Abstract
Lisianthus (Eustoma grandiflorum) is an important ornamental plant ranking in the top 10 cut flowers worldwide. During 2016–2017, root and stem rot was a major problem that limited the production of lisianthus in Fujian Province, China. Disease symptoms include yellowing of the leaves and root and stem rot leading to wilting. The aim of this study was to isolate and identify the causal pathogen. Based on symptoms on host plants, morphological characteristics, sequence analysis of the internal transcribed spacer rRNA region (ITS) and the gene encoding translation elongation factor-1α (TEF), and pathogenicity tests, the pathogen was identified as Fusarium solani (Mart.) Sacc. Isolates recovered from inoculated, diseased lisianthus plants had a similar morphology to the fungal isolates used to inoculate the plants, which completed Koch’s postulates. To our knowledge, this is the first report of F. solani causing root and stem rot on lisianthus in China.
Résumé
Le lisianthus (Eustoma grandiflorum) est une plante ornementale de grande importance, puisqu’il se classe parmi les 10 fleurs coupées les plus populaires au monde. En 2016–2017, la pourriture des racines et de la tige s’est révélée hautement problématique, limitant la production de lisianthus dans la province du Fujian, en Chine. Les symptômes de la maladie incluent le jaunissement des feuilles ainsi que la pourriture des racines et de la tige engendrant le flétrissement. Le but de cette étude était d’isoler et d’identifier l’agent pathogène causal. En se basant sur les symptômes des plantes hôtes, sur les caractéristiques morphologiques, sur l’analyse de la région de l’espaceur transcrit interne de l’ARNr (ITS) et du gène codant le facteur d’élongation de la traduction-1 α (TEF) ainsi que sur des tests de pathogénicité, l’agent pathogène a été identifié en tant que Fusarium solani (Mart.) Sacc. La morphologie des isolats récupérés des plants de lisianthus inoculés et malades était semblable à celle des isolats fongiques utilisés pour inoculer les plants, ce qui vérifiait les postulats de Koch. À notre connaissance, il s’agit du premier rapport de la pourriture de la tige et des racines causée par F. solani chez le lisianthus en Chine.
Introduction
Lisianthus (Eustoma grandiflorum (Raf.) Shinn.) is an important ornamental plant which is ranked in the top 10 cut flowers worldwide because of its elegant rose-like flowers of bright and pastel colours, and long post-harvest life (Bertoldo et al., Citation2015). Lisianthus is also widely used as a flowering potted and bedding plant (Harbaugh, Citation2007). Currently, greenhouse production of lisianthus exceeds 260 ha in China (Yang et al., Citation2016). Lisianthus can be affected by several pathogens, including Fusarium oxysporum (Yuan et al., Citation2010), F. avenaceum (Nalim et al., Citation2009), F. solani (Wolcan et al., Citation2001), Peronospora chlorae (Yang et al., Citation2015), Meloidogyne arenaria (Neves et al., Citation2016), Pothos latent virus (Chen et al., Citation2016) and Tomato yellow leaf curl virus (Kil, Citation2014).
During 2016 and 2017, root and stem rot became a principal disease limiting lisianthus production in Sanming City in Fujian Province, China. The lower leaves of naturally infected plants in the greenhouse turned yellow to brown in colour, the roots and stem bases rotted, and wilting eventually occurred (). The aim of this study was to isolate and identify the causal pathogen associated with root and stem rot of lisianthus in Sanming, China.
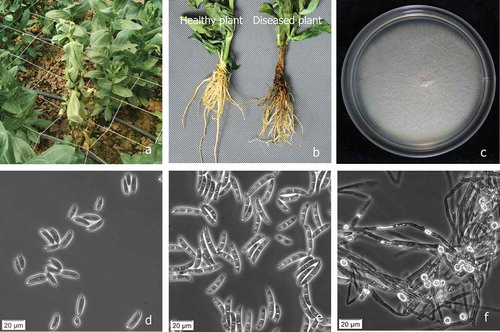
Fig. 1 (Colour online) Disease symptoms on lisianthus plant and morphological features of Fusarium solani. a, Fusarium wilt of lisianthus plants infected by F. solani in the field in Fujian Province, China. b, Healthy plant (left), diseased plant with brown and rotting infected roots and stem (right). c, Colony of F. solani grown on potato dextrose agar at 25°C for 7 days. d, Microconidia. e, Macroconidia. f, Chlamydospores. Scale bars: d, e, f = 20 μm.
Materials and methods
Isolation and characterization of the pathogen
Ten diseased lisianthus plants were collected from March to August 2017 in different greenhouses in Qingliu County, Sanming City, Fujian Province, China (116.82E, 26.18N). Eight pieces of diseased root and stem tissue were used for isolation from each plant. Infected tissue samples (5 × 3 × 1 mm) were surface-sterilized by immersion in a 10% bleach solution (containing 12% sodium hypochlorite and 0.03% sodium hydroxide) for 8 min and rinsed 2–3 times with sterile distilled water. All sterilized samples were transferred onto potato dextrose agar (PDA) amended with 100 mg L−1 streptomycin sulphate in Petri dishes and incubated at 25°C with a 12 h light-dark cycle for 5 days. The resulting fungal cultures were purified by using a single-spore technique onto fresh 2% water agar followed by subculturing on new PDA dishes. The single spore isolates were transferred to Spezieller Nährstoffarmer Agar (SNA) plates for 7–10 days at 25°C with a 12 h light-dark cycle (Leslie & Summerell, Citation2006). Cultures on PDA were used for the observation of colony characteristics, and cultures on SNA plates for the observation of conidial morphology. The shape, length and width of 50 conidia and chlamydospores of three representative isolates were observed using a phase contrast microscope at 400× magnification. Three representative isolates – FJAT-31 352, FJAT-31 353 and FJAT-31 354 – were used for pathogenicity testing and molecular identification.
Pathogenicity testing
Pathogenicity of isolates was tested on 2-month-old potted lisianthus plants cultivar ‘F1 Arena’ obtained from a commercial nursery. Spores of each isolate were collected from 14-day-old PDA cultures maintained at 28°C and suspended in sterilized water at 2 × 106 conidia mL−1 as determined using a hemocytometer. The resulting suspension was filtered through four layers of sterile cheesecloth to remove mycelial fragments. Thirty lisianthus seedlings were inoculated with each fungal isolate by dipping the roots into a conidial suspension for 2 h, and another 30 lisianthus seedlings were treated with sterile water as controls. All treated plants were potted in autoclaved commercial cultivation substrates (Jiangping Biology Substrate Technology Corporation Ltd, Xiamen, China) and maintained at 25/20°C and 70/40% relative humidity in a growth chamber with a 16 h day–8 h night cycle. Plants were observed every two days after inoculation for symptom appearance. Isolations were made from any resulting lesions on the inoculated plants, and the morphological and cultural characteristics of any isolated fungi were compared with the original fungal isolates used as inoculum. The re-isolated fungi were further identified using molecular methods. The pathogenicity tests for the three isolates were repeated three times.
Molecular identification
Total DNA of each of the three isolates was extracted from fresh aerial mycelia grown on PDA plates using a Genomic DNA Kit (Tiangen, Beijing, China), following the manufacturer’s instructions. The complete internal transcribed spacer region of ribosomal DNA (rDNA-ITS) and the translation elongation factor 1α gene (TEF) were amplified using primers ITS4/ITS5 (White et al., Citation1990) and EF-1/EF-2 (Geiser et al., Citation2004), respectively. DNA amplification was performed in a reaction mixture containing 25 μL 2× Taq PCR MasterMix (containing 20 mM Tris-HCl, 100 mM KCl, 3 mM MgCl2, 50 μM dNTP each, 0.1U Polynerase μL−1 and ddH2O; Tiangen, Beijing, China), 0.5 μM of each primer, 10 ng DNA template, and autoclaved ddH2O to bring the reaction volume to 50 μL. PCR amplification was performed in a C1000 thermal cycler system (Bio-Rad, USA). The following cycle parameters were used for ITS amplification: an initial denaturation step at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 60 s, annealing for 60 s at 55°C, and elongation at 72°C for 60 s, with a final elongation step at 72°C for 10 min. The following cycle parameters were used for TEF amplification: an initial denaturation step at 94°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing for 60 s at 53°C, and elongation at 72°C for 60 s, with a final elongation step at 72°C for 5 min. Amplification products were examined by electrophoresis in 1.2% agarose gels and visualized with a UV transilluminator. PCR products were purified with a Gel Extraction Kit (Tiangen, Beijing, China) and cloned into the pGEM-T Easy vector (Promega, Madison, WI) according to the manufacturer’s instructions. PCR products were sequenced by a commercial sequencing service provider (Lifetech, Shanghai, China). Sequences of ITS and TEF were compared with those in the GenBank database and Fusarium MLST database (http://www.westerdijkinstitute.nl/fusarium) (O’Donnell et al., Citation2010), respectively. Phylogenetic analysis of ITS and TEF sequence data was conducted by neighbour-joining methods using MEGA version 6.0 (Tamura et al., Citation2013), and sequence distance was calculated based on the Maximum Composite Likelihood model. Confidence intervals were estimated using bootstrap analysis with 1000 replications.
Results
Isolation and characterization of the pathogen
A total of 67 fungal cultures of similar morphology were consistently isolated from basal stems and roots of 10 diseased lisianthus plants, and 52 pure cultures derived from single spores were obtained. Colonies of the fungal isolates grown on PDA at 25 ± 2°C for 7 days appeared as white, cottony mycelia (). Microconidia were generally single celled, oval to kidney-shaped, 6.7–15.8 μm × 2.2–4.5 μm, with zero to one septa (). Macroconidia were abundant, cylindrical, slightly curved, with blunt and rounded apical cells and foot-shaped or notched basal cells, 25.5–38.5 μm × 3.0–4.8 μm, with three to five septa (). Chlamydospores were terminal or intercalary, solitary, in pairs or in chains, and 8.6–12.5 μm (). Based on the morphological and culture characteristics, the fungus was identified as Fusarium solani (Mart.) Sacc. as described by Booth (Citation1971).
Pathogenicity testing
Disease symptoms began with leaf yellowing and stunting by 25 days after inoculation. Root rot and ultimately wilting occurred by 32 days after inoculation. The incidence of diseased plants was 100% for FJAT-31 352 and FJAT-31 354. Symptoms on the inoculated plants were similar to those on diseased plants in the field. Fungi isolated from diseased root and stem tissues of inoculated symptomatic plants were confirmed as F. solani by morphological characteristics and molecular identification. Seedlings treated with isolate FJAT-31 353 and control seedlings treated with sterile water did not develop symptoms, and no fungi were isolated from them (). The same results were obtained in all three repetitions of the pathogenicity tests. Thus, these results satisfied Koch’s postulates.
Fig. 2 (Colour online) Disease symptoms on lisianthus plants after artificial inoculation with Fusarium solani. a, Disease symptoms with leaf yellowing, stunting and wilting; b, Diseases caused by isolates FJAT-31 352 and FJAT-31 354, but not by isolate FJAT-31 353. The 2-month-old lisianthus seedlings were inoculated with the isolates by dipping the roots into a conidial suspension (2 × 106 conidia mL−1), while control seedlings were treated with sterile water. All treated plants were potted in autoclaved commercial cultivation substrates and maintained at 25/20°C and 70/40% relative humidity (day/night) in a growth chamber with a 16 h day-8 h night cycle.

Molecular identification
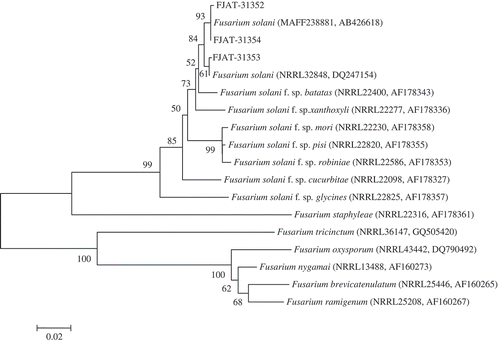
DNA sequence data generated for three isolates used in this study were deposited in GenBank under accession numbers MG183706, MG183707 and MG183708, respectively, for ITS, and accession numbers MG183710, MG183711 and MG183712, respectively, for TEF. A BLAST search of the ITS sequence in GenBank yielded 100% nucleotide identity with the sequences of strains of F. solani. Further comparisons for the TEF sequence, using the Fusarium MLST database, showed similarity above 99% to the sequences of strains of F. solani. For further understanding of interspecies relationships, phylogenetic trees were constructed for the ITS and TEF sequences. The representative isolates FJAT-31 352, FJAT-31 353 and FJAT-31 354 were placed within a monophyletic clade comprising reference isolates of F. solani species with above 99% bootstrap support, and formed a separate clade distinct from other species of Fusarium (, ). Based on molecular data, the fungus was confirmed as F. solani (Mart.) Sacc.
Fig. 3 Phylogenetic tree constructed with sequences of internal transcribed spacer ribosomal DNA (rDNA) region (ITS) of isolates FJAT-31 352, FJAT-31 353 and FJAT-31 354 obtained in this study, and gene sequences of the closest known relatives of F. solani, inferred by the neighbour-joining method and reference sequences retrieved from GenBank database. The per cent bootstrap support values (1000 replications, ≥50%) are shown in the branches. Bar = number of nucleotide substitutions per site. NRRL = ARS Culture Collection National Center for Agricultural Utilization Research, Peoria, Illinois, USA. BBA = Federal Biological Research Centre for Agriculture and Forestry, Braunschweig, Germany. MAFF = Ministry of Agriculture, Forestry and Fisheries, Tokyo, Japan.
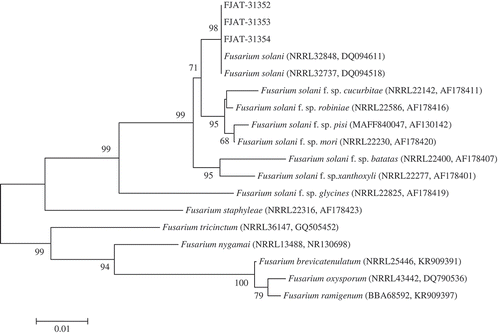
Fig. 4 Phylogenetic tree constructed with sequences of partial translation elongation factor-1α (TEF) of isolates FJAT-31 352, FJAT-31 353 and FJAT-31 354 obtained in this study, and gene sequences of the closest known relatives of F. solani, inferred by the neighbour-joining method and reference sequences retrieved from GenBank and Fusarium MLST database. The per cent bootstrap support values (1000 replications, ≥50%) are shown in the branches. Bar = number of nucleotide substitutions per site. NRRL = ARS Culture Collection National Center for Agricultural Utilization Research, Peoria, Illinois, USA. MAFF = Ministry of Agriculture, Forestry and Fisheries, Tokyo, Japan.
Discussion
Fusarium is a large genus of ubiquitous, ascomycetous fungi that includes many important plant pathogens and mycotoxin producers, as well as saprophytes and endophytes. Species identification is very important for determining the ecological roles of Fusarium species and for diagnosing the diseases they cause. Worldwide, lisianthus is subject to destructive wilt diseases caused by several Fusarium species, such as F. avenaceum, F. oxysporum and F. solani. Fusarium avenaceum can cause root, crown and stem rot of lisianthus with mortality rates frequently reaching 70% in California and Florida (Koike et al., Citation1996; Nalim et al., Citation2009). Fusarium oxysporum f. sp. eustomae can also severely attack lisianthus and has caused a damaging wilt disease in Korea and Italy (Hahm, Citation1998; Yuan et al., Citation2010). Affected plants are wilted, stunted, and show chlorotic leaves and red-brown vascular discolouration. Fusarium solani is reported to cause root rot, crown rot, stem rot, damping-off and stunting of lisianthus in Argentina (Wolcan et al., Citation2001), South Africa (Truter & Wehner, Citation2004) and Janpan (Tomioka et al., Citation2011). Although the symptoms are similar, these diseases are caused by different species of Fusarium in different regions of the world. In this study, the fungus causing root and stem rot on lisianthus was identified as F. solani, based on disease symptoms, morphological characteristics, molecular identification and pathogenicity testing. To our knowledge, this is the first report of F. solani causing root and stem rot on lisianthus in China. Fusarium solani is a cosmopolitan soilborne pathogen with a wide and diverse host range (Bogale et al., Citation2009). The disease on lisianthus can be considered a potentially new and serious threat to this ornamental plant in China. Because this plant is of great economic importance, our results will assist in focusing integrated management strategies such as disinfestation of soils, chemical fungicides and biological control for reducing the damage caused by this pathogen.
Additional information
Funding
References
- Bertoldo C, Gilardi G, Spadaro D, Gullino ML, Garibaldi A. 2015. Genetic diversity and virulence of Italian strains of Fusarium oxysporum, isolated from Eustoma grandiflorum. Eur J Plant Pathol. 141:83–97.
- Bogale M, Steenkamp ET, Wingfield MJ, Wingfield BD. 2009. Diverse Fusarium solani isolates colonise agricultural environments in Ethiopia. Eur J Plant Pathol. 124:369–378.
- Booth C. 1971. The genus Fusarium. Kew (Surrey (England)): CMI Press.
- Chen Y, Chang Y, Chao H. 2016. Identification and characterization of Pothos latent virus causing necrotic ringspots and line patterns on lisianthus (Eustoma grandiflorum) in taiwan. J Phytopathol. 164:650–658.
- Geiser DM, Jimenez-Gasco MM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 110:473–479.
- Hahm YI. 1998. Occurrence of Fusarium wilt on lisianthus (Eustoma grandiflorum) caused by Fusarium oxysporum f. sp. eustomae. Kor J Plant Pathol. 14:188–190.
- Harbaugh BK. 2007. Lisianthus. In: Anderson APNO, editor. Flower breeding and genetics. Netherlands: Springer; p. 644–663.
- Kil EJ. 2014. First report of Tomato yellow leaf curl virus infecting eustoma (Eustoma grandiflorum) in Korea. Plant Dis. 98:1163.
- Koike ST, Gordon TR, Lindow SE. 1996. Crown rot of Eustoma caused by Fusarium avenaceum in California. Plant Dis. 80:1429.
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Ames: Blackwell Publishing; p. 388.
- Nalim FA, Elmer WH, Mcgovern RJ, Geiser DM. 2009. Multilocus phylogenetic diversity of Fusarium avenaceum pathogenic on lisianthus. Phytopathology. 99:462–468.
- Neves CG, Bellé C, Nascimento MB, Grolli PR, Gomes CB, Barros DRD. 2016. First report of Meloidogyne arenaria on lisianthus (Eustoma grandiflorum) in Brazil. Plant Dis. 101:511.
- O’Donnell K, Sutton DA, Rinaldi MG, Sarver BA, Balajee SA, Schroers HJ, Summerbell RC, Robert VA, Crous PW, Zhang N, et al. 2010. Internet-accessible DNA sequence database for identifying Fusaria from human and animal infections. J Clin Microbiol. 48:3708–3718.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tomioka K, Hirooka Y, Takezaki A, Aoki T, Sato T. 2011. Fusarium root rot of prairie gentian caused by a species belonging to the Fusarium solani species complex. J Gen Plant Pathol. 77:132–135.
- Truter M, Wehner FC. 2004. Crown and root infection of lisianthus caused by Fusarium solani in South Africa. Plant Dis. 88:573.
- White TJ, Burns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York (NY): Academic Press; p. 315–322.
- Wolcan S, Lori G, Ronco L. 2001. First report of Fusarium solani causing stunt on lisianthus. Plant Dis. 85:443.
- Yang XM, Zhang YP, Wang LH, Wang JH, Zhang LF, Li JZ, Qu SP. 2016. First report of downy mildew caused by Peronospora chlorae on lisianthus (Eustoma grandiflorum) in China. Plant Dis. 100:650.
- Yuan L, Angelo G, Marialodovica G. 2010. Genetic variability analysis and molecular detection of Fusarium oxysporum f. sp. eustomae isolated from Eustoma grandiflorum in northern Italy. J Phytopathol. 158:546–553.
