Wang, Y., Zhao, C., Tang, H. & Chen, J. (2020). Chicken bromodomain-containing protein 2 interacts with the Newcastle disease virus matrix protein and promotes viral replication. Veterinary Researh, 51, 120.
- Duan, Z., Hu, Z., Zhu, J., Xu, H., Chen, J., Liu, H., Hu, S. & Liu, X. (2014). Mutations in the FPIV motif of Newcastle disease virus matrix protein attenuate virus replication and reduce virus budding. Archives of Virology, 159, 1813–1819.
- Duan, Z., Ji, X., Xu, H., Zhao, J., Ruan, Y. & Chen, J. (2015). Identification of a genotype VIId Newcastle disease virus isolated from Sansui sheldrake ducks in Guizhou province, China. Genome Announcements, 3, e00161–15.
- Duan, Z., Li, J., Zhu, J., Chen, J., Xu, H., Wang, Y., Liu, H., Hu, S. & Liu, X. (2014). A single amino acid mutation, R42A, in the Newcastle disease virus matrix protein abrogates its nuclear localization and attenuates viral replication and pathogenicity. Journal of General Virology, 95, 1067–1073.
- Duan, Z., Song, Q., Wang, Y., He, L., Chen, J., Zhu, Y., Hu, S. & Liu, X. (2013). Characterization of signal sequences determining the nuclear export of Newcastle disease virus matrix protein. Archives of Virology, 158, 2589–2595.
- Duan, Z., Xu, H., Ji, X., Zhao, J., Xu, H., Hu, Y., Deng, S., Hu, S. & Liu, X. (2018). Importin α5 negatively regulates importin β1-mediated nuclear import of Newcastle disease virus matrix protein and viral replication and pathogenicity in chicken fibroblasts. Virulence, 9, 783–803.
- Duan, Z., Yuan, C., Han, Y., Zhou, L., Zhao, J., Ruan, Y., Chen, J., Ni, M. & Ji, X. (2020). TMT-based quantitative proteomics analysis reveals the attenuated replication mechanism of Newcastle disease virus caused by nuclear localization signal mutation in viral matrix protein. Virulence, 11, 607–635.
- Forster, A., Maertens, G.N., Farrell, P.J. & Bajorek, M. (2015). Dimerization of matrix protein is required for budding of respiratory syncytial virus. Journal of Virology, 89, 4624–4635.
- Ghildyal, R., Baulch-Brown, C., Mills, J. & Meanger, J. (2003). The matrix protein of human respiratory syncytial virus localises to the nucleus of infected cells and inhibits transcription. Archives of Virology, 148, 1419–1429.
- Guan, J., Han, S., Wu, J., Zhang, Y., Bai, M., Abdullah, S.W., Sun, S. & Guo, H. (2021). Ribosomal protein L13 participates in innate immune response induced by foot-and-mouth disease virus. Frontiers in Immunology, 12, 616402.
- Gupta, K.C. & Ono, E. (1997). Stimulation of Sendai virus C’ protein synthesis by cycloheximide. Biochemistry Journal, 321, 811–818.
- Hightower, L.E. & Bratt, M.A. (1974). Protein synthesis in Newcastle disease virus-infected chicken embryo cells. Journal of Virology, 13, 788–800.
- Hu, J., Zhu, W., Li, Y., Guan, Q., Yan, H., Yu, J., Fu, Z., Lu, X. & Tian, J. (2017). SWATH-based quantitative proteomics reveals the mechanism of enhanced Bombyx mori nucleopolyhedrovirus-resistance in silkworm reared on UV-B treated mulberry leaves. Proteomics, 17.
- Ke, Z., Strauss, J.D., Hampton, C.M., Brindley, M.A., Dillard, R.S., Leon, F., Lamb, K.M., Plemper, R.K. & Wright, E.R. (2018). Promotion of virus assembly and organization by the measles virus matrix protein. Nature Communications, 9, 1736.
- Leh, V., Yot, P. & Keller, M. (2000). The cauliflower mosaic virus translational transactivator interacts with the 60S ribosomal subunit protein L18 of Arabidopsis thaliana. Virology, 266, 1–7.
- Li, S. (2019). Regulation of ribosomal proteins on viral infection. Cells, 8, 508.
- Li, S., Li, X. & Zhou, Y. (2018). Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Research, 247, 15–20.
- Li, X., Bi, X., An, M., Xia, Z. & Wu, Y. (2020). iTRAQ-Based proteomic analysis of watermelon fruits in response to cucumber Green mottle mosaic virus infection. International Journal of Molecular Sciences, 21, 2541.
- Li, X., Li, X., Cao, H., Wang, Y. & Zheng, S.J. (2013). Engagement of new castle disease virus (NDV) matrix (M) protein with charged multivesicular body protein (CHMP) 4 facilitates viral replication. Virus Research, 171, 80–88.
- Lu, H., Li, W., Noble, W.S., Payan, D. & Anderson, D.C. (2004). Riboproteomics of the hepatitis C virus internal ribosomal entry site. Journal of Proteome Research, 3, 949–957.
- Miller, C.M., Selvam, S. & Fuchs, G. (2021). Fatal attraction: the roles of ribosomal proteins in the viral life cycle. Wiley Interdisciplinary Reviews RNA, 12, e1613.
- Molouki, A., Hsu, Y.T., Jahanshiri, F., Abdullah, S., Rosli, R. & Yusoff, K. (2011). The matrix (M) protein of Newcastle disease virus binds to human bax through its BH3 domain. Virology Journal, 8, 385.
- Pantua, H.D., McGinnes, L.W., Peeples, M.E. & Morrison, T.G. (2006). Requirements for the assembly and release of Newcastle disease virus-like particles. Journal of Virology, 80, 11062–11073.
- Peeples, M.E., Wang, C., Gupta, K.C. & Coleman, N. (1992). Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. Journal of Virology, 66, 3263–3269.
- Peeters, B.P., Gruijthuijsen, Y.K., de Leeuw, O.S. & Gielkens, A.L. (2000). Genome replication of Newcastle disease virus: involvement of the rule-of-six. Archives of Virology, 145, 1829–1845.
- Rajani, K.R., Pettit Kneller, E.L., McKenzie, M.O., Horita, D.A., Chou, J.W. & Lyles, D.S. (2012). Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathogens, 8, e1002929.
- Raux, H., Obiang, L., Richard, N., Harper, F., Blondel, D. & Gaudin, Y. (2010). The matrix protein of vesicular stomatitis virus binds dynamin for efficient viral assembly. Journal of Virology, 84, 12609–12618.
- Ren, X., Xue, C., Kong, Q., Zhang, C., Bi, Y. & Cao, Y. (2012). Proteomic analysis of purified Newcastle disease virus particles. Proteome Science, 10, 32.
- Rofeal, M. & Abd El-Malek, F. (2020). Ribosomal proteins as a possible tool for blocking SARS-COV 2 virus replication for a potential prospective treatment. Medical Hypotheses, 143, 109904.
- Shah, M., Bharadwaj, M.S.K., Gupta, A., Kumar, R. & Kumar, S. (2019). Chicken viperin inhibits Newcastle disease virus infection in vitro: a possible interaction with the viral matrix protein. Cytokine, 120, 28–40.
- Spurgers, K.B., Alefantis, T., Peyser, B.D., Ruthel, G.T., Bergeron, A.A., Costantino, J.A., Enterlein, S., Kota, K.P., Boltz, R.C., Aman, M.J., Delvecchio, V.G. & Bavari, S. (2010). Identification of essential filovirion-associated host factors by serial proteomic analysis and RNAi screen. Molecular & Cellular Proteomics, 9, 2690–2703.
- Wang, B., Duan, X., Fu, M., Liu, Y., Wang, Y., Li, X., Cao, H. & Zheng, S.J. (2018). The association of ribosomal protein L18 (RPL18) with infectious bursal disease virus viral protein VP3 enhances viral replication. Virus Research, 245, 69–79.
- Xu, H., Duan, Z., Chen, Y., Liu, J., Cheng, X., Liu, J., Zhu, J., Wang, X., Liu, X., Hu, S. & Liu, X. (2016). Simultaneous mutation of G275A and P276A in the matrix protein of Newcastle disease virus decreases virus replication and budding. Archives of Virology, 161, 3527–3533.
- Yu, X., Shahriari, S., Li, H.M. & Ghildyal, R. (2016). Measles virus matrix protein inhibits host cell transcription. PLoS One, 11, e0161360.
- Zhan, Y., Yu, S.Q., Yang, S., Qiu, X.S., Meng, C.C., Tan, L., Song, C.P., Liao, Y., Liu, W.W., Sun, Y.J. & Ding, C. (2020). Newcastle disease virus infection activates PI3 K/Akt/mTOR and p38 MAPK/Mnk1 pathways to benefit viral mRNA translation via interaction of the viral NP protein and host eIF4E. PLoS Pathogens, 16, e1008610.
- Zhu, Z., Li, W., Zhang, X., Wang, C., Gao, L., Yang, F., Cao, W., Li, K., Tian, H., Liu, X., Zhang, K. & Zheng, H. (2020). Foot-and-mouth disease virus capsid protein VP1 interacts with host ribosomal protein SA to maintain activation of the MAPK signal pathway and promote virus peplication. Journal of Virology, 94, e01350–19.
