ABSTRACT
In recent years, reports of chicken astrovirus (CAstV) infections in commercial birds have increased, affecting the poultry industries in Brazil and worldwide. The objective of this study was to use molecular methods to characterize the CAstV found in one breeding and three different incubation companies that reported increased embryonic mortality and the appearance of birds with white chick syndrome. RT-qPCR with SYBR green chemistry was used to determine the presence of the virus in the faeces of breeders, organs of neonatal chickens, and unhatched embryos. By sequencing the ORF2 gene that codes for the viral capsid, the strains responsible for these clinical signs were characterized using strains previously reported in Brazil, North America, Europe, and Asia. The percentage of identity of the amino acid sequences compared with those from group A was less than 41% (37.01% to 40.52%) and the identity with those from subgroups Bi, Bii, and Biii was less than 90% (81.81% to 89.85%). Therefore, the sequences were characterized within subgroup Biv with identity greater than 95% (95.26% to 99.59%), together with CAstV strains previously found in Brazil, Canada, and the United States. Using antigenicity prediction tools, 14 highly conserved peptides located on the surface of the capsid protein were considered potentially responsible for inducing the immune response in the host. Our data provide important information related to the increase in embryo mortality in vertically-infected birds, reinforcing the potential association of white chick syndrome with CAstV Biv subgroup.
RESEARCH HIGHLIGHTS
CAstV infections were found in farms and incubators with increased embryo mortality.
Brazilian CAstV Biv strains were associated with white chick syndrome.
Antigenic peptides were predicted on the surface of the capsid protein.
Introduction
Chicken astrovirus (CAstV) is a virus associated with hatchery disease and digestive problems, potentially originated by vertical and horizontal (faecal–oral) routes, respectively, especially in young birds (Pantin-Jackwood et al., Citation2008; Koo et al., Citation2013; Kang et al., Citation2018). CAstV is mainly transmitted by the faecal–oral route (horizontal) but it is thought that some strains can also be transmitted from the parents (vertical) as dead-in-shell embryos and 1-day-old chickens were detected with high viral loads (McIlwaine et al., Citation2021). Like other astroviruses, CAstV is highly resistant to inactivation by disinfectants and can be persistent in poultry houses and hatcheries (Smyth, Citation2017). In recent years, new information has emerged on CAstV strains that present a characteristic sign in hatched chicks called “white chick syndrome” (WCS). This disease is mostly observed in 1-day-old chicks and presents as a characteristic discolouration of the feathers and green-to-bronze discoloured liver lesions (Smyth et al., Citation2013; Raji & Omar, Citation2022). Some clinical signs are similar to those in runting-stunting syndrome (RSS) including runting, lesions in kidneys and liver, poor productive performance, weakness, and increased mortality (Smyth et al., Citation2013; Sajewicz-Krukowska et al., Citation2016; Long et al., Citation2018). WCS is also called “white chick hatchery disease” because of its detrimental impact on hatchability, which can be reduced by values between 4% and 68% (Smyth et al., Citation2013; Sajewicz-Krukowska et al., Citation2016). These particularities have given importance to the epidemiology related to this virus due to the significant economic losses in the poultry industry, especially in companies related to broiler chicken growth (Koo et al., Citation2013; De la Torre, Nuñez et al., Citation2018). WCS was reported in several countries including Poland, Brazil, Canada, and the US, although it seems to have a wider global distribution (McIlwaine et al., Citation2021). Despite the absence of prevalence studies of WCS caused by CAstV, it was estimated the annual case incidence rate in breeder flocks is between 2.5% and 18.3% (Long et al., Citation2018).
Classically, CAstV have been associated with RSS along with other enteric viruses (avian orthoreovirus (ARV), chicken parvovirus (ChPV), avian nephritis virus (ANV), avian rotavirus (ARtV), fowl adenovirus (FAdV), and infectious bronchitis virus) (Smyth, Citation2017; De la Torre, Nuñez et al., Citation2018). Because of that, a single aetiological agent of RSS is still undetermined. On the other hand, astroviruses are amongst the most prevalent viruses in young chickens with enteric disease, with up to 86–100% of chicken flocks infected (Pantin-Jackwood et al., Citation2008; Saif et al., Citation2019). However, they also can be isolated from healthy birds leading to questions about their specific role in disease pathogenicity (Saif et al., Citation2019).
Taxonomically, CAstV belongs to the Astroviridae family, with two genera: Avastrovirus (1, 2 and 3) and Mamastrovirus (1–19). Avastrovirus 1 corresponds to the turkey astrovirus (TAstV-1 and TAstV-2); Avastrovirus 2 to ANV and CAstV; and Avastrovirus 3 to duck astrovirus (DAstV-1 and DAstV-2) (Walker et al., Citation2021). CAstV is a small, nonenveloped virus with icosahedral symmetry and a genome that consists of a positive-sense, single-stranded RNA of approximately 7.5 kb length (Sajewicz-Krukowska & Domanska-Blicharz, Citation2016). Within the genome, there are three open reading frames: ORF1a and ORF1b, which encode the nonstructural proteins of the virus, including a protease and the RNA-dependent RNA polymerase, respectively, and ORF2, which encodes the structural protein of the viral capsid (Patel et al., Citation2017). Based on protein structure and amino acid sequence, CAstV is organized into two groups (A and B), and, according to their genetic similarity, each group is divided into subgroups Ai, Aii, Aiii, Bi, Bii, Biii, and Biv. All groups and subgroups are related to enteric problems; however, subgroup Biv is related to hatchery disease, such as WCS (Smyth, Citation2017). Nevertheless, an Aiii isolate was also reported to be associated with WCS (Sajewicz-Krukowska et al., Citation2016)
In this study, we characterized CAstV isolates found in some Brazilian incubator companies that reported a significant increase in embryonic mortality and the appearance of white chicks after hatching. Based on the complete sequencing of ORF2, we predicted peptides with potential antigenic characteristics located on the surface of the capsid protein.
Materials and methods
Sample collection
Through the diagnostic service of the Laboratory Avian Diseases, School of Veterinary Medicine – University of São Paulo, we received samples of faeces and organs of neonatal chickens and dead embryos from a breeder flock and incubator companies, respectively, from different regions of Brazil. Samples consisted of a pool from a specific origin (faeces or organs). The first company (C1) developed a CAstV monitoring system that consisted of sampling the faeces distributed around the floor of each shed and correlated the hatching rate with the presence/absence of CAstV. In this case, the sample included 20 fresh faeces drops collected from the tested shed. The other companies (C2, C3, C4) reported an increased cumulative mortality rate of up to 10% from the incubation period until hatching, and the emergence of white chicks as typical signs of CAstV infections. In these cases, each sample included specific organs that had been collected from 20 different chicks from the same flock. Progenitors of the flocks were between 28 and 32 weeks of age and the vaccination programme included immunization against chicken infectious anaemia virus, fowlpox virus, infectious bursal disease virus, infectious bronchitis virus, Marek’s disease virus, and Newcastle disease virus. Additional information on the samples is given in . Each sample was homogenized with sterile scissors and stored at −20°C until processing.
Table 1. Information of the samples used in this study.
Molecular detection of CAstV
A ∼750-mg aliquot of each sample (one for each company) was suspended in phosphate-buffered saline (PBS) pH 7.2 at a 1:1 ratio, frozen and thawed three times for cell disruption, and centrifuged for 30 min at 12,000x g at 4°C. Up to 200 µl of supernatant was collected for manual RNA extraction using the MagMAX™ Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific Baltics, Vilnius, Lithuania) according to the manufacturer’s instructions. Reverse transcription (RT) was performed with the M-MLV Reverse Transcriptase kit (Life Technologies, Carlsbad, CA, USA) using custom random primers synthesized by Invitrogen Brazil, Ltd., São Paulo, Brazil. Molecular detection was performed by RT-qPCR using SYBR Green chemistry with PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific Baltics) and the primers Forward: GCYGCTGCTGAAGAWATACAG and Reverse: CATCCCTCTACCAGATTTTCTGAAA, as described previously (Smyth et al., Citation2010; Palomino-Tapia et al., Citation2020). The referenced RT-qPCR targets a 70-bp fragment within the 3’ end of ORF1b and the intergenic region between ORF1b and ORF2 (Smyth et al., Citation2010). The reactions were conducted in the fast run mode of a QuantStudio 3 instrument (Applied Biosystems, Austin, TX, USA), with an initial step at 50°C for 2 min for uracil-DNA glycosylase activation, followed by a second step at 95°C for 2 min for DNA polymerase activation, and 40 cycles at 95°C for 15 s for denaturation and primer annealing/DNA amplification at 60°C for 30 s. A melting curve was generated with the standard steps from the QuantStudio 3 system. The presence of concomitant enteric viruses infections was tested with qPCR assays by using previously published primers for ANV (Todd et al., Citation2010), ARV (De la Torre et al., Citation2021), ARtV-A (De la Torre, Astolfi-Ferreira et al., Citation2018), ChPV (Nuñez et al., Citation2018), and FAdV (Günes et al., Citation2012). These qPCR procedures were performed with PowerUpTM SYBRTM Green Master Mix (Applied Biosystems) as previously reported (Chacón et al., Citation2022).
CAstV characterization
The ORF2 fragment encoding the viral capsid was amplified by RT–PCR using the same cDNA from a previous RT reaction and Platinum™ SuperFi™ DNA Polymerase (Invitrogen, Carlsbad, CA, USA). The primers used for this reaction were those described by Smyth et al. (Citation2012), which target the whole ORF2 segment (∼2.2 kb): CAstV_PRECAP (5’-TAGAGGGATGGACCGAAATATAGCAGC-3’) and CAstV_POSTCAP (5’-TGCAGCTGTACCCTCGATCCTA-3’). Thermocycling conditions included an initial denaturation at 95°C for 5 min, followed by 38 cycles of denaturation at 95°C for 1 min, annealing at 62°C for 30 s, and amplification at 72°C for 2.5 min, with a final elongation at 72°C for 10 min. PCR products were electrophoresed in a 1% ultrapure agarose gel with SYBR™ Safe DNA Gel Stain (Invitrogen). The ∼2.2 kb band was purified with the GPX™ PCR DNA and Gel Band Purification kit (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer’s instructions. Purified DNA was then cloned using the NEB® PCR Cloning Kit (New England Biolabs, Ipswich, MA, USA), and DNA sequencing of the insert was performed using the primer walking approach by the Sanger technique with the Big Dye Terminator V3.1 Cycle Sequencing Kit (Invitrogen) (Berg & Olaisen, Citation1994). DNA sequences were obtained using a ABI 3730 DNA Analyzer (Applied Biosystems), assembled in CLC Main Workbench V7.9.1 (QIAGEN Digital Insights, Redwood City, CA, USA) and aligned using ClustalX 2.1. A phylogenetic tree was constructed with MEGA 7 software, and a similarity matrix was inferred in CLC Main Workbench software. All sequences used in the phylogenetic analysis and the similarity matrix were downloaded from GenBank (National Center for Biotechnology Information, Bethesda, MD, USA).
Antigenic site prediction
To predict the peptides with possible antigenic characteristics within the viral capsid, the deduced amino acid sequences of our four strains were analysed using the EMBOSS antigenic tool (https://www.bioinformatics.nl/cgi-bin/emboss/antigenic). A 3D viral capsid protein was modelled with the Swiss Model Server (Biasini et al., Citation2014), and the predicted antigenic sites were identified and coloured on the surface of the virtual protein with PyMOL software (The PyMOL Molecular Graphics System, Version 2.0.3, Schrödinger, LLC, New York, NY, USA). The predicted antigenic were compared between all the available Biv subgroup sequences and plotted with WebLogo (Crooks et al., Citation2004).
Results
Molecular detection of CAstV and concomitant viruses
CAstV was detected in all the samples with cycle threshold (CT) values as follows: USP BR-1051 (CT = 23.08), USP BR-1204 (CT = 27.03), USP BR-1220 (CT = 21.30), and USP BR-1276 (CT = 16.94).
Also, the presence of concomitant enteric viruses was examined. Cases USP BR-1051 and USP BR-1220 tested negative for all of these viruses. In the case of USP BR-1204, ANV (CT = 34.70), ChPV (CT = 30.90) and ARV (CT = 36.63) were detected. For USP BR-1276, only FAdV was detected (CT = 35.37).
CAstV characterization
The deduced amino acid sequences were translated from the complete nucleotide sequences of ORF2, with a total product size of 739 amino acids, including the terminal codon. The alignment of the amino acid sequences was performed with 45 representative sequences of the A and B groups of CAstV. The similarity matrix was inferred with the previous alignment, showing that there was a significant percentage difference between the sequences of this study and the sequences belonging to the subgroups Ai (38.82% to 40.52%), Aii (37.01% to 40.34%), and Aiii (37.15% to 39.19%). The sequences in subgroups Bi (85.93% to 89.72%), Bii (81.81% to 85.35%), Biii (87.58% to 89.43%), and Biv (95.26–99.59%) were more closely related. The details of the similarity ranges are described in and the complete identity matrix in Table S1 (online supplemental data). The phylogenetic tree placed the studied strains into the subgroup Biv with a 100% bootstrap support (). The subgroup Biv includes a previous Brazilian strain associated with WCS (Nuñez et al., Citation2016), 14 Canadian strains associated with WCS (Palomino-Tapia et al., Citation2020) and two USA strains referred to as aetiological agents of RSS (Kang et al., Citation2018). The obtained sequences were submitted to GenBank under the accession numbers MN413615-MN413618.
Figure 1. Phylogenetic tree including CAstV Brazilian strains and representative nucleotide sequences of each CAstV group and subgroup. The tree was inferred using MEGA 7 software (Pennsylvania State University, PA, USA) with the neighbor-joining method and 1000 bootstrap replicates. The evolutionary distances were computed using the Jukes-Cantor method. Avian nephritis virus was used as external group. Black circles show the strains used in this study.
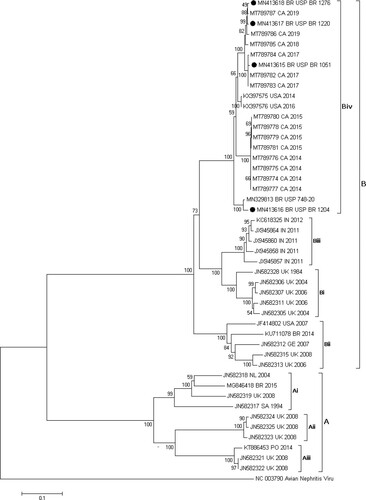
Table 2. Maximum and minimum percentages of identity between the amino acid sequences from this study and the sequences from each subgroup of CAstV.
Prediction of antigenic sites
The EMBOSS antigenic tool returned 14 peptides located on the capsid protein. The potential antigenic peptides were shared and placed in the same sites for the four studied sequences and revealed a high degree of conservation between all strains of the Biv subgroup (). However, two of these (YQTVTSAVPVQQGPLVLTQI and QPRYLVGP) presented a variant peptide because of a non-synonymous mutation in one site. In addition to those, the peptide DVYVMYS showed variation in one site when all the Biv sequences were compared (). The peptides were identified with numbers 1–14 according to their prediction score and were also identified throughout the protein structure, showing that they were indeed located on the surface of the protein and confirming the possibility that they are part of the antigenic regions of the viral capsid ().
Figure 2. Surface diagram from all four angles of view of the CAstV capsid protein with 14 predicted antigenic peptides according to the EMBOSS antigenic tool prediction. All numbers and colours represent the peptides described in . Predictions were based on the four sequenced strains of the present study.
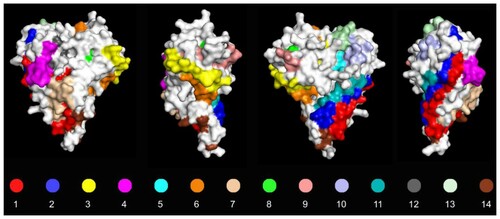
Table 3. EMBOSS antigenic predicted peptides within the ORF2 capsid protein.
Discussion
Chicken astrovirus has been associated with enteric problems in young birds, and a specific group of CAstV show a peculiar characteristic consisting of the appearance of white chicks after hatching (Nuñez et al., Citation2016). Chicken farms have been affected by reduced weight gain and growth retardation, and hatching companies have suffered decreased productivity due to low hatchability and embryonic mortality (Long et al., Citation2017; Kang et al., Citation2018). Due to these typical clinical signs reported by the three companies in this study, CAstV infection was the main suspicious aetiological agent, and the significant viral load values estimated by the qPCR assay suggest the potential involvement of CAstV. On the other hand, concomitant viruses were detected with low viral load, suggesting a secondary role for those viruses.
The capsid protein contains the antigenic determinants responsible for cell tropism and antigenic properties and includes regions within the amino acid sequence that are used for the differentiation of CAstV strains (Pantin-Jackwood et al., Citation2011; Smyth et al., Citation2012). Thus, we used this approach to characterize the strains present in these clinical cases reported in our laboratory. This type of analysis was also used by Espinoza et al. (Citation2016), with samples of 30- to 32-week-old breeders and laying hens from Rio Grande do Sul, where a strain was identified as part of group B, subgroup Bii of CAstV. In a second study, Nuñez et al. (Citation2020) demonstrated the presence of a strain related to the appearance of white chickens from a local incubator. This characterization classified the strain within group B, subgroup Biv, in accordance with molecular characterization based on ORF2 sequences (Smyth, Citation2017; Palomino-Tapia et al., Citation2020; McIlwaine et al., Citation2021). In our study, we classified the four strains into the same group B, subgroup Biv. In this way, we can contribute to previous results that certain CAstV strains circulating in Brazil not only cause phenotypic changes and digestive alterations in birds but are also associated with important effects on hatchability and embryonic death. Our results are in concordance with previous studies reporting outbreaks of WCS in Finland, Norway and Canada linked to Biv strains of CAstV (Smyth, Citation2017; Palomino-Tapia et al., Citation2020). To date, most sequenced strains of the Biv subgroup have been found to be associated with WCS. However, two strains from the USA also share a branch with a common ancestor of our strains, and they were reported as aetiological agents for only RSS, with no information about the emergence of white chicks. On the other hand, an Aiii isolate was also reported to be associated with WCS (Sajewicz-Krukowska et al., Citation2016). Thus, it is unclear whether the strains that produce WCS and enteric problems are also present in chicks without WCS. There also exists the possibility of recombination events involving Biv and other non-associated WCS subgroups as previously reported (Palomino-Tapia et al., Citation2020). Further studies are important to sequence and characterize additional CAstV strains associated with WCS from different countries to reaffirm the link with the Biv subgroup and to test the role of additional factors like breed, age, transmission, viral load, and coinfections.
The prediction of peptides with antigenic characteristics was based on looking for regions with hydrophobic residues, i.e. Cys, Leu, and Val, which in experimental assays have been shown to be part of antigenic sites when they are located on the surface of the protein (Kolaskar & Tongaonkar, Citation1990). This information will be useful in designing strategies for the development of vaccines and serological diagnostics for the strains circulating in the Brazilian territory.
In Brazil, there are different strains of CAstV associated with WCS, which has generated problems of dwarfism, malabsorption syndrome, and decreased production parameters. Additionally, we have also been able to relate the presence of this virus in incubators with low hatchability and increased embryonic mortality, which suggests the need for the development of vaccines to reduce the effects of CAstV on the Brazilian poultry industry.
Ethical statement
This study was approved by the Ethics Commission on Animal Use of the School of Veterinary Medicine, University of São Paulo (FMVZUSP), under CEUAVET protocol no. 1727010620.
Supplemental Material
Download MS Word (59.2 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Berg, E.S. & Olaisen, B. (1994). Hybrid PCR sequencing: sequencing of PCR products using a universal primer. BioTechniques, 17, 896–901.
- Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., Kiefer, F., Gallo Cassarino, T., Bertoni, M., Bordoli, L. & Schwede, T. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42, W252–W258.
- Chacón, R.D., Sedano-Herrera, B., Alfaro-Espinoza, E.R., Quispe, W.U., Liñan-Torres, A., De la Torre, D., de Oliveira, A., Astolfi-Ferreira, C.S. & Ferreira, A.J.P. (2022). Complete genome characterization of reticuloendotheliosis virus detected in chickens with multiple viral coinfections. Viruses, 14, 798.
- Crooks, G.E., Hon, G., Chandonia, J.-M. & Brenner, S.E. (2004). Weblogo: a sequence logo generator. Genome Research, 14, 1188–1190.
- De la Torre, D.I., Nuñez, L.F., Astolfi-Ferreira, C.S. & Piantino Ferreira, A.J. (2018). Enteric virus diversity examined by molecular methods in Brazilian poultry flocks. Veterinary Sciences, 5, E38.
- De la Torre, D., Astolfi-Ferreira, C.S., Chacon, R.D. & Piantino Ferreira, A.J. (2018). Sensitive SYBR green-real time PCR for the detection and quantitation of avian rotavirus A. Veterinary Sciences, 6, E2.
- De la Torre, D., Astolfi-Ferreira, C.S., Chacón, R.D., Puga, B. & Piantino Ferreira, A.J. (2021). Emerging new avian reovirus variants from cases of enteric disorders and arthritis/tenosynovitis in Brazilian poultry flocks. British Poultry Science, 62, 361–372.
- Espinoza, L.L., Beserra, L.A.R., Soares, R.M. & Gregori, F. (2016). Turkey astrovirus type 1 (TAstV-1) and chicken astrovirus (CAstV) detection in Brazilian chicken flocks. Avian Diseases, 60, 681–687.
- Günes, A., Marek, A., Grafl, B., Berger, E. & Hess, M. (2012). Real-time PCR assay for universal detection and quantitation of all five species of fowl adenoviruses (FAdV-A to FAdV-E). Journal of Virological Methods, 183, 147–153.
- Kang, K.-I., Linnemann, E., Icard, A.H., Durairaj, V., Mundt, E. & Sellers, H.S. (2018). Chicken astrovirus as an aetiological agent of runting-stunting syndrome in broiler chickens. Journal of General Virology, 99, 512–524.
- Kolaskar, A.S. & Tongaonkar, P.C. (1990). A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Letters, 276, 172–174.
- Koo, B.S., Lee, H.R., Jeon, E.O., Han, M.S., Min, K.C., Lee, S.B. & Mo, I.P. (2013). Molecular survey of enteric viruses in commercial chicken farms in Korea with a history of enteritis. Poultry Science, 92, 2876–2885.
- Long, K.E., Hastie, G.M., Ojkić, D. & Brash, M.L. (2017). Economic impacts of white chick syndrome in Ontario, Canada. Avian Diseases, 61, 402–408.
- Long, K.E., Ouckama, R.M., Weisz, A., Brash, M.L. & Ojkić, D. (2018). White chick syndrome associated with chicken astrovirus in Ontario, Canada. Avian Diseases, 62, 247–258.
- McIlwaine, K., Law, C.J., Lemon, K., Grant, I.R. & Smyth, V.J. (2021). A review of the emerging white chick hatchery disease. Viruses, 13, 2435.
- Nuñez, L.F.N., Santander Parra, S.H., Carranza, C., Astolfi-Ferreira, C.S., Buim, M.R. & Piantino Ferreira, A.J. (2016). Detection and molecular characterization of chicken astrovirus associated with chicks that have an unusual condition known as “white chicks” in Brazil. Poultry Science, 95, 1262–1270.
- Nuñez, L.F., Santander-Parra, S.H., Chaible, L., De la Torre, D.I., Buim, M.R., Murakami, A., Zaidan Dagli, M.L., Astolfi-Ferreira, C.S. & Piantino Ferreira, A.J. (2018). Development of a sensitive real-time fast-qPCR based on SYBR® Green for detection and quantification of chicken parvovirus (ChPV). Veterinary Sciences, 5, E69.
- Nuñez, L.F.N., Santander-Parra, S.H., Kyriakidis, N.C., Astolfi-Ferreira, C.S., Buim, M.R., De la Torre, D. & Piantino Ferreira, A.J. (2020). Molecular characterization and determination of relative cytokine expression in naturally infected day-old chicks with chicken astrovirus associated to white chick syndrome. Animals, 10, E1195.
- Palomino-Tapia, V., Mitevski, D., Inglis, T., van der Meer, F., Martin, E., Brash, M., Provost, C., Gagnon, C.A. & Abdul-Careem, M.F. (2020). Chicken astrovirus (CAstV) Molecular studies reveal evidence of multiple past recombination events in sequences originated from clinical samples of white chick syndrome (WCS) in western Canada. Viruses, 12, E1096.
- Pantin-Jackwood, M.J., Day, J.M., Jackwood, M.W. & Spackman, E. (2008). Enteric viruses detected by molecular methods in commercial chicken and turkey flocks in the United States between 2005 and 2006. Avian Diseases, 52, 235–244.
- Pantin-Jackwood, M.J., Strother, K.O., Mundt, E., Zsak, L., Day, J.M. & Spackman, E. (2011). Molecular characterization of avian astroviruses. Archives of Virology, 156, 235–244.
- Patel, A.K., Pandit, R.J., Thakkar, J.R., Hinsu, A.T., Pandey, V.C., Pal, J.K., Prajapati, K.S., Jakhesara, S.J. & Joshi, C.G. (2017). Complete genome sequence analysis of chicken astrovirus isolate from India. Veterinary Research Communications, 41, 67–75.
- Raji, A.A. & Omar, A.R. (2022). An insight into the molecular characteristics and associated pathology of chicken astroviruses. Viruses, 14, 722.
- Saif, Y.M., Guy, J.S., Day, J.M., Cattoli, G. & Hayhow, C.S. (2019). Viral enteric infections. In D.E. Swayne, M. Boulianne, C.M. Logue, L.R. McDougald, V. Nair, D.L. Suarez, S. de Wit, T. Grimes, D. Johnson, M. Kromm, T.Y. Prajitno, I. Rubinoff & G. Zavala (Eds.), Diseases of Poultry 14th edn (pp. 401–445). Hoboken, NJ: Wiley-Blackwell.
- Sajewicz-Krukowska, J., Pać, K., Lisowska, A., Pikuła, A., Minta, Z., Króliczewska, B. & Domańska-Blicharz, K. (2016). Astrovirus-induced “white chicks” condition - field observation, virus detection and preliminary characterization. Avian Pathology, 45, 2–12.
- Sajewicz-Krukowska, J. & Domanska-Blicharz, K. (2016). Nearly full-length genome sequence of a novel astrovirus isolated from chickens with “white chicks” condition. Archives of Virology, 161, 2581–2587.
- Smyth, V.J., Jewhurst, H.L., Wilkinson, D.S., Adair, B.M., Gordon, A.W. & Todd, D. (2010). Development and evaluation of real-time TaqMan® RT-PCR assays for the detection of avian nephritis virus and chicken astrovirus in chickens. Avian Pathology, 39, 467–474.
- Smyth, V.J., Todd, D., Trudgett, J., Lee, A. & Welsh, M.D. (2012). Capsid protein sequence diversity of chicken astrovirus. Avian Pathology, 41, 151–159.
- Smyth, V., Trudgett, J., Wylie, M., Jewhurst, H., Conway, B., Welsh, M., Kaukonen, E. & Perko-Mäkelä, P. (2013). Chicken astrovirus detected in hatchability problems associated with “white chicks”. The Veterinary Record, 173, 403–404.
- Smyth, V.J. (2017). A review of the strain diversity and pathogenesis of chicken astrovirus. Viruses, 9, E29.
- Todd, D., Trudgett, J., McNeilly, F., McBride, N., Donnelly, B., Smyth, V.J., Jewhurst, H.L. & Adair, B.M. (2010). Development and application of an RT-PCR test for detecting avian nephritis virus. Avian Pathology, 39, 207–213.
- Walker, P.J., Siddell, S.G., Lefkowitz, E.J., Mushegian, A.R., Adriaenssens, E.M., Alfenas-Zerbini, P., Davison, A.J., Dempsey, D.M., Dutilh, B.E., García, M.L., Harrach, B., Harrison, R.L., Hendrickson, R.C., Junglen, S., Knowles, N.J., Krupovic, M., Kuhn, J.H., Lambert, A.J., Łobocka, M., Nibert, M.L., Oksanen, H.M., Orton, R.J., Robertson, D.L., Rubino, L., Sabanadzovic, S., Simmonds, P., Smith, D.B., Suzuki, N., Van Dooerslaer, K., Vandamme, A.-M., Varsani, A. & Zerbini, F.M. (2021). Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Archives of Virology, 166, 2633–2648.
