Abstract
Cotton leaf curl Multan virus (CLCuMuV), in conjunction with cotton leaf curl Multan betasatellite (CLCuMuB), causes cotton leaf curl disease in South Asia. CLCuMuV-CLCuMuB was first found in 2006 to be associated with a leaf curl disease of Hibiscus rosa-sinensis in China. Recently, CLCuMuV-CLCuMuB has become prevalent in H. rosa-sinensis in the southern part of the country. Here, we identified CLCuMuV-CLCuMuB in H. sabdariffa for the first time. To our knowledge, H. sabdariffa is the fourth seed-propagated plant in China to be affected by CLCuMuV-CLCuMuB.
Résumé
Le virus de la frisolée du cotonnier de la région de Multan (CLCuMuV), en conjonction avec le betasatellite de la frisolée de la feuille du cotonnier (CLCuMuB), cause la frisolée de la feuille du cotonnier dans le sud de l’Asie. CLCuMuV-CLCuMuB a été détecté pour la première fois, en 2006, alors qu’il était associé à une frisolée d’Hibiscus rosa-sinensis en Chine. Récemment, CLCuMuV-CLCuMuB s’est répandu chez Hibiscus rosa-sinensis dans le sud du pays. Ici, nous avons identifié CLCuMuV-CLCuMuB pour la première fois chez H. sabdariffa. À notre connaissance, il s’agit de la quatrième espèce de plante propagée par semence en Chine qui est touchée par CLCuMuV-CLCuMuB.
Introduction
Cotton leaf curl disease has been reported in Africa, as well as in several countries in Asia, such as Pakistan, India and China (Sattar et al., Citation2013). In Asia, the disease is caused by a number of distinct begomoviruses, including Cotton leaf curl Multan virus (CLCuMuV), in conjunction with a disease-specific betasatellite, Cotton leaf curl Multan betasatellite (CLCuMB) (Sattar et al., Citation2013). CLCuMuV-CLCuMuB were first identified to be associated with a leaf curl disease of Hibiscus rosa-sinensis in Guangzhou, Guangdong province of China in 2006 (Mao et al., Citation2008; Du et al., Citation2015). In recent years, this virus complex was found to be prevalent in H. rosa-sinensis in many provinces of south China, including Guangdong, Guangxi, Hainan, Yunnan, Fujian and Jiangsu (Du et al., Citation2015; authors, personal observations). Besides H. rosa-sinensis, the virus complex was also detected in okra (H. esculentus) (Dong et al., Citation2012), cotton (Gossypium hirsutum) (Cai et al., Citation2010) and kenaf (H. cannabinus) (Tang et al., Citation2015) in Guangdong, Guangxi and Hainan provinces, respectively. Here, we report the first detection of CLCuMuV-CLCuMuB in H. sabdariffa in Fujian province of China.
Materials and methods
CLCuMuV-CLCuMuB detection and sequencing
Samples of leaves from symptomatic and healthy-appearing H. sabdariffa shrubs were obtained in mid-September 2015, where a stunting disease was found in a garden of Fujian Agriculture and Forestry University, Fuzhou, Fujian province, China. About 70% of the plants were affected. Upon close observation, additional symptoms, including vein swelling, leaf curling and yellowing, were observed. Conspicuous enations on the veins were observed on the undersides of some leaves as shown in . These symptoms suggested that the plants might have been infected by a begomovirus. Three samples each from healthy and symptomatic shrubs were collected. DNA was extracted using the CTAB procedure (Biswas et al., Citation2013). The degenerate primer pair AV494/Dep3 (5ʹ-GANGSATGHGTRCADGCCATATA-3ʹ/5ʹ-GCCYATRTAYAGRAAGCCMAG-3ʹ), which amplifies a conserved 570-bp region on the begomovirus, was used to confirm the presence of a begomovirus in the symptomatic samples (He et al., Citation2007). The PCR amplicon was electrophoresed at 5 V cm−1 through 1% (w/v) agarose gels in 1× Tris Acetic acid EDTA (TAE) electrophoresis buffer. The PCR band of the expected size were recovered, purified, ligated to a T vector (pMD18-T, TAKARA, Dalian, China) and transformed to E. coli DH5α. After growing overnight on Luria-Bertani (LB) agar plates supplemented with 50 µg mL−1 of ampicillin at 37°C, three positive colonies were randomly selected and sequenced for each transformation. A primer pair specific to CLCuMuV (CLCuMuVF and CLCuMuVR 5ʹCAACAGGCATGGACAAACAG3ʹ and 5ʹ-CCAATACGATGGGTCAAACC-3ʹ) was used to confirm the presence of CLCuMuV in symptomatic H. sabdariffa. To sequence CLCuMuV and CLCuMuB infecting H. sabdariffa, rolling circle amplification (RCA) was conducted to enrich circular DNAs. The circular DNAs were amplified using β01/02 (Briddon et al., Citation2002) or a back-to-back primer pair designed based on the 570-bp sequences obtained above. The fragments obtained from the PCR were cloned by using a T-vector (pMD18-T, TAKARA, Dalian, China). Three colonies each for CLCuMuV and CLCuMuB were submitted to Beijing Genomics Institute (BGI, Shenzhen, China) for Sanger sequencing and the sequences were assembled using DNASTAR Lasergene 10 (DNASTAR Inc.). For both CLCuMuV and CLCuMuB, a consensus sequence each was deposited in GenBank. The accession numbers KY992859 and KY992858 have been assigned to CLCuMuV and CLCuMuB, respectively.
Phylogenetic analysis
To understand the origin of the CLCuMuV-CLCuMuB infecting H. sabdariffa, related viral sequences were retrieved from the NCBI non-redundant nucleotide database. Phylogenetic trees were constructed using the aligned viral sequences. The trees were inferred using the Neighbour-joining method. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analysed (Felsenstein, Citation1985). Evolutionary analyses were conducted in MEGA7 (Kumar et al., Citation2016).
Results and discussion
A degenerate PCR was conducted to amplify a conserved region on the genome of the suspected begomovirus infecting H. sabdariffa. PCR amplicons of the expected size (about 570 bp) were obtained from symptomatic but not from asymptomatic plant samples. A BlastN search of the NCBI non-redundant nucleotide database revealed more than 99% identity of the PCR amplicons to a corresponding fragment of CLCuMuV occurring in China (JX861210) (Du et al., Citation2015). PCR using a primer pair designed according to the obtained sequence was used to confirm the presence of CLCuMuV from 10 other symptomatic samples. PCR fragments of the expected size (197 bp) were obtained from all 10 samples (data not shown). In contrast, no PCR amplicon was obtained from asymptomatic plant samples. These results indicated that the H. sabdariffa showing begomovirus-like symptoms might have been caused by CLCuMuV.
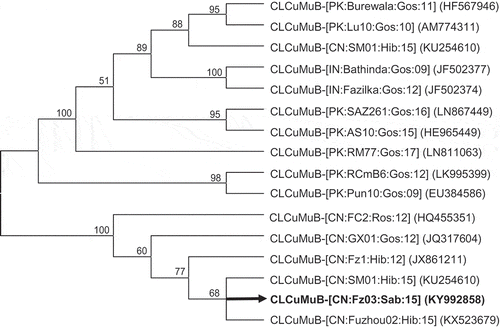
Full-length CLCuMuV and CLCuMuB infecting H. sabdariffa were obtained by PCR as described in the Materials and methods. Sequence analysis revealed that the CLCuMuV and CLCuMuB isolates were more than 99% identical to their counterparts occurring in China, but shared a nucleotide sequence identity of less than 94% with those found in South Asia. In phylogenetic trees constructed with the aligned genomic sequences of CLCuMuV () and CLCuMuB (), CLCuMuV occurring in China, including the new isolate identified here, formed a monophyletic clade together with a few of their counterparts occurring in South Asia. However, Chinese CLCuMuV formed a distinct branch within this clade. Similarly, CLCuMuB occurring in China, including the new isolate identified here, clustered and formed a monophyletic clade relative to their counterparts from South Asia.
Fig. 2 Phylogenetic relationships of the newly identified Cotton leaf curl Multan virus (CLCuMuV) affecting H. sabdariffa (black and thickened line) with other CLCuMuV. The numbers near the branches indicate the percentage of bootstrap replicates supporting the branch. Nodes with bootstrap supports (BP) < 50% are collapsed. Virus isolates are presented according to the latest guidelines (talk.ictvonline.org/ictv_wikis/m/files_gemini/5120.aspx) and their accession numbers in NCBI are provided in brackets.
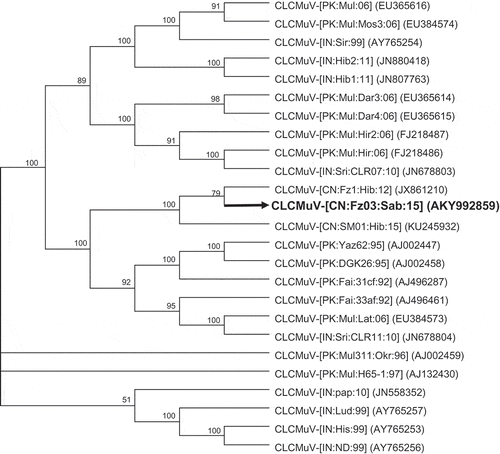
Fig. 3 Relationship of the Fujian isolate of Cotton leaf curl Multan betasatellite (CLCuMuB, black and thickened line) with other CLCuMuBs. The numbers near the branches indicate the percentage of bootstrap replicates supporting the branch. Nodes with bootstrap supports (BP) < 50% are collapsed. CLCuMuB isolates are presented according to the latest guidelines (talk.ictvonline.org/ictv_wikis/m/files_gemini/5120.aspx) and their accession numbers in NCBI are provided in brackets.
In summary, we identified a new host of CLCuMuV. To our knowledge, H. sabdariffa is the sixth malvaceous plant species and the fourth seed-propagated malvaceous plant species to be infected by CLCuMuV in China. Moreover, H. sabdariffa is the first seed-propagated plant found to be infected by CLCuMuV in Fujian province. The increasing number of malvaceous plants, particularly the seed-propagated malvaceous plants found to be infected by CLCuMuV in China, underscores the potential threat of this virus to malvaceous plants in south Asia as well as outside of the region.
Additional information
Funding
References
- Biswas C, Dey P, Satpathy S, Sarkar SK, Bera A, Mahapatra BS. 2013. A simple method of DNA isolation from jute (Corchorus olitorius) seed suitable for PCR-based detection of the pathogen Macrophomina phaseolina (Tassi) Goid. Lett Appl Microbiol. 56:105–110.
- Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. 2002. Universal primers for the PCR-mediated amplification of DNA β. Mol Biotechnol. 20:315–318.
- Cai JH, Xie K, Lin L, Qin BX, Chen BS, Meng JR, Liu YL. 2010. Cotton leaf curl Multan virus newly reported to be associated with cotton leaf curl disease in China. Plant Pathol. 59:794–795.
- Dong D, Zhu YH, He ZF, Chai ZX, She XM, Luo FF. 2012. Molecular characterization of Cotton leaf curl Multan virus and the associated satellite DNA infecting okra in Guangdong. J South China Agric Univ. 33:33–39.
- Du ZG, Tang Y, He ZF, She X. 2015. High genetic homogeneity points to a single introduction event responsible for invasion of Cotton leaf curl Multan virus and its associated betasatellite into China. Virol J. 12:163.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
- He ZF, Yu H, Mao MJ, Luo FF, Lin YH, Wang ST. 2007. Tomato yellow leaf curl disease in Guangdong is caused by Tomato leaf curl Taiwan virus. Chin J Agric Biotech. 4:127–131.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Mao MJ, He ZF, Yu H, Li HP. 2008. Molecular characterization of Cotton leaf curl Multan virus and its satellite DNA that infects Hibiscus rosa-sinensis. Chin J Virol. 24:64–68.
- Sattar MN, Kvarnheden A, Saeed M, Briddon RW. 2013. Cotton leaf curl disease – an emerging threat to cotton production worldwide. J Gen Virol. 94:695–710.
- Tang YF, He ZF, Du ZG, She XM, Lan GB. 2015. Detection and identification of the pathogen causing kenaf (Hibiscus cannabinus) leaf curl disease in Hainan Province of China. Acta Phytopathol Sin. 45:561568.

