Abstract
A new disease was observed on jujube (Ziziphus jujuba Mill.) in Awti county, Xinjiang, China, in 2014. The symptoms on infected leaves consisted of spots which were tan in the centre, with black pycnidia, surrounded by dark-brown to black margins, and yellow halos. Infected fruit became dark brown to black, and were covered by small pycnidia. The causal agent was isolated from infected leaves and fruit. Pathogenicity tests showed that the fungus could infect jujube fruits, which developed the same symptoms under artificial inoculation conditions as those observed in the field. The fungus was identified based on morphological and cultural characteristics as Nothophoma quercina (Syd) Q. Chen & L. Cai. Identification was confirmed by combining multi-locus phylogenetic analyses based on the internal transcribed spacer regions (ITS 1 and 2) and intervening 5.8S nrDNA, and partial gene regions of β-tubulin (TUB 2). This is the first report of brown spot of jujube caused by Nothophoma quercina (Phoma fungicola) in China.
Résumé
En 2014, une nouvelle maladie a été observée sur le jujubier (Ziziphus jujuba Mill.) dans le comté d’Awti, dans la province chinoise du Xinjiang. Les symptômes sur les feuilles infectées consistaient en taches dont le centre était brun clair, comportant des pycnides noires, et entouré de marges brun foncé à noires et d’aréoles jaunes. Les fruits infectés passaient du brun foncé au noir et étaient recouverts de petites pycnides. L’agent causal a été isolé à partir de feuilles et de fruits infectés. Les tests de pathogénicité ont montré que le champignon pouvait infecter les fruits du jujubier qui affichaient les mêmes symptômes à la suite d’inoculations artificielles que ceux observés au champ. Le champignon a été identifié en tant que Nothophoma quercina (Syd) Q. Chen & L. Cai, en se basant sur ses caractéristiques morphologiques et culturales. L’identification a été confirmée en combinant les analyses phylogénétiques multilocus basées sur les régions de l’espaceur transcrit interne (ITS1 et 2) et de l’ADNrn 5.8S intermédiaire, les séquences nucléotidiques de la grande sous-unité (LSU) partielle de l’ADNrn (28S) et les régions géniques partielles de la β-tubuline (TUB 2). Il s’agit de la première mention de la tache brune trouvée sur le jujubier en Chine, causée par Nothophoma quercina (Phoma fungicola).
Introduction
Jujube (Ziziphus jujuba Mill.), an important fruit tree in China, is becoming increasingly popular because of its high therapeutic value, health benefits and wide adaptation to different soil pH and drought conditions (Liu Citation2009). In recent years, jujube has been grown widely in four prefectures in southern Xinjiang, China, and has become a major crop accounting for more than 80% of farmers’ annual income. At present, the four prefectures have the largest production base of jujube in the world. However, with the increased planting area, the use of different types of cultivation in different ecological microclimates, and the frequent introduction of new breeding materials, some secondary diseases have gradually become more important, and some new diseases have emerged, such as black spot of jujube and jujube fruit Erwinia rot.
Brown spots on the leaves and fruit of jujube were observed in a jujube yard in Awti (Xinjiang, China) in 2014. The symptoms consisted of spots which were tan in the centre, with black pycnidia, surrounded by dark-brown to black margins, and yellow halos on infected leaves (). Infected fruit became dark brown to black, covered by small pycnidia (). In 2015, the disease was found in 6% of the surveyed jujube fields, with an average incidence of 24%, which caused significant yield loss. The aim of this study was to identify the pathogen causing jujube brown spot in Xinjiang, China.
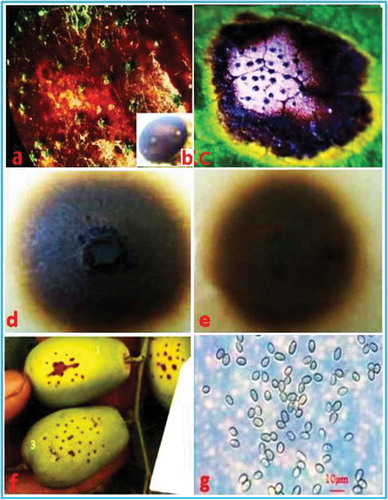
Fig. 1 (Colour online) Symptoms of brown spot of jujube caused by natural infections and inoculations with Nothophoma quercina in the field. (a) Natural infections on fruit of field-grown plants. (b) Enlarged view of a pycnidium of N. quercina. (c) Natural infections on leaves of field-grown plants. (d) Characteristics of a colony of N. quercina on the upper surface of oat agar (OA). (e) Reverse characteristics of a colony of N. quercina on OA. (f) Chlorotic lesions on fruit surfaces 5–7 days after artificial inoculation. (g) Conidia on OA culture. Bar: g = 10 µm.
Materials and methods
Fungal isolation and morphology
Diseased leaves and fruit (20 leaves and 15 fruit samples) were collected from jujube fields in Awti county, Xinjiang, China in 2015. Diseased tissues were cut into pieces (2 mm × 3 mm) and sterilized by submersion in 3% NaOCl for 1 min. The sterilized samples were rinsed three times with sterile distilled water, and incubated on potato dextrose agar (PDA) at 25°C in darkness for 7 days. Isolates were purified by single spore culturing according to Roustaee et al. (Citation2000) and subcultured on oatmeal agar (OA) at 25°C with a 12 h photoperiod, in order to characterize fungal growth and morphology.
Pathogenicity test
Three single-spore isolates originating from three different fields were used to determine pathogenicity. The pathogenicity test was conducted on 4-year-old jujube trees in the field. A 25 µL spore suspension containing 1 × 106 spores mL−1 was inoculated onto the fruit of jujube (50 fruits per isolate) using a micropipette technique (Roustaee et al. Citation2000). The size of inoculated fruit was 1.5–2.0 cm in diameter. Inoculated fruit were wrapped with Parafilm for 48 h. Fifty fruit were treated with sterilized water as a control. After 3 weeks, pathogenicity of the isolates was assessed by presence or absence of symptoms of brown spot. The pathogen was re-isolated and compared with the original isolate. The experiment was repeated three times for each isolate during July–September 2015.
Molecular identification
Genomic DNA of the seven single-spore isolates originating from three different fields in Awti county, Xinjiang, including the three single-spore isolates used in the pathogenicity test, was extracted using a DNA quick extraction kit (Shanghai Biological Engineering Co., China). The LSU region was amplified with the primer pair LR0R (Rehner & Samuels Citation1994) and LR7 (Vilgalys & Hester Citation1990), the ITS region with V9G (de Hoog & Gerrits van den Ende Citation1998) and ITS4 (White et al. Citation1990), the TUB2 region with primers bt2a and bt2b (Glass & Donaldson Citation1995). The PCR amplifications were performed in a total volume of 25 μL containing 2.5 μL 10× EasyTaq Buffer (200 mM Tris-HCl, pH 8.4, 200 mM KCl, 100 mM (NH4)2SO4, and 20 mM MgSO4; TransGen Biotech, Beijing, China), 25 μM dNTPs, 0.1 μM of each primer, 0.5 U Taq DNA polymerase and 5 ng genomic DNA. PCR conditions for LSU, ITS and TUB2 were set as follows: an initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation, annealing and extension, and a final extension step at 72°C for 10 min. For the LSU amplification, the 35 cycles consisted of 45 s at 95°C, 45 s at 50°C and 1 min at 72°C; for the ITS 30 s at 94°C, 30 s at 52°C and 90 s at 72°C; and for the TUB2 region 30 s at 94°C, 30 s at 57°C and 45 s at 72°C. The amplified products were resolved by electrophoresis on a 1.5% agarose gel. The target band was retrieved with DNA Fragment Quick Purification Kit (Shanghai Biological Engineering Co., China) and sequenced (Shanghai Biological Engineering Co., China). The products were used to search the nucleotide database of GenBank using the program Blastn. The nucleotide sequences (21 characters for LSU, 26 for ITS and 23 for TUB 2) from different isolates representing different Phoma species were selected from GenBank based on the results of morphological observations of our isolates (Nothophoma quercina) and used for phylogenetic analysis. Multiple sequence alignment was carried out using Clustal W (Thompson et al. Citation1994), and manually adjusted as required. Phylogenetic analysis was constructed utilizing the Maximum likelihood method, in the Jukes–Cantor model (MEGA version 5), with a bootstrap of 1000 replicates (Tamura et al. Citation2011). Nucleotide sequences (LSU, ITS and TUB 2) of Sclerotinia sclerotiorum were used as the out-group taxon.
Results and discussion
Fungal isolation and morphology
The same fungus was consistently isolated from naturally infected fruit and leaves of jujube (–c), and the frequency of isolation from diseased tissue segments was 78%. The colonies (, e) on OA were greenish olivaceous to olivaceous, with pure white aerial mycelium. The aerial mycelium had a floccose to woolly appearance. Pycnidia (, b) were solitary, globose, peroblate to suboblate. Pycnidia (n = 50) were 65.0–135.0 µm in diameter. Conidia (n = 50; ) were variable in shape and size, subglobose to oval or obtuse, 4.3–8.5 × 3.0–4.8 µm in diameter.
Pathogenicity test
Symptoms of brown spot were observed on all of the inoculated jujube fruits, but not on the non-inoculated control jujube fruits. Small chlorotic lesions () appeared around the wounds of the fruit at 5–7 days after inoculation. The symptoms on the inoculated fruit were similar to those observed in the field during the next growing stage. The fungus was re-isolated from all infected fruit for each isolate, and the re-isolated fungal cultures were identical to the original isolate based on the cultural and morphological characteristics.
Molecular identification
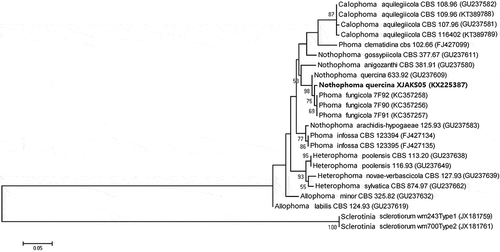
The target band, with a predicted size of 295 bp for TUB2, 1321 bp for LSU and 510 bp for the ITS region, was obtained from all seven single spore isolates (–c). The searches revealed that the sequences of each gene (ITS, LSU and TUB 2) from the seven isolates shared 100% identity, and the target sequences from ITS, LSU and TUB 2 of one representative isolate (XJAKS05) were deposited in GenBank (accession nos. KX225387, KX645663, KX645664, respectively). In the phylogenetic analysis, the target sequence for the ITS region (GenBank accession no. KX645664) was clustered in a distinct clade with those of Nothophoma species isolates retrieved from GenBank, with 84% bootstrap value and 53% bootstrap value with those of Nothophoma quercina (). Since the locus phylogenies of LSU displayed low resolution at both the generic and species level using the sequence (GenBank accession no. KX645663), the data is not shown. The sequence for the TUB 2 (GenBank accession no. KX225387) was clustered in a distinct clade with those of Nothophoma quercina isolates retrieved from GenBank, with 98% bootstrap value (). The fungus was identified as N. quercina (Syd.) Q. Chen & L. Cai, based on the morphological, cultural characteristics and phylogenetic trees generated using the ITS and TUB 2 sequence analysis.
Fig. 2 (Colour online) Agarose gel showing amplification of TUB 2, LSU and ITS gene fragments from seven isolates of Nothophoma quercina, respectively. (a) TUB 2; (b) LSU; (c) ITS. Lanes 1–7: isolates of No. quercina. Ck: negative control. M: DNA Ladder Plus (Shanghai Biotechnology Co., China).
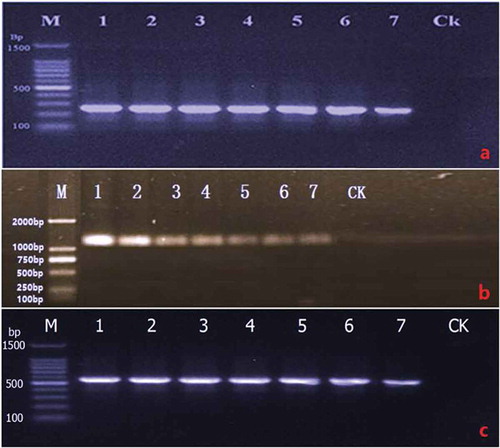
Fig. 3 Phylogenetic tree constructed with the nucleotide sequences of the ITS gene of isolates from 17 different Phoma species, including one new isolate from this study (KX645664), retrieved from GenBank. Nucleotide sequences of the ITS gene of Sclerotinia sclerotiorum were used as the out-group taxon. The bar indicates nucleotide substitutions per site. Numbers of bootstrap support values ≥50% based on 1000 replicates.

Fig. 4 Phylogenetic tree constructed with the nucleotide sequences of the TUB 2 gene of isolates from 12 different Phoma species, including one new isolate from this study (KX225387), retrieved from GenBank. Nucleotide sequences of the TUB2 gene of Sclerotinia sclerotiorum were used as the out-group taxon. The bar indicates nucleotide substitutions per site. Numbers of bootstrap support values ≥50% based on 1000 replicates.
The genus Phoma is a polyphyletic group with species scattered in at least six families in Pleosporales (Aveskamp et al. Citation2008). Historically, the genus was separated into nine sections based on morphological characters, which proved to be artificial and not reflective of their natural evolutionary history. A recently published study by Chen et al. (Citation2015) refined the classification within the genus based on the molecular phylogeny of four loci (LSU, ITS, RPB2, TUB 2) and erected multiple new genera. We used the LSU, ITS and TUB2 genes in our study but attempts to amplify the RPB2 gene failed.
Phoma fungicola belongs to the genus Nothophoma, and the current name is Nothophoma quercina (Phoma fungicola). Phoma fungicola has been reported to cause stem blight and fruit blight of Pistachio in Arizona (Chen et al. Citation2013), and dieback of olive trees in Tunisia (Krid Hadj Taieb et al. Citation2014). Here, we report that the brown spot of jujube, caused by N. quercina, is a new disease, not previously reported in China. To our knowledge, it has also not been reported elsewhere.
Additional information
Funding
References
- Aveskamp MM, de Gruyter J, Crous PW. 2008. Biology and recent developments in the systematic of Phoma, a complex genus of major quarantine significance. Fungal Div. 31:1–18.
- Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. 2015. Resolving the Phoma enigma. Stud Mycol. 82:137–217.
- Chen SF, Morgan DP, Michailides TJ. 2013. First report of Phoma fungicola associated with stem canker and fruit blight of pistachio in Arizona. J Plant Pathol. 95:450.
- De Hoog GS, Gerrits van den Ende AHG. 1998. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 41:183–189.
- Glass NL, Donaldson G. 1995. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 61:1323–133.
- Krid Hadj Taieb S, Ma T, Hammami I, Rhouma A. 2014. First report of dieback of olive trees caused by Phoma fungicola in Tunisia. J Plant Pathol. 96:S4.117.
- Liu MJ. 2009. Germplasm resources and production of Chinese jujube in China. Acta Hort Sin. 840:25–31.
- Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98:625–634.
- Roustaee A, Costes S, Dechamp-Guillaume G, Barrault G. 2000. Phenotypic variability of Leptosphaeria lindquistii (Anamorph: Phoma macdonaldii) a fungal pathogen of sunflower. Plant Pathol. 49:227–234.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymati-cally amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 172:4238–4246.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; p. 315–322.
