Abstract
Coconut palms in the south coastal littoral of Grand-Lahou have long been affected by the Côte d’Ivoire lethal yellowing (CILY), associated with subgroup 16SrXXII-B ‘Candidatus Phytoplasma palmicola’-related strains. Recently, foliar discolouration and yellowing symptoms were observed in oil palms (Elaeis guineensis), Rônier palms (Borasus aethiopium), and raffia palms (Raphia vinifera) located in the vicinity of coconut palms (Cocos nucifera) showing CILY-like symptoms in coconut-growing villages of Grand-Lahou. Immature leaf samples were randomly collected from symptom-bearing palms, including coconut palms. Total DNA was extracted and used as a template for a nested PCR assay with universal primers that target the phytoplasma 16S rRNA gene, and the translocation protein (secA) genes. The R16F2n/R2 and SecA amplicons from representative samples of each palm species were purified, cloned, and sequenced. R16F2n/R2 sequences were over 99% identical to phytoplasmas enclosed in group 16SrXXII. Based on iPhyclassifier analyses with 17 restriction enzymes, the phytoplasmas found in C. nucifera, E. guineensis, B. aethiopium and R. vinifera were identical to each other. However, a unique Sau3AI RFLP pattern was observed, and similarity coefficients were lower than 0.97, and so a new 16SrXXII subgroup, 16SrXXII-C, was assigned. Phylogeny based on both R16F2n/R2 and SecA sequences supported this RFLP classification. The presence of the subgroup 16SrXXII-C in the same area where coconut palms are severely infected by the subgroup 16rSXXII-B could lead to different epidemiological constraints that may affect the severity of the coconut disease. Thus, the 16SrXXII-C phytoplasma poses a new threat for other Arecaceae species, which may impact CILY management and control in Grand-Lahou.
Résumé
Les cocotiers du littoral de la Côte d’Ivoire situés à Grand-Lahou sont depuis longtemps touchés par la maladie du jaunissement mortel (JMCI), auquel le phytoplasme associé est ‘Candidatus Phytoplasma palmicola’ du sous-groupe 16SrXXII-B. Récemment, des symptômes de décoloration et de jaunissement foliaires ont été observés sur les elaeis (Elaeis guineensis), sur les rôniers (Borasus aethiopium) et sur les raphia (Raphia vinifera) présents dans le voisinage des cocotiers (Cocos nucifera) infectés par le JMCI dans les cocoteraies de Grand-Lahou. Des échantillons de feuilles immatures symptomatiques ont été collectés de façon aléatoire sur des palmiers, rhonier, raphia et cocotiers. L’ADN total en a été extrait et utilisé comme matrice pour les PCR avec les amorces universels et spécifiques ciblant l’ARNr 16S et les gènes du phytoplasme de la protéine de translocation (secA). Les amplicons R16F2n/R2 et SecA issus d’échantillons représentatifs de chaque espèce de palmaceae ont été purifiés, clonés et séquencés. Les séquences de R16F2n/R2 étaient identiques à plus de 99% aux phytoplasmes appartenant au groupe 16SrXXII. En se basant sur les analyses effectuées par iPhyclassifier avec 17 enzymes de restriction, les phytoplasmes trouvés chez C. nucifera, E. guineensis, B. aethiopium et R. vinifera étaient identiques. Toutefois, un profile Sau3AI RFLP unique a été observé et les indices de simililarité étaient inférieurs à 0.97, Ce résultat justifie par conséquent la présence d’un nouveau sous-groupe de 16SrXXII, nommé 16SrXXII-C. La phylogénie basée sur les séquences de R16F2n/R2 et de SecA confirme la classification obtenue par la RFLP. L’identification du sous-groupe 16SrXXII-C dans la zone où les cocotiers sont déjà infectés par le sous-groupe 16SrXXII-B pourrait engendrer différentes contraintes épidémiologiques aggravant de la maladie du JMCI chez les cocotiers. La présence du phytoplasme 16SrXXII-C constitue une nouvelle menace pour les autres espèces d’arécacées, et peut influencer la gestion de la lutte contre le JMCI à Grand-Lahou.
Introduction
In Côte d’Ivoire, coconut palms (Cocos nucifera L.) are mainly cultivated on ~50 000 hectares in the south coastal littoral (Konan Konan et al. Citation2013). Côte d’Ivoire is the top African country exporting coconut oil from copra to Europe and West Africa, which is the major source of income for around 85 000 coconut smallholder farmers in Grand-Lahou (Arocha Rosete et al. Citation2017). Côte d’Ivoire produces on average 1 564 000 million tonnes of oil palm seeds per year and exports about 292 278 tonnes of oil (Béhi et al. Citation2002). A total of 170 000 ha of oil palms are located in the village areas where nearly 40,000 producers manage the plantations (Sangaré et al. Citation2009).
Three other palm species in the Arecaceae family are important for the production of either palm wine or palm oil (Béhi et al. Citation2002), and include the oil palm (Elaeis guineensis Jacq.), Rônier palm (Borassus aethiopum Mart.) and raffia palm (Raphia vinifera P. Beauv.).
These species are used as ornamental palms, and also for the local production of the traditional fermented beverage consumed by the farmers, palm wine, known as ‘bangui’ in Dioula or ‘nzan’ in Baoulé. Alladatin (Citation2011) has estimated a total annual contribution of the R. vinifera sector at 192 429 633 CFA (US$385 000), bringing relatively large incomes. These palm species are widely grown in the south littoral coastal area of Grand-Lahou, where the Côte d’Ivoire lethal yellowing (CILY) has currently devastated over 400 ha of coconut groves, and is threatening more than 7000 ha of the coconut-growing village area of Grand-Lahou (Arocha Rosete et al. Citation2017). Lethal yellowing (LY) diseases associated with phytoplasmas are the major threat for the coconut crop and for nearly 40 other palm species in Central and North America and the Caribbean, Africa and Asia (Danyo Citation2011). Phytoplasmas are cell wall-less bacteria of the class Mollicutes, transmitted by sap-feeding leafhoppers, psyllids or fulgoroids of the order Hemiptera (Duduk & Bertaccini Citation2011).
Restriction fragment length polymorphism (RFLP) of the 16S rRNA gene sequences (1.2 kb) amplified by polymerase chain reaction (PCR) using phytoplasma universal primers (Gundersen & Lee Citation1996; Lee et al. Citation2000) has allowed phytoplasmas associated with LY to be assigned to the following sub-groups of group 16SrIV: Coconut Lethal Yellows subgroups 16SrIV-A, 16SrIV-B, 16SrIV-C, 16SrIV-D, 16SrIV-E and 16SrIV-F, which occur in North and Central America and the Caribbean, and Africa (Lee et al. Citation2000; Wei et al. Citation2007; Harrison et al. Citation2008, Citation2014; Zhao et al. Citation2009; Gurr et al. Citation2016). Other phytoplasma groups associated with coconut LY-like diseases include 16SrXI ‘Candidatus Phytoplasma oryzae’, 16SrXIV ‘Ca. P. cynodontis’, ‘Ca. P. malaysianum’ in Asia (Nejat et al. Citation2009, Citation2013), and a ‘Ca. P. pini’-related in Mozambique (Bila et al. Citation2015b). In West Africa, ‘Ca. P. palmicola’ was the main phytoplasma detected, in which two ribosomal subgroups were identified, 16SrXXII-A and 16SrXXII-B (Harrison et al. Citation2014).
Non-ribosomal single-copy genes such as secA, which encodes the ATP-dependent energy generator in the bacterial precursor protein translocation cascade system (SecA), has been widely used to cross-confirm the phytoplasma classification based on the 16S rRNA gene (Hodgetts et al. Citation2008; Bila et al. Citation2015b). Based on SecA sequence analysis, Bila et al. (Citation2015b) separated phytoplasmas affecting coconut palms into at least three distinct groups, previously described by Hodgetts et al. (Citation2008), reflecting the strains' geographic origins: an Americas group typified by LY, an East African group typified by Tanzanian LD, and a West African group typified by lethal decline (LD) from Nigeria and Cape St. Paul Wilt Disease (CSPWD) from Ghana.
The aim of the present study was to determine whether other palm species could pose any threat for the spread of CILY disease in Grand-Lahou. The work included PCR screening for the presence of phytoplasma, as well as RFLP and sequence analysis based on two phytoplasma genes – the 16S rRNA and translocation protein (secA) – in samples collected from coconut palms showing CILY-like symptoms and other nearby-grown palm species.
Materials and methods
Sample collection and total DNA extraction
Palms were randomly surveyed in three CILY-affected villages of Grand-Lahou: Doudougbazou (located 45 km from Grand-Lahou), Yaokro (43 km from Grand-Lahou) and Braffedon (18 km from Grand-Lahou). Distances between Braffedon and Yaokro villages are 55 km, while that between Yaokro and Doudougbazou villages is just 2 km. In each village, palms belonging to E. guineensis, B. aethiopium and R. vinifera were located in a perimeter of ~300 to 400 metres from the coconut groves where coconut palms were sampled. Immature leaves from around the apical meristem were collected from palms exhibiting foliar discolouration and necrosis of the lower (oldest leaves), and from C. nucifera palms showing CILY symptoms () similar to those described for disease stages 2 and 3 (Arocha Rosete et al. Citation2017). Leaf samples were also collected from two symptomless palms of each species. For each palm species, total DNA was extracted (FAST DNA Spin Kit, MP Biomedicals, USA) from 200 mg of leaf tissue, and used as a template for PCR testing. DNA concentration was measured using a QuantiFluor® dsDNA System and the QuantiFluor®-ST Handheld Fluorometer (Promega, USA). Total DNA from a coconut palm infected with LYM, 16SrXXII-A strain (‘Ca. P. palmicola’) provided by Dr Bila from Mozambique, and DNA extracts from coconut palms from Ghana, provided by Dr Yankey, were used as positive controls.
Polymerase chain reaction (PCR)
Two phytoplasma genes were amplified from the palm tissue samples – the 16S rRNA and the secA. All the PCR products (5 µL) were separated in a 1.5% agarose gel and visualized with SYBR Safe DNA Gel Stain (Invitrogen, USA) in an Alpha Imager (Alpha Innotech, USA). Fifty nanograms of the total DNA extract was used as a template for all PCR reactions, in a final volume of 50 μL (DreamTaq Green PCR Master Mix, ThermoFisher Scientific, Canada) containing 0.4 μM of each primer. Amplicons obtained with universal primers R16mF1/R1 (Gundersen & Lee Citation1996) that amplify the phytoplasma 16S rRNA gene only were nested with primers R16F2n/R2 (Gundersen & Lee Citation1996) and U5/U3 (Lorenz et al. Citation1995). For the amplification of the secA gene, primers SecAfor1⁄SecArev3 were nested with primers SecAfor5⁄SecArev2 as described by Dickinson & Hodgetts (Citation2013). The direct PCR product was diluted 30-fold and used as a DNA template for the nested PCR. Samples yielding negative results to SecA amplification were tested with CoxF3 and CoxB3 primers that amplify the plant cytochrome oxidase (cox) to assess for PCR inhibitors (Dickinson & Hodgetts Citation2013). For the primer combination R16mF1/R1 and R16F2n/R2, the PCR protocol was 35 cycles of 1 min (2 min for the initial denaturation) at 94ºC, 2 min at 50ºC and 3 min (10 min for the final extension) at 72ºC; and for fU5/rU3, 35 cycles of 94°C for 30 s (2 min for the initial denaturation), 53°C for 1 min and 72°C for 1 min; and 1 cycle at 72°C for 10 min. For primers SecAfor1⁄SecArev3 and SecAfor5⁄SecArev2, PCR cycling was 30 cycles of 94°C for 30 s (2 min for the initial denaturation), 53°C for 1 min and 72°C for 1 min; and 1 cycle at 72°C for 10 min (Dickinson & Hodgetts Citation2013).
Sequencing, restriction fragment length polymorphism (RFLP) and phylogenetic analyses
R16F2n/R2, fU5/rU3 and SecA amplicons representing each palm species per village were purified (E.Z.N.A. Cycle Pure, Omega Bio-tek, USA), cloned according to the manufacturer’s instructions (p-GEMT Easy Vector Systems, Promega, USA), and sequenced bi-directionally using M13F/M13R primers (Centre for the Analysis of Genome Evolution and Function, University of Toronto). Sequences were compared with those available in GenBank (https://www.ncbi.nlm.nih.gov) by BLAST (Altschul et al. Citation1990). MEGA version 4.0 (Tamura et al. Citation2007) was used to align the sequences and construct the phylogenetic trees using the neighbour-joining method and the Tamura-Nei substitution model with default values and 1000 replicates for bootstrap analysis. Acholeplasma laidlawii was used as the outgroup to root the phylogenetic tree. One consensus each of R16F2n/R2 and SecA sequence were respectively selected corresponding to each palm species and deposited in GenBank.
The R16F2n/R2 sequences were analysed through iPhyClassifier (Zhao et al. Citation2009) for the delineation of group and subgroup with restriction endonucleases RsaI, HaeIII, AluI, TaqI, HinfI, BamHI, BfaI, BstUI, DraI, EcoRI, HhaI, HpaI, HpaII, KpnI, Sau3AI, MseI and SspI. All R16F2n/R2 amplicons were also subjected to RFLP with the restriction enzymes yielding unique RFLP profiles. For this, 10 µL (corresponding to ~200 ng) of each R16F2n/R2 PCR amplicon were digested with the selected restriction endonucleases (New England Biolabs, Canada), following manufacturer’s recommendations. RFLP profiles were visualized in a 3% agarose gel stained with SYBRR Safe DNA Gel Stain (Invitrogen, USA) in a gel documenter (Alpha Innotech, USA). The U5/U3 and SecA sequences were subjected to virtual restriction analysis (pDRAW32 software, Acaclone, http://www.acaclone.com) to identify signature restriction profiles.
Results
PCR amplification from palm species
A total of 111 palms were sampled from the three villages in Grand-Lahou, of which 106 samples were PCR positive for the primer pair U5/U3 (). For the primers R16F2n/R2, 42 palms were PCR positive, while for primers SecAfor5/rev2, 100 samples yielded amplicons (). No amplicons were obtained with SecAfor5/rev2 nested primers for two samples of E. guineensis, two of R. vinifera and nine of C. nucifera. These 13 samples showed amplicons of the expected length after being tested with the cox primers (data not shown), ruling out the possibility of PCR amplification inhibition.
Table 1. Results of PCR testing of palm species E. guineensis, B. aethiopium, R. vinifera and C. nucifera surveyed in Grand-Lahou for the presence of phytoplasma DNA.
In silico and actual RFLP analysis
Twenty-four R16F2n/R2 amplicons were selected for cloning and sequencing (). These included amplicons from E. guineensis (four from Doudougbazou, four from Yaokro and one from Braffedon); B. aethiopium (one from Doudougbazou, one from Yaokro and three from Braffedon); R. vinifera (one from Yaokro and four from Braffedon); and C. nucifera (three randomly selected amplicons from each village Doudougbazou, Yaokro and Braffedon), for a total of 24 out of 42 R16F2n/R2 amplicons. The iPhyclassifier analysis of the R16F2n/R2 sequences confirmed that the phytoplasma detected in E. guineensis, B. aethiopium, R. vinifera and C. nucifera was a member of group 16SrXXII. The comparison of the R16F2n/R2 sequences with those of phytoplasmas in subgroups 16SrXXII-B and 16SrXXII-A yielded similarity coefficient values lower than 0.97 ().
Table 2. Sequence similarities based on the 16S rRNA gene sequences of the 16SrXXII-C phytoplasma and phytoplasma strains in 16SrXXII and 16SrIV groups/subgroups.
The virtual restriction analyses of the 24 R16F2n/R2 sequences with iPhyclassifier showed that the Sau3AI profile of the phytoplasma detected in Ivoirian palms and C. nucifera was unique when compared with the Sau3AI pattern obtained from R16F2n/R2 sequences of the 16SrXXII-A and 16SrXXII-B phytoplasma representatives (). This was also confirmed through actual RFLP analysis (), where the Sau3AI RFLP patterns (c. 750, 280 and 120 bp) were all identical to each other for the 24 R16F2n/R2 sequences. The 18 remaining R16F2n/R2 amplicons were also subjected to actual RFLP with the Sau3AI restriction enzyme, which yielded the same Sau3AI RFLP profile previously obtained through virtual and actual RFLP from the 24 R16F2n/R2 amplicons sequenced. These results confirmed that the phytoplasma infecting E. guineensis, B. aethiopium, R. vinifera and C. nucifera is as a member of a new subgroup of group 16SrXXII, which was termed as 16SrXXII-C. Thirty U5/U3 amplicons and 30 SecA amplicons corresponding to E. guineensis (three from each of the villages Doudougbazou, Yaokro and Braffedon); B. aethiopium (one from Doudougbazou, two from Yaokro and three from Braffedon); R. vinifera (three from Yaokro and three from Braffedon); and C. nucifera (three from each of the villages Doudougbazou, Yaokro and Braffedon) were selected for cloning and sequencing. The U5/U3 and SecA sequences selected corresponded to the same samples for which the R16F2n/R2 amplicons were sequenced. Since the aim of the study was to have a total of three amplicons per palm species per village, new samples were selected to complete the total number in those cases where only one or two samples per palm species per village were available. The virtual restriction analysis for the U5/U3 sequences yielded no signature profile. However, all the SecA sequences yielded a unique virtual RFLP profile with the restriction enzyme HpyCH4III (a) that distinguished the new subgroup 16SrXXII-C from the subgroups 16SrXXII-B and 16SrXXII-A. The same HpyCH4III profile (about 380 and 80 bp) was obtained from the remaining 70 SecA amplicons when subjected to actual RFLP with the HpyCH4III () restriction enzyme.
Fig. 2 Virtual Sau3AI profile obtained from the iPhyClassifier analysis of the R16F2n/R2 sequences from palms and 16SrXXII representatives. Lanes MW: Molecular weight marker (NEB 100 bp DNA Ladder), Lane 1: R. vinifera (GenBank accession number, AC: KY711302), representing the same profile for B. aethiopium (AC: KY711392), E. guineensis (AC: KY711391) and C. nucifera (AC: KY767914), Lane 2: LYM phytoplasma (AC: KP938848, 16SrXXII-A); Lane 3: CILY phytoplasma (AC: KC999037, 16SrXXII-B).
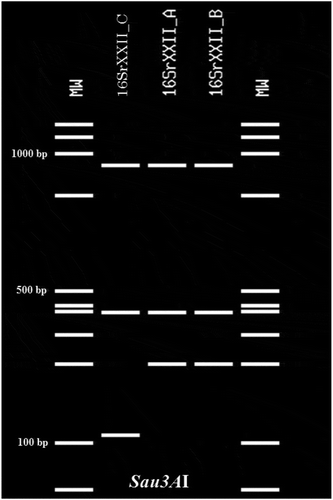
Fig. 3 Sau3AI profile obtained from the R16F2n/R2 amplicons of the phytoplasmas identified in E. guineensis, B. aethiopium, R. vinifera and C. nucifera palms, and positive controls of subgroups 16SrXXII-A (LYM), and 16SrXXII-B (CSPWD, CILY). Lane M: Molecular weight marker (Quick Load 100 bp DNA Ladder, New England BioLabs); Lane 1, R. vinifera (16SrXXII-C); Lane 2, B. aethiopium (16SrXXII-C); Lane 3, E. guineensis (16SrXXII-C); Lane 4, C. nucifera (16SrXXII-C); Lane 5, CILY phytoplasma (16SrXXII-B, Côte d’Ivoire); Lane 6, CSPWD phytoplasma (16SrXXII-B, Ghana); Lane 7, LYM phytoplasma (16SrXXII-A).
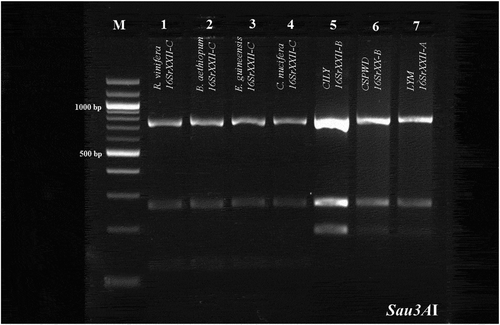
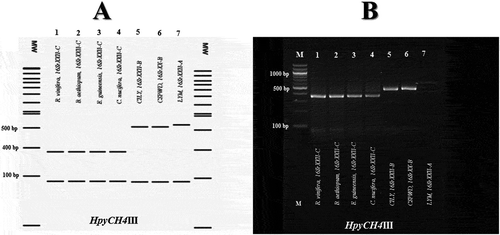
Fig. 4 Virtual (a) and actual (b) HpyCH4III profiles obtained from the SecA amplicons of the phytoplasmas identified in E. guineensis, B. aethiopium, R. vinifera and C. nucifera palms, and phytoplasma strains in subgroups 16SrXXII-A (LYM), and 16SrXXII-B (CSPWD, CILY). Lanes MW (A): NEB 100 bp DNA ladder. Lane M (B): Molecular weight marker (Quick Load 100 bp DNA Ladder, New England BioLabs); Lanes 1, R. vinifera (16SrXXII-C); Lanes 2, B. aethiopium (16SrXXII-C); Lanes 3, E. guineensis (16SrXXII-C); Lanes 4, C. nucifera (16SrXXII-C); Lanes 5, CILY phytoplasma (16SrXXII-B, Côte d’Ivoire); Lanes 6, CSPWD phytoplasma (16SrXXII-B, Ghana); Lanes 7, LYM phytoplasma (16SrXXII-A).
Sequence and phylogenetic analysis
SecA and R16F2n/R2 sequences obtained from E. guineensis, B. aethiopium, R. vinifera and C. nucifera in each of the three villages were 100% identical, respectively, to each other. BLAST comparison revealed that the phytoplasma found in E. guineensis, B. aethiopium, R. vinifera and C. nucifera exhibited over 99% of sequence identity of their R16F2n/R2 and SecA sequences with those of phytoplasmas classified in group 16SrXXII-B. The R16F2n/R2 sequence exhibited a highest score of 99.68% with that of the CILY phytoplasma (GenBank accession number KC999037). Sequence identities shared by phytoplasma subgroups within the group 16SrIV ranged from 95.6 to 99.4%, which coincide with the sequence identities values shown by the phytoplasma subgroups within the group 16SrXXII ranging from 95.6 to 99.2% ().
Out of the 24 R16F2n/R2 and 30 SecA sequences, one consensus sequence was respectively selected corresponding to each palm species and deposited in GenBank since no signature differences were observed indicating rRNA operon heterogeneity. The assigned GenBank accession numbers for R16F2n/R2 sequences are shown in , and those for SecA sequences were: E. guineensis (AC: KY769401), B. aethiopium (AC: KY769400), R. vinifera (AC: KY769399) and C. nucifera (AC: KY769402).
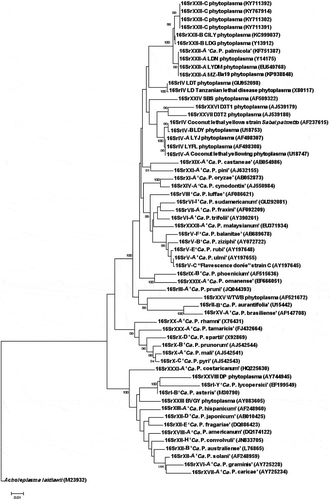
The R16F2n/R2 sequences of the phytoplasma detected in E. guineensis, B. aethiopium, R. vinifera and C. nucifera compared with those of phytoplasma members of subgroups 16SrXXII-A and 16SrXXII-B revealed four unique substitutions, A/G8, G/A13, G/A1174 and A/T1218 when compared with those of the phytoplasma members of groups 16SrIV and 16SrXXII, subgroups -A and -B. Phylogenetic trees based on both R16F2n/R2 () and SecA sequences (data not shown) supported both the in silico and actual RFLP results, as well as the sequence comparison results. The E. guineensis, B. aethiopium, R. vinifera and C. nucifera phytoplasmas grouped in the same cluster including phytoplasmas affecting coconut and palm species from Central America and the Caribbean, North America, and West and East Africa from groups 16SrIV and 16SrXXII. However, the phytoplasma found in coconut and palms in Gran-Lahou was separated as an individual cluster, well supported branch, confirming its designation as a new subgroup, 16SrXXII-C.
Fig. 5 Phylogenetic tree based on the R16F2n/R2 sequences of the 16SrXXII-C phytoplasmas and selected 16Sr phytoplasma groups. ‘Ca. P. sp.’: ‘Candidatus Phytoplasma sp.’; CILY: Côte d’Ivoire Lethal Yellowing; CSPWD, Cape St. Paul Wilt Disease; LDG: Lethal Decline (Ghana); LYDM: Lethal Yellowing Disease (Mozambique); LDN: Lethal Decline (Nigeria); LDT: Lethal Decline (Tanzania); SBS: Sorghum Bunchy Shoot; D3T1 and D3T2: phytoplasmas from sugarcane; C. palmata: Carludovica palmata; LDY: Lethal Decline (Yucatan); LYFL: Lethal Yellowing (Florida); LYJ: Lethal Yellowing (Jamaica); BVGY: Buckland Valley Grapevine Yellows; A. laidlawii, outgroup. GenBank accession numbers shown above branches. Bootstrap values shown are greater than 85%. Branch lengths are proportional to the number of inferred state transformations. Bar indicates 0.01 substitutions per nucleotide position.
Discussion
The RFLP typing of PCR-amplified 16S rRNA gene sequences coupled with the RFLP analysis and similarity coefficient is one of the best approaches to classify phytoplasmas into 16Sr groups and subgroups (Duduk & Bertaccini Citation2011). A threshold similarity coefficient of 0.97 was established for a new subgroup delineation, where as few as one restriction site difference can distinguish a new subgroup (Zhao et al. Citation2009; Zhao & Davis Citation2016). Thus, a similarity coefficient of 0.97 or less shown by the strains under study derived from their virtual R16F2n/R2 RFLP pattern when compared with those of 16S rRNA genes of all described representatives of the given 16SrXXII group, supports the assignment to a new subgroup based on RFLP differential pattern (Zhao et al. Citation2009; Zhao & Davis Citation2016).
The assignment of the phytoplasma from E. guineensis, B. aethiopium, R. vinifera and C. nucifera to a new subgroup, 16SrXXII-C, was also confirmed through the analysis grouped in a single cluster enclosing the phytoplasmas detected in E. guineensis, B. aethiopium, R. vinifera and C. nucifera, separate from the CILY and CSPWD phytoplasma strains (data not shown). Results were also supported by the fact that a signature HpyCH4III profile on the SecA sequences distinguished the 16SrXXII-C phytoplasma present in E. guineensis, B. aethiopium, R. vinifera and C. nucifera from phytoplasmas classified within the other two phytoplasma subgroups (16SrXXII-B and 16SrXXII-A) affecting coconut palms in Ghana and Mozambique, respectively. Other phytoplasmas have been successfully detected and characterized on the basis of the secA gene sequences (Hodgetts et al. Citation2008; Valiunas et al. Citation2015). The secA gene has been used to distinguish among phytoplasmas in group 16SrIV (Lethal Yellowing) in palms (Ntushelo et al. Citation2013), as well as for the CSPWD phytoplasma in coconut trees in Ghana (Yankey et al. Citation2014), and ‘Ca. P. palmicola’ (16SrXXII-A) and subgroup 16SrIV-C in coconut and other palms in Mozambique (Bila et al. Citation2015b). The secA gene is a tool to cross-confirm the phytoplasma identity, but also, as a single copy gene in the phytoplasma genome, its confirmation of differential sequence supports the presence of a new phytoplasma subgroup.
Over 35 palm species have been shown to harbour phytoplasmas of group 16SrIV in Mexico, Texas and Florida (Harrison et al. Citation2002, Citation2008; Vázquez-Euán et al. Citation2011; Ntushelo et al. Citation2013; Gurr et al. Citation2016). Six different subgroups 16SrIV-A, -B, -C, -D, -E and -F (Wei et al. Citation2007; Zhao et al. Citation2009), determined by RFLP approach based on less than 0.97 RFLP similarity coefficient values have been associated with LY-like diseases in palm species in different geographic locations. In North and Central America, and the Caribbean, LY affecting coconut and other palm species is associated with phytoplasma subgroup 16SrIV-A (Harrison et al. Citation2002, Citation2008; Gurr et al. Citation2016), while the subgroup 16SrIV-B is responsible for LD in coconut palms of Yucatan, Mexico (Harrison et al. Citation2002), and in coyol palms in Honduras (Roca et al. Citation2006). LD is also associated with the subgroup 16SrIV-C in Kenya and Tanzania (Sullivan & Harrison Citation2013; Córdova et al. 2014), subgroup 16SrIV-D in palms in USA, Mexico and Puerto Rico (Gurr et al. Citation2016), subgroup 16SrIV-E in coconut in Dominican Republic (Martinez et al. Citation2008), and 16SrIV-F in Washingtonia robusta in Florida (Harrison et al. Citation2008), and P. dactylifera (Ntushelo et al. Citation2013). Phytoplasmas associated with LY diseases in palms have also been found in other plant species outside the Arecaceae family such as Emelia forsbegii, Synedrella nodiflora and Stachytarpheta jamaicensis in Jamaica (Brown et al. Citation2008; Brown & McLaughlin Citation2011), and Cynodon dactylon in Malaysia (Nejat & Vadamalai Citation2010).
Other phytoplasma groups have been found in palm hosts in South America, Asia and Africa. In Colombia, a 16SrI phytoplasma was associated with oil palm lethal wilt (Alvarez et al. Citation2014), whereas in India, 16SrXI ‘Ca. P. oryzae’ was associated with the yellow leaf disease of areca palm, Areca catechu L. (Ramaswamy et al. Citation2013), and in Malaysia, a LY-type phytoplasma has been identified from E. guineensis (Nejat & Vadamalai Citation2010). More recently, Bila et al. (Citation2015a) identified B. aethiopum and E. guineensis as potential alternate hosts of the 16SrXXII-A phytoplasma associated with LY in Mozambique.
The group 16SrXXII has also been found in palm and non-palm species. The subgroup -A embraces the ‘Awka Wilt’ phytoplasma strain from Nigeria, which infects coconut palms, and the novel ‘Candidatus Phytoplasma’ species, ‘Ca. P. palmicola’ from Mozambique (Harrison et al. Citation2014). The subgroup -A has already been reported from other palm species than coconut such as B. aethiopum and E. guinneensis in Mozambique (Bila et al. Citation2015a). The subgroup -B is present in coconut palms in Ghana; however, its Ivoirian strain, CILY phytoplasma, has been reported in coconut palms (Konan Konan et al. Citation2013), and in a number of non-coconut plant hosts, which belong to five different botanical families in Côte d’Ivoire (Arocha Rosete et al. Citation2016). These results suggest that the CILY phytoplasma, which devastated the coconut groves in Grand-Lahou may require a secondary host after the coconut primary host succumbs to the disease, an hypothesis that has also been stated for the 16SrXXII-A phytoplasma associated with LY in Mozambique (Bila et al. Citation2015a).
The 16SrXXII-C phytoplasma was associated with two different groups of symptoms exhibited by different palm species. It was detected from E. guineensis, B. aethiopium and R. vinifera palms exhibiting foliar discolouration of the lowermost (oldest leaves), and from C. nucifera palms showing CILY-like symptoms. In all cases, the E. guineensis, B. aethiopium and R. vinifera palm species surveyed were located nearby the coconut palms within a perimeter less than 500 m. The findings of the present work are in agreement with those from Bila et al. (Citation2015b), who identified the subgroup 16SrXXII-A in two different palm species exhibiting specific symptoms; in this case, B. aethiopum palms with symptoms of a skirt-shaped brown discolouration (necrosis) of the older leaves, and E. guineensis palms with brown discolouration of the mature and spear leaves. Further studies are needed to unveil the epidemiological factors that may correlate the presence of a particular subgroup with the occurrence of distinct symptoms in different palm species in a particular geolocation.
Two phytoplasma subgroups are currently present in Grand-Lahou, the 16SrXXII-B associated with CILY in coconut palms (Arocha Rosete et al. Citation2017), and the new subgroup 16SrXXII-C (this study) associated with foliar discolouration in E. guineensis, B. aethiopium and R. vinifera palms, and also detected in coconut palms with CILY-like symptoms. It is not clear how much the 16rSXXII-B or the 16SrXXII-C phytoplasmas may contribute to disease development. Nevertheless, the presence of the subgroup 16SrXXII-C in the same area where coconut palms are severely infected by the subgroup 16SrXXII-B phytoplasma may imply different epidemiological constraints that may trigger the severity of the disease. Thus, both 16rSXXII-B and 16SrXXII-C phytoplasmas pose new threats for other Arecaceae species, which may impact on the further disease management and control in Grand-Lahou.
References
- Alladatin J. 2011. The raffia exploitation in the hlanzoun swampy forest: from a contribution to the socio-economic development and natural resources damages. http://etudescaribeennes.revues.org/5556
- Altschul S, Gish W, Miller W, Meyers E, Lipman D. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Alvarez E, Mejía JF, Contaldo N, Paltrinieri S, Duduk B, Bertaccini A. 2014. ‘Candidatus Phytoplasma asteris’ strains associated with oil palm lethal wilt in Colombia. Plant Dis. 98:311–318.
- Arocha Rosete Y, Diallo HA, Konan Konan JL, Aep K, Séka K, Kra KD, Toualy MN, Kwadjo KE, Daramcoum WAMP, Beugré NI, et al. 2016. Detection and identification of the coconut lethal yellowing phytoplasma in weeds growing in coconut farms in Côte d’Ivoire. Can J Plant Pathol. 38:164–173.
- Arocha Rosete Y, Diallo HA, Konan Konan JL, Yankey N, Saleh M, Pilet F, Contaldo N, Paltrinieri S, Bertaccini A, Scott J. 2017. Detection and differentiation of the coconut lethal yellowing phytoplasma in coconut-growing villages of Grand-Lahou, Côte d’Ivoire. Ann Appl Biol. doi:10.1111/aab.12333
- Béhi Y, Mollet M, Girardin O, Sorg JP, Herzog F. 2002. Le vin de palme, aliment et source de revenu pour les populations rurales en Côte d’Ivoire. Schweiz. Z. Forstwes. 153:123–129.
- Bila J, Högberg N, Mondjana A, Samils B. 2015a. African fan palm (Borassus aethiopum) and oil palm (Elaeis guineensis) are alternate hosts of coconut lethal yellowing phytoplasma in Mozambique. Afr J Biotech. 14:3359–3365.
- Bila J, Mondjana A, Samils B, Högberg N. 2015b. High diversity, expanding populations and purifying selection in phytoplasmas causing coconut lethal yellowing in Mozambique. Plant Pathol. 64:597–604.
- Brown SE, Been BO, McLaughlin WA. 2008. First report of the presence of the lethal yellowing group (16Sr IV) of phytoplasmas in the weeds Emilia fosbergii and Synedrella nodiflora in Jamaica. Plant Pathol. 57: 770.
- Brown SE, McLaughlin WA. 2011. Identification of lethal yellowing group (16SrIV) of phytoplasmas in the weeds Stachytarpheta jamaicensis, macroptilium lathyroides and cleome rutidosperma in Jamaica. Phytopathogenic Mollicutes. 1:27–34.
- Danyo G. 2011. Review of scientific research into the Cape Saint Paul wilt disease (CSPWD) of coconut in Ghana. Afr J Agr Res. 6:4567–4578.
- Dickinson M, Hodgetts J. 2013. PCR analysis of phytoplasmas based on the secA gene. In: Dickinson M, Hodgetts J, editors. Phytoplasmas: Methods and protocols. New York Heidelberg Dordrecht London, UK: Humana Press, Springer; p. 205–217.
- Duduk B, Bertaccini A. 2011. Phytoplasma classification: taxonomy based on 16S ribosomal gene, is it enough? Phytopathogenic Mollicutes. 1:3–13.
- Gundersen DE, Lee IM. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Medit. 35:144–151.
- Gurr GM, Johnson AC, Ash GJ, Wilson BAL, Ero MM, Pilotti CA, Dewhurst CF, You MS. 2016. Coconut lethal yellowing diseases: a phytoplasma threat to palms of global economic and social significance. Front Plant Sci. 7:1521.
- Harrison N, Davis RE, Oropeza C, Helmick E, Narvaez M, Eden-Green S, Dollet M, Dickinson M. 2014. ‘Candidatus Phytoplasma palmicola’, a novel taxon associated with a lethal yellowing-type disease (LYD) of coconut (Cocos nucifera L.) in Mozambique. Int J Syst Evol Microbiol. 64:1890–1899.
- Harrison NA, Myrie W, Jones P, Carpio ML, Castillo M, Doyle MM, Oropeza P. 2002. 16S rRNA interoperon sequence heterogeneity distinguishes strain populations of palm lethal yellowing phytoplasma in the Caribbean region. Ann Appl Biol. 141:183–193.
- Harrison NN, Helmick EE, Elliot M. 2008. Lethal yellowing-type diseases of palms associated with phytoplamas newly identified in Florida, USA. Ann Appl Biol. 153:85–94.
- Hodgetts J, Boonham N, Mumford R, Harrison N, Dickinson M. 2008. Phytoplasma phylogenetics based on analysis of secA and 23S rRNA gene sequences for improved resolution of candidate species of ‘Candidatus Phytoplasma’. Int J Syst Evol Microbiol. 58:1826–1837.
- Konan Konan JL, Diallo AH, Benabid R, Michelutti R, Arocha-Rosete Y. 2013. First report on the molecular identification of a phytoplasma associated with a lethal yellowing disease in Côte d’Ivoire. New Dis Rep. 28:3.
- Lee I-M, Davis RE, Gundersen-Rindal DE. 2000. Phytoplasma: Phytopathogenic mollicutes. Ann Rev Microbiol. 54:221–255.
- Lorenz KH, Schneider B, Arhens U, Seemüller E. 1995. Detection of the apple proliferationand pear decline phytoplasma by PCR amplification of ribosomal and nonribosomal DNA. Phytopathology. 85:771–776.
- Martinez RT, Narvaez M, Fabre S, Harrison NA, Oropeza C, Dollet M, Hichez E. 2008. Coconut lethal yellowing on the southern coast of the Dominican Republic is associated with a new 16SrIV group phytoplasma. Plant Pathol. 57:366–376.
- Nejat N, Sijam K, Abdullah SNA, Vadamalai G, Dickinson M. 2009. Phytoplasmas associated with disease of coconut in Malaysia: phylogenetic groups and host plant species. Plant Pathol. 58:1152–1160.
- Nejat N, Vadamalai G. 2010. Phytoplasma detection in coconut palm and other tropical crops. Plant Pathol J. 9:112–121.
- Nejat N, Vadamalai G, Davis RE, Harrison NA, Sijam K, Dickinson M. 2013. ‘Candidatus Phytoplasma malaysianum’, a novel taxon associated with virescence and phyllody of Madagascar periwinkle (Catharanthus roseus). Int J Syst Evol Microbiol. 63:540–548.
- Ntushelo K, Elliott MM, Harrison N. 2013. Palm yellows phytoplasmas and their genetic classification. Afr J Biotech. 12:3376–3382.
- Ramaswamy R, Nair S, Soumya VP, Thomas GV. 2013. Phylogenetic analysis identifies a ‘Candidatus Phytoplasma oryzae’-related strain associated with yellow leaf disease of areca palm (Areca catechu L.) in India. Int J Syst Evol Microbiol. 63:1376–1382.
- Roca MM, Castillo MG, Harrison NA, Oropeza C. 2006. First report of a 16SrIV group phytoplasma associated with declining coyol palms in Honduras. Plant Dis. 90: 526.
- Sangaré A, Koffi EM, Akamou FM, Fall C. 2009. Alassane État des ressources phytogénétiques pour l’alimentation et l’agriculture: Second rapport national. Ministère de l’agriculture. 22.
- Sullivan M, Harrison N. 2013. CPHST Pest Datasheet for ‘Candidatus Phytoplasma palmae’ and related strains. [Internet]. [revised 2013 June; cited 2013 Nov 14]. USDA-APHIS-PPQ-CPHST. Available from http://caps.ceris.purdue.edu.
- Tamura K, Dudley J, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4. Mol Biol Evol. 24:1596–1599.
- Valiunas D, Jomantiene R, Ivanauskas A, Urbonaite I, Sneideris D, Davis RE. 2015. Molecular identification of phytoplasmas infecting diseased pine trees in the UNESCO-protected Curonian spit of Lithuania. Forests. 6:2469–2483.
- Vázquez-Euán R, Harrison N, Narvaez M, Oropeza C. 2011. Occurrence of a 16SrIV group phytoplasma not previously associated with palm species in Yucatan, Mexico. Plant Dis. 95:256–262.
- Wei W, Davis RE, Lee IM, Zhao Y. 2007. Computer-simulated RFLP analysis of 16S rRNA genes: Identification of ten new phytoplasma groups. Int J Syst Evol Microbiol. 57: 1855–1867.
- Yankey EN, Swarbrick PJ, Nipah JO, Quaicoe RN, Dickinson MJ. 2014. Detection of the cape St. Paul Wilt phytoplasma in coconut palms in Ghana through the combination of visual symptoms assessment and molecular diagnosis using secA gene based assay. J Plant Pathol. 96:281–285.
- Zhao Y, Davis RE. 2016. Criteria for phytoplasma 16Sr group/subgroup delineation and the need of a platform for proper registration of new groups and subgroups. Int J Syst Evol Microbiol. 66:2121–2123.
- Zhao Y, Wei W, Lee IM, Shao J, Suo X, Davis RE. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X disease phytoplasma group (16SrIII). Int J Syst Evol Microbiol. 59:2582–2593.

