Abstract
Isolates of Apple chlorotic leaf spot virus (ACLSV) were collected from three species of pear (genus Pyrus) growing in Shandong and Hubei Provinces, China. The complete genome of one isolate (JB) and near-full-length genomes (30 nucleotides at 5′ end excluded) of two isolates (KMS and YH) were determined. These three isolates showed an overall nucleotide identity of 87.3–100%, and had the highest identity of 83.0% to an isolate MO-5 infecting apple in Japan and 74.9–75.8% identity to two recently reported ACLSV isolates from peach plants grown in China. The three ACLSV isolates from pear were mostly similar to ‘B6 type’ by having signature sites (S40-V59 or M59-Y75-T130-L184) in their coat protein. This is the first comparative study on the complete genomic sequences of ACLSV isolates infecting pear.
Résumé
Des isolats du virus de la tache chlorotique du pommier (VTCP) ont été collectés sur trois espèces de poiriers (genre Pyrus) poussant dans les provinces du Shandong et du Hubei, en Chine. La séquence génomique complète d’un isolat (JB) et les séquences génomiques presque complètes (à l’exception de 30 nucléotides situés à l’extrémité 5′) de deux isolats (KMS et YH) ont été déterminées. Ces trois isolats ont affiché un taux d’identité globale en nucléotides de 87.3 à 100% et possédaient le taux le plus élevé (83.0%) relativement à un isolat (MO-5) infectant des pommiers au Japon, ainsi qu’un taux d’identité de 74.9 à 75.8% par rapport à deux isolats du VTCP signalés récemment sur des pêchers cultivés en Chine. Les trois isolats du VTCP provenant des poiriers ressemblaient surtout au « type B6 » à cause des sites distinctifs (S40-V59 ou M 59-Y75-T130-L184) décelés dans la protéine de leur enveloppe. Il s’agit de la première étude comparative traitant des séquences génomiques complètes des isolats du VTCP qui infectent le poirier.
Introduction
Apple chlorotic leaf spot virus (ACLSV), the type species of the genus Trichovirus in the family Betaflexiviridae (Martelli et al. Citation1994; King et al. Citation2012), occurs worldwide and infects many pome and stone fruit tree species in the family Rosaceae, including apple, pear, peach, almond, plum, apricot and cherry (Nemeth Citation1986). Infection by ACLSV in commercially cultivated apple trees is usually symptomless. However, in some pear varieties, ACLSV induces severe leaf malformation and chlorotic rings or line patterns referred to as pear ring pattern mosaic. Moreover, a synergistic infection by the mixture of ACLSV, Apple stem grooving virus (ASGV) and/or Apple stem pitting virus (ASPV) can cause apple and pear top-working disease when grafted onto susceptible rootstocks (Yanase et al. Citation1979; Desvignes & Boyé Citation1989). In stone fruit trees, ACLSV induces severe symptoms, including dark green sunken mottle in peach, rosette formation and graft incompatibility in apricot, pseudopox and bark split in plum and bark split in cherry (Dunez et al. Citation1972; Desvignes & Boyé Citation1989; Cieślińska et al. Citation1995; Jelkmann & Kunze Citation1995).
The complete genome sequence of ACLSV has been determined for 11 isolates from plum (P863 and PBM1) (German et al. Citation1990; Jelkmann Citation1996), apple (P-205, A4, B6, RC and MO-5) (Sato et al. Citation1993; Yaegashi et al. Citation2007; Dhir et al. Citation2013), cherry (Bal1) (German et al. Citation1997) and peach (TaTao5, Z1 and Z3) (Marini et al. Citation2008; Niu et al. Citation2012), respectively. The genome consists of 7474–7555 nucleotides (nt), excluding the poly (A) tail at its 3′ end, and contains three overlapping open reading frames (ORFs 1, 2 and 3) encoding a 216 kDa protein involved in replication (Rep), a 50 kDa movement protein (MP) and a 22 kDa coat protein (CP), respectively.
Early studies showed distinct serological and biological characteristics among ACLSV isolates from different sources (Barba & Clark Citation1986; Cieślińska et al. Citation1995; Hong & Wang Citation1999). Most of these studies focused on apple and stone fruit isolates. A recent study from our laboratory indicated that CP genes of isolates of ACLSV originating from sand pear (Pyrus pyrifolia) were also highly divergent (Song et al. Citation2011). To better understand the molecular characteristics of ACLSV from pear, the complete or near-full-length genomic sequences of three ACLSV isolates from three different pear species grown in China were determined for the first time in this study.
Materials and methods
Plant materials
Plant shoots with leaves were collected from three pear varieties: Pyrus communis ‘Early Red’ and P. bretschneideri ‘Jingbai’ from Shandong province in northern China, and P. pyrifolia ‘Yuanhuang’ from Hubei province in central China. The presence of ACLSV in these samples was confirmed by RT-PCR using primer pair 216f/216r (GGTGAGAGGCTCTATTCACATCTTG/GGAGCTTTTCACCCCAGCAATTGG) designed basing on conserved regions of the CP gene.
RNA extraction and reverse transcription-polymerase chain reaction
Total RNA was extracted from 0.1 g leaf material with a cetyltriethyl ammonium bromide (CTAB) method (Li et al. Citation2008). Reverse transcription was performed at 37 °C for 1.5 h using 3 µL of total RNA and 1 µL of random primer in a 20 µL reaction volume with Maloney murine leukaemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI) according to the manufacturer’s protocols. For the amplification of ACLSV genomes, primer sets () were designed by aligning all ACLSV genomic sequences available in GenBank. Their targeted amplicons were overlapped by more than 100 bp. The 5′ RACE was attempted using an Invitrogen GeneRacer Kit (Invitrogen, USA) according to the manufacturer’s instructions. PCR reactions were performed in a 25 µL volume with reaction mixtures containing 2.5 µL of 10 × PCR buffer, 3 µL of cDNA, 2 mM of each dNTP, 0.5 mM of each primer, one unit of rTaq DNA polymerase (TaKaRa, Dalian, China) and a proper volume of ddH2O. The PCR reaction conditions were as follows: 94 °C for 3 min, followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 45 s at 50–54 °C (depending on primer pairs used in distinct reactions), extension for 2 min at 72 °C, and a final extension for 10 min at 72 °C.
Table 1. Primers used for the amplification of the genomes of ACLSV isolates KMS, YH and JB.
Cloning, sequencing and sequence analysis
The PCR products were gel-purified and ligated into pMD18-T vector (TaKaRa, Dalian, China). Four positive clones of each product were sequenced at Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd, Shanghai, China.
Multiple sequence alignment was performed using the Clustal X program (http://www.clustal.org) (Chenna et al. Citation2003). Phylogenetic trees were derived by importing the aligned sequences (produced with Clustal X) into MEGA5 and constructed using the neighbour-joining method with 1000 bootstrap replicates (Tamura et al. Citation2011). The sequence sources and GenBank accession numbers of the ACLSV isolates referred for sequence analysis are listed in .
Table 2. Origin and GenBank accession numbers of full-length sequences of Apple chlorotic leaf spot virus isolates used for sequence alignment and phylogenetic analyses.
Results and discussion
The complete genome sequence of ACLSV JB from P. bretschneideri ‘Jingbai’ (GenBank accession KC935956) consisted of 7560 nts (excluding a poly (A) tail), and the partial genome sequences (about 30 nt at 5′ end excluded) of both ACLSV KMS from Pyrus communis ‘Early Red’ and ACLSV YH from P. pyrifolia ‘Yuanhuang’ consisted of 7528 nts (excluding a poly (A) tail) (). These sequences were deposited in the GenBank database with accession numbers KC935954, KC935955 and KC935956, respectively. Although isolates KMS and YH were obtained from different pear species, they had highly similar genome sequences with 96.8% identity at the nt level, and over 98% amino acid (aa) identities between their encoded proteins. The isolate JB shared only 88.3–88.8% nt identities and 92.4–98.5% aa identities with isolates KMS and YH. The genome nucleotide sequence identities among these three isolates and all ACLSV isolates available in GenBank ranged from 68.0% (TaTao5) to 83.0% (MO-5). The complete genomes of these three ACLSV pear isolates showed an overall 74.9–75.8% nt identity to the two recently reported ACLSV genomes originating from peach trees grown in China (Niu et al. Citation2012). The genomic organization of KMS, YH and JB was the same as that of previously described ACLSV isolates, and consisted of three ORFs. The ORF1 and ORF2 of the three isolates consisted of 5637 nts and 1377 nts, respectively, although they varied among different ACLSV isolates. The 5′-UTRs of KMS and YH were 100% identical, and 93.4% and 91.6% identical to those of JB and MO-5, and only 58.1–81.2% identical to the corresponding regions of other isolates. The ORF1-, ORF2- and ORF3-encoded proteins showed overall 73.7–90.5%, 58.7–88.9% and 73.6–94.3% aa identities to those of published isolates, and as reported previously, the coat protein appeared to be the most conserved protein among ACLSV isolates by sharing over 90% aa identities among those fully sequenced ACLSV isolates except for two divergent isolates (Ball and TaTao5).
Table 3. Sequence comparison of the complete genome and different genomic regions between ACLSV JB and isolates KMS, YH and 10 ACLSV isolates available in GenBank.
The hyper-variable segment downstream of the methyl-transferase domain (ORF1) was described previously (German et al. Citation1997). The multiple sequence alignment of the replicase of ACLSV isolates revealed that the region at sites 468–670 aa was highly divergent (). In this region, JB, KMS and YH were 73.8–92.8% identical to each other, but were only 29–38.8% identical to isolate TaTao5 and 35.5–62.4% identical to other isolates. Meanwhile, it was found that several conserved aa were present in three ACLSV pear isolates JB, YH and KMS and some other conserved aa were present in those three pear isolates and an apple isolate MO-5 from Japan. These results indicated that the sequence in the hyper-variable region might be related to the phylogenetic evolution of the virus.
Fig. 1 (Colour online) Amino acid sequence alignments of the highly variable region at sites 468–670 aa of replicase among ACLSV isolates. The conserved amino acids among ACLSV isolates JB, YH and KMS are marked with red boxes, and amino acids conserved among these three ACLSV isolates and an apple isolate MO-5 are marked with black boxes.

Previously, the aa motif (S40-L59-Y75-T130-L184) for ‘B6 type’ ACLSV isolates was proposed (Yaegashi et al. Citation2007). Multiple alignment of the ACLSV CP aa sequences revealed that the two Chinese pear isolates KMS and YH had the same aa combination (S40-V59-Y75-T130-L184), but isolate JB had aa M at site 59 (S40-M59-Y75-A130-L184) as that of a kuerle isolate, which were identified from Kuerle Fragrant pear (Pyrus sinkiangensis Yü) grown specially at the Kuerle region in Xinjiang in the north-western China (Cai et al. Citation2005). Therefore, these three Chinese pear isolates were mostly similar to ‘B6 type’ ACLSV isolates. Moreover, isolates JB, KMS and YH together with a kuerle isolate and isolate MO-5 in the same sub-group II-b had a specific aa combination S73-D82-L83-G98 (data not shown).
The phylogenetic trees generated from both the genome and hyper-variable Rep segment (at sites 468–670 aa) sequences of ACLSV pear isolates JB, KMS and YH together with 11 completely sequenced isolates available in GenBank consistently revealed three distinct clades (). The three Chinese pear ACLSV isolates formed one clade along with an apple isolate MO-5 identified in Japan. The results indicated that the hyper-variable Rep segment might be considered as an important molecular marker of different ACLSV isolates. However, comparable biological properties related to the variation remain to be elucidated. The tree based on the aa sequences of ACLSV CPs also showed that the three Chinese pear ACLSV isolates were closely related to MO-5 and a kuerle isolate. A previous study from our laboratory revealed that most of the ACLSV isolates from sand pear fell into group I represented by ACLSV isolates from Prunus spp. (Song et al. Citation2011). Therefore, our results suggest that the ACLSV population from pear trees grown in China at least consisted of divergent isolates in two separate phylogenetic groups basing on their CP sequences. Moreover, in the postulated epitope regions of ACLSV (Song et al. Citation2011), those three isolates had aa residues similar to those in isolate P205, indicating that they might have the serological reactivity similar to that of ACLSV isolates in the sub-clade A and clade II represented by isolate P205.
Fig. 2 Unrooted Maximum likelihood trees constructed from nucleotide sequences of the genome (A), and amino acid sequences of partial replicase at sites 468–670 aa (B) and full-length CP (C) of Apple chlorotic leaf spot virus (ACLSV). The ACLSV isolates available in GenBank are followed by their hosts and accession numbers. The Grapevine berry inner necrosis virus (GINV, accession no. NC-015220), a member of the genus Trichovirus, was used as outgroup in the genome sequences-based tree. The numbers at the nodes indicate the percentage of 1000 bootstraps occurring in this group. Values below 80% are suppressed. The ACLSV isolates sequenced in this study are indicated in bold. The bar represents 0.01 substitutions per site.
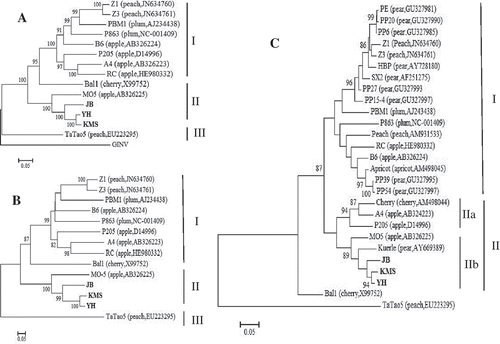
This study represents the first report on complete genomic sequences of ACLSV isolates from pear. Our results indicated that the molecular characteristics of ACLSV isolates might be related to their host of origin (Al Rwahnih et al. Citation2004). Until now, no natural vector transmission for the virus has been found; consequently, cross-infection of ACLSV isolates from different host species, especially species in a different genus, should rarely happen. Since the seedlings of P. xerophila and P. calleryana used widely as the rootstocks of pear trees in China are virus-free, the very closely related sequences of ACLSV isolates in different pear species should not have originated from infected rootstock plants. Transmission of the same isolates among different pear varieties or species, and the mixed infection of molecular variants, could occur through mechanical transmission during pruning. A previously reported highly divergent ACLSV isolate TaTao5 from the propagation material of a peach clone might have a different origin (Marini et al. Citation2008). Further studies on the viral population structure may help gain new insight into the evolution of this virus.
Acknowledgements
We are grateful to Professor Aiming Wang, Southern Crop Protection and Food Research Centre, Agriculture and Agri-Food Canada, for critical review of the manuscript. This work was supported financially by the Chinese Ministry of Agriculture, the agricultural projects (201203076-03 and 200903004) and earmarked fund for Pear Modern Agro-industry Technology Research System (nycytx-29-08).
References
- Al Rwahnih MA, Turturo C, Minafra A, Saldarelli P, Myrta A, Pallás V. 2004. Molecular variability of Apple chlorotic leaf spot virus in different hosts and geographical regions. J Plant Pathol. 86:117–122.
- Barba M, Clark MF. 1986. Detection of strains of Apple chlorotic leafspot virus by F(ab)2-based indirect ELISA. Acta Hortic. 193:297–304.
- Cai Y, Xiang BC, Xi DH, Li SX, Du YJ. 2005. Cloning and prokaryotic expression of CP gene of Apple chlorotic leaf spot virus Kuerle isolate and preparation of its specific antiserum. J Agric Biotechnol. 13:533–537.
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500. doi:10.1093/nar/gkg500
- Cieślińska M, Malinowski T, Zawadzka BJ. 1995. Studies on several strains of Apple chlorotic leaf spot virus (ACLSV) isolated from different fruit tree species. Acta Hortic. 386:63–71.
- Desvignes JC, Boyé R. 1989. Different diseases caused by the Chlorotic leaf spot virus on the fruit trees. Acta Hortic. 235:31–38.
- Dhir S, Zaidi AA, Hallan V. 2013. Molecular characterization and recombination analysis of the complete genome of Apple chlorotic leaf spot virus. J Phytopathol. 161:704–712. doi:10.1111/jph.12121
- Dunez J, Marenaud G, Delbos RP, Lansac M. 1972. Variability of symptom induced by the Apple chlorotic leaf spot virus (CLSV) - a type of CLSV probably responsible for bark split disease of prune trees. Plant Dis Rep. 56:293–295.
- German S, Bergey B, Delbos RP, Candresse T, Dunez J. 1997. Complete nucleotide sequence of the genome of a severe cherry isolate of Apple chlorotic leaf spot trichovirus (ACLSV). Arch Virol. 142:833–841. doi:10.1007/s007050050122
- German S, Candresse T, Lanneau M, Huet JC, Pernollet JC, Dunez J. 1990. Nucleotide sequence and genomic organization of Apple chlorotic leaf spot closterovirus. Virology. 179:104–112. doi:10.1016/0042-6822(90)90279-Z
- Hong N, Wang GP. 1999. The biological and biochemical characterization of Apple chlorotic leaf spot virus. Acta Phytopathol Sin. 29:77–81.
- Jelkmann W. 1996. The nucleotide sequence of a strain of Apple chlorotic leaf spot virus (ACLSV) responsible for plum pseudopox and its relation to an apple and plum bark split strain. Phytopathology. 86:101–101.
- Jelkmann W, Kunze L. 1995. Plum pseudopox in German prune after infection with an isolate of Apple chlorotic leafspot virus causing plum line pattern. Acta Hortic. 386:122–125.
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2012. Virus taxonomy: classification and nomenclature of viruses. Ninth Report of the International Committee on Taxonomy of Viruses. London: Elsevier-Academic Press.
- Li R, Mock R, Huang Q, Abad J, Hartung J, Kinard G. 2008. A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J Virol Methods. 154:48–55. doi:10.1016/j.jviromet.2008.09.008
- Marini DB, Gibson PG, Scott SW. 2008. The complete nucleotide sequence of an isolate of Apple chlorotic leaf spot virus from peach (Prunus persica (L.) Batch). Arch Virol. 153:1003–1005. doi:10.1007/s00705-008-0076-z
- Martelli GP, Candresse T, Namba S. 1994. Trichovirus, a new genus of plant viruses. Arch Virol. 134:451–455. doi:10.1007/BF01310583
- Nemeth M. 1986. Virus, mycoplasma and rickettsia diseases of fruit trees. The Netherlands and AkademiaiKiado, Hungary: Martinus Nijhoff.
- Niu FQ, Pan S, Wu ZJ, Jiang DM, Li SF. 2012. Complete nucleotide sequences of the genomes of two isolates of Apple chlorotic leaf spot virus from peach (Prunus persica) in China. Arch Virol. 157:783–786. doi:10.1007/s00705-011-1195-5
- Sato K, Yoshikawa N, Takahashi T. 1993. Complete nucleotide sequence of the genome of an apple isolate of Apple chlorotic leaf spot virus. J Gen Virol. 74:1927–1931. doi:10.1099/0022-1317-74-9-1927
- Song YS, Hong N, Wang LP, Hu HJ, Tian R, Xu WX, Ding F, Wang GP. 2011. Molecular and serological diversity in Apple chlorotic leaf spot virus from sand pear (Pyrus pyrifolia) in China. Eur J Plant Pathol. 130:183–196. doi:10.1007/s10658-011-9744-z
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. doi:10.1093/molbev/msr121
- Yaegashi H, Isogai M, Tajima H, Sano T, Yoshikawa N. 2007. Combinations of two amino acids (Ala40 and Phe75 or Ser40 and Tyr75) in the coat protein of Apple chlorotic leaf spot virus are crucial for infectivity. J Gen Virol. 88:2611–2618. doi:10.1099/vir.0.82984-0
- Yanase H, Yamaguchi A, Mink GI, Sawamura K. 1979. Back transmission of Apple chlorotic leafspot virus (type strain) to apple and production of apple topworking disease symptoms in Maruba Kaido (Malus prunifolia Borkh. var. ringo Asami). Jpn J Phytopathol. 45:369–374. doi:10.3186/jjphytopath.45.369
