Abstract
Severe infection by powdery mildew was observed on California poppy (Eschscholzia californica) plants in Texcoco, State of Mexico, during spring of 2014 to 2017. Symptoms included colonies of white to greyish mildew-like growth on the abaxial and adaxial surfaces of the leaves. The identification of the fungal species was performed by examination of morphological structures using light microscopy and scanning electron microscope (SEM), as well as sequence analysis of the 5ʹ-end of 28S rRNA gene and the internal transcribed spacer (ITS) region of rDNA. Using the combination of morphological characterization and a phylogenetic analysis using Bayesian inference, the fungal agent was identified as Erysiphe eschscholziae. Pathogenicity tests were conducted on leaves of California poppy plants, and Koch’s postulates were fulfilled. This is the first report of E. eschscholziae causing powdery mildew on Eschscholzia californica in Mexico.
Résumé
À partir du printemps de 2014, et ce, jusqu’à 2017, une grave infection causée par l’oïdium a été observée sur des plants de pavot de la Californie (Eschscholzia californica) à Texcoco, dans l’État de Mexico. Les symptômes incluaient des colonies blanchâtres à grisâtres, semblables à de la moisissure, se développant sur les surfaces abaxiales et adaxiales des feuilles. L’identification de l’espèce fongique découle de l’examen des structures morphologiques par microscopie optique et microscopie à balayage, de même que par analyse de la séquence de l’extrémité 5ʹ du gène 28S de l’ARNr et de la région de l’espaceur transcrit interne de l’ADNr. En se basant sur la caractérisation morphologique combinée à une analyse phylogénétique reposant sur l’inférence bayésienne, l’agent fongique a été identifié en tant qu’Erysiphe eschscholziae. Des tests de pathogénicité ont été effectués sur les feuilles de pavot de la Californie et les postulats de Koch ont été vérifiés. Il s’agit de la première mention d’E. eschscholziae causant l’oïdium chez Eschscholzia californica au Mexique.
Introduction
California poppy (Eschscholzia californica Cham.), belonging to the Papaveraceae family, is a native plant to the USA and northern Mexico. This plant is mainly used as an ornamental in gardens, but it also has medicinal properties due to the different types of alkaloids produced by the plant (Lim, Citation2014).
The powdery mildews (Erysiphales) represent a large group of common, obligate plant pathogens of cosmopolitan distribution (Braun & Cook, Citation2012). These obligate parasites occur on many herbaceous perennials and annuals, as well as on woody ornamental plants and trees. The typical symptom caused by these fungi is white powdery colonies which are mainly on the upper surface of the leaves, but also on stems and flowers. Sometimes the colonies coalesce to completely coat an infected plant organ such as a leaf (Gleason et al., Citation2009).
Braun (Citation1999) and Braun et al. (Citation2002) indicated the need to conduct more research involving the exploration of geographic regions where little is known about powdery mildews, as well as detailed examinations of anamorphs using light microscopy (LM) and scanning electron microscopy (SEM), in order to extend knowledge of Erysiphales and their taxonomy. Similarly, Cook & Braun (Citation2009) proposed a key using conidial germination patterns as an additional character to help identify the former subgenera of Oidium. Subsequently, Braun & Cook (Citation2012) revealed features on the surfaces of conidia that reinforced differences observable by LM and reported additional features seen with the SEM for identification of Erysiphales when crucial characters are not clear using LM. These features allow differentiation of the genera and diverse species of anamorphs within the Erysiphales, without the need to characterize the chasmothecia of the teleomorph or sexual stage. However, there are complexes of Erysiphales species that can only be separated or distinguished by the combination of morphological characterization and phylogenetic analysis of DNA sequence data (Braun et al., Citation2002; Braun & Cook, Citation2012; Bereczky et al., Citation2015; Pastirčáková et al., Citation2016; Scholler et al., Citation2016).
Erysiphe cruciferarum Opiz ex L. Junell, Leveillula taurica (Lév.) G. Arnaud (Braun & Cook, Citation2012), E. polygoni DC., Golovinomyces cichoracearum (DC.) V.P. Heluta and Oidium sp. (Farr & Rossman, Citation2018) have been previously reported on Es. californica, with E. cruciferarum being the species most commonly found worldwide. However, a recent analysis of ITS and 28S sequences from the Swiss collections of powdery mildews on Es. californica concluded that they formed a separate group from those of E. cruciferarum, and represent a species complex consisting of more morphologically indistinguishable lineages specialized to various plant families or groups of hosts (Pastirčáková et al., Citation2016). Previously, Oidium sp. was identified using morphological characteristics on Es. californica in Mexico (Yáñez-Morales et al., Citation2009).
Plants of California poppy growing in a flower garden in Texcoco, State of Mexico, Mexico, were severely infected with powdery mildew during the spring months of 2014 to 2017. The aim of this study was to identify the causal agent of powdery mildew of Es. californica based on morphological characterization and phylogenetic analyses, as well as pathogenicity tests.
Materials and methods
Sampling
Four plants (one per year) of Es. californica showing symptoms of powdery mildew were collected in Texcoco, State of Mexico, during spring of 2014 to 2017. Diseased plants exhibited colonies of white to greyish mildew-like growth on the abaxial and adaxial surfaces of the leaves (). A voucher specimen was deposited in the Herbarium of the Department of Agricultural Parasitology at the Chapingo Autonomous University under accession number UACH-140.
Morphological characterization
Morphological characterization of the fungus was conducted using light microscopy (LM) and scanning electron microscopy (SEM). Semi-permanent preparations of mycelia and sporulating structures were made in a drop of 85% lactic acid by collecting the fungus from an infected leaf and observing using an Eclipse Ni-U light microscope (Nikon Corporation®, Japan) equipped with a Moticam 580 camera (Motic®, China). The morphological characteristics of 100 conidia, 30 conidiophores, 20 hyphal appressoria and 20 germ tubes were recorded. The identification of the fungus was conducted using detailed descriptions reported by Pastirčáková et al. (Citation2016).
For SEM studies, small pieces of leaves (5 mm2) with sporulating colonies were fixed in vials with 3% glutaraldehyde (Electron Microscopy Sciences®, USA) and stored at 4°C for 18 h, to be processed following the standard procedures reported by Bozzola & Russell (Citation1992). The previously fixed pieces were fixed a second time for 24 h in 3% glutaraldehyde, washed three times in 0.1 M Sorensen’s phosphate buffer, pH 7.2, for 20 min each and were dehydrated in a graded ethanol series (30, 40, 50, 60, 70, 80, 90 and 96%) for 20 min each, and two times in 100% ethanol for 30 min each. The specimens were further dehydrated in a Sandri-780A critical point dryer (Tousimis Research Corporation®, USA) with CO2. The desiccated specimens were placed on slides using double copper adhesive tape, and coated with a gold layer of 500Ǻ thickness by placing them in an Ion Sputter JFC-1100 ionizer (Jeol®, Japan) for 4 min. The specimens were examined under a JSM-6390 scanning electron microscope (Jeol®, Japan) operated at an accelerating voltage of 15 kV.
DNA extraction, PCR amplification and sequencing
Mycelium, conidiophores and conidia were scraped from the surface of an infected leaf. Genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen®, USA) following the manufacturer’s instructions. DNA concentrations were quantified using a NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific®, USA).
The 5ʹ-end of the 28S rRNA gene (including the domains D1 and D2), and ITS region (including the 5.8S) of rDNA, were amplified by polymerase chain reaction (PCR). The amplification of the 28S rRNA gene was performed by a nested PCR using the primers sets PM3 (Takamatsu & Kano, Citation2001)/TW14 (Mori et al., Citation2000) and NL1/TW14 (Mori et al., Citation2000) for the first and second reactions, respectively. For amplification of the ITS region, the primers ITS1/ITS4 were used (White et al., Citation1990). PCR was conducted in a Bio-Rad C1000 thermocycler (Bio-Rad Laboratories®, USA) using the following conditions: an initial denaturation step at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 52°C for the 28S gene and 55ºC for ITS region for 30 s, extension at 72°C for 60 s and a final extension step at 72°C for 10 min. The amplified PCR products were purified using a QIAquick PCR Purification Kit (Qiagen®, USA) and were sent to Macrogen (www.macrogen.com) for sequencing.
Phylogenetic analysis
Forward and reverse sequences were assembled using the Staden Package (Staden et al., Citation1998). For each locus analysed, GenBank sequences from Erysiphe species reported on Papaveraceaea family plants (Pastirčáková et al., Citation2016) were downloaded. Multiple sequence alignments for each locus independently were performed using ClustalX v. 1.81 (Thompson et al., Citation1997) and manually adjusted. Alignment of each locus was loaded in Sequence Matrix v.1.8 (Vaidya et al., Citation2011) to build the concatenated matrix. The phylogeny for each locus (28S and ITS) and for the concatenated matrix were inferred under the Bayesian analyses. Bayesian phylogenetic estimates were done in MrBayes 3.2.6 (Ronquist et al., Citation2012) implemented on CIPRES Science Gateway portal (https://www.phylo.org/portal2/home.action), based on the GTR nucleotide substitution model. Bayesian analysis with four runs, four MCMC chains per run, Markov chains were run over 100 000 000 generations and trees and parameter values sampled every 10 000 generations. In addition, Maximum Parsimony phylogenetic analyses were conducted using MEGA 5 (Tamura et al., Citation2011) with partial deletion of gaps, substitution models proposed by this program and 1000 bootstrap replicates. The resulting 28S and ITS sequences were deposited in GenBank under accession numbers MG832602 and MF582577, respectively.
Pathogenicity test
Pathogenicity was demonstrated through inoculation by gently dusting conidia from infected leaves onto healthy leaves of 10 one-month-old California poppy plants. Five non-inoculated plants served as controls. Plants were incubated in a moist chamber for 48 h, and then maintained in a greenhouse at temperatures ranging from 20 to 25°C. The pathogenicity test was performed twice.
Results and discussion
Microscopic examination revealed amphigenous mycelium, which was hyaline, septate, smooth, thin-walled and 4.6–8.2 μm wide. Hyphal appressoria were lobed or simple and opposite in pairs or single. Conidiophores were erect, simple, 67–114 × 8.2–9.3 μm, with foot-cells straight or flexuous in the basal half, followed by 1–2 shorter cells or longer cells (). Conidia formed singly and were cylindrical to ellipsoid-doliiform, hyaline, 32.6–45.8 × 12.3–17.7 μm, without fibrosin bodies (). Germ tubes were subterminal, terminating in a lobed appressorium. Chasmothecia were not observed. Morphological characteristics of the fungus were in full agreement with the previous description of Erysiphe eschscholziae Pastirč. & Jankovics by Pastirčáková et al. (Citation2016).
Fig. 2 (Colour online) Structures of Erysiphe eschscholziae on Eschscholzia californica observed using light microscopy (a–c) and scanning electron microscopy (d–g). a, Conidiophore with single conidium. b–c, Conidia. d–e, Hyphal appressoria. f, Conidiophore with single conidium. g, Conidia with ‘angular/rectangular’ wrinkling pattern. Scale bars (a, c) = 20 μm; (b, g) = 10 μm; (d, e) = 5 μm; (f) = 50 μm.
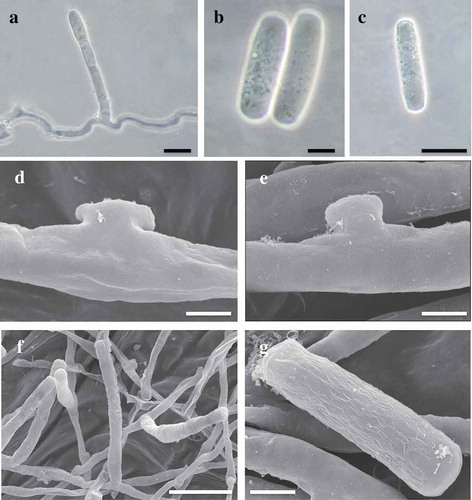
Hyphal appressoria observed under SEM were conspicuous, lobed or simple, smooth to somewhat scaly, without pattern (). Conidia matured singly (), as also revealed by light microscopy (). The outer walls of turgid conidia were coarse and scaly, with a fibrillar septum and a conspicuous rim. When wrinkled, outer walls developed an angular/rectangular pattern of ornamentation (). A pattern of rods surrounding a central complex with ear-like lobes typical of the Pseudoidium anamorph of Erysiphe sec. Erysiphe (Cook et al., Citation1997; Braun & Cook, Citation2012) was difficult to detect because of the orientation of the conidium.
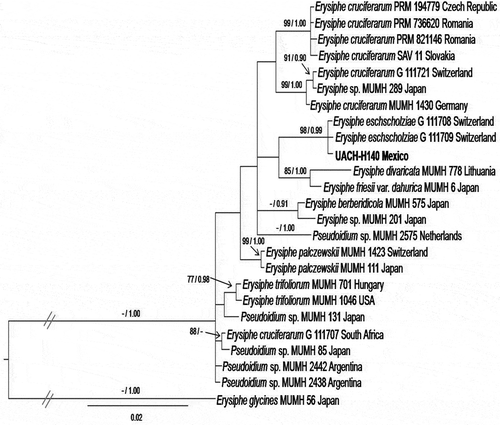
The combined sequences of the ITS region and 28S gene of isolate UACH-140 from Es. californica from Mexico clustered with those of isolates G-111708 and G-111709 of E. eschscholziae from Switzerland ().
Fig. 3 Bayesian tree based on combined internal transcribed spacer (ITS) region and 28S rRNA gene sequences. The isolate UACH-140 of powdery mildew obtained from Eschscholzia californica in this study is shown in bold type. Bootstrap values by the Maximum parsimony method and probabilities by the Bayesian analysis are shown on the respective branches. Bootstrap values below 70% and posterior probabilities below 0.90 are not shown. The sequences of Erysiphe glycines MUMH-56 were used as an outgroup.
All inoculated plants developed powdery mildew colonies after 11 days, whereas the control plants remained symptomless. The fungus present on the inoculated plants was identical morphologically to that originally observed on diseased plants, fulfilling Koch’s postulates.
Erysiphe eschscholziae is morphologically similar to, but distinguishable from, the asexual stage of E. cruciferarum. According to a recent analysis of ITS and 28S sequences from the Swiss Pseudoidium collections on Es. californica, it was concluded that E. eschscholziae formed a separate group from E. cruciferarum. On the other hand, E. cruciferarum is a commonly reported species infecting this ornamental plant and represents a species complex consisting of more morphologically indistinguishable lineages specialized to various plant families or groups of hosts, including those of the Papaveraceae (Pastirčáková et al., Citation2016).
Pastirčáková et al. (Citation2016) described the species E. eschscholziae based on a revision of herbarium specimens originally designated as E. cruciferarum by Bolay (Citation2005) and confirmed that E. cruciferarum is not able to infect Es. californica. Therefore, collections recorded as E. cruciferarum and Oidium sp. on Es. californica in Australia, Tasmania (Amano, Citation1986), Romania (Eliade, Citation1990), Japan (Nomura, Citation1997), South Africa (Crous et al., Citation2000), the USA (Glawe, Citation2006), Germany (Schmidt & Scholler, Citation2011) and the UK (Pastirčáková et al., Citation2016) need to be re-examined using molecular tools. A re-examination may show that some specimens are E. eschscholziae. On the other hand, Erysiphe polygoni, also a species with Pseudoidium anamorph, has been reported on Es. californica and Es. cristata from the USA (French, Citation1989) and India (Sarbhoy et al., Citation1971), respectively. These records also need a re-examination using DNA sequence data.
The first report of powdery mildew on Es. californica in Mexico was in 2008, but that specimen was identified as Oidium sp. using only morphological characteristics (Yáñez-Morales et al., Citation2009). Our results confirm the presence of powdery mildew on California poppy in Mexico. However, to our knowledge, the current study represents the second report of the occurrence of E. eschscholziae worldwide and is the first confirmed report of this fungus causing powdery mildew of California poppy in Mexico and North America.
Acknowledgements
The authors are grateful to Greta Hanako Rosas Saito for providing technical assistance with the SEM.
References
- Amano K. 1986. Host range and geographical distribution of the powdery mildew fungi. Tokyo (Japan): Japan Scientific Societies Press.
- Bereczky Z, Pintye A, Csontos P, Braun U, Kiss L. 2015. Does the parasite follow its host? Occurrence of morphologically barely distinguishable powdery mildew anamorphs on Oenothera spp. in different parts of the world. Mycoscience. 56:267–272.
- Bolay A. 2005. Les oïdiums de Suisse (Erysiphacées). Cryptogam Helv. 20:1–176.
- Bozzola JJ, Russell LD. 1992. Electron Microscopy: principles and Techniques for Biologists. Boston (MA): Jones and Bartlett Publishers.
- Braun U. 1999. Some critical notes on the classification and the generic concept of the Erysiphaceae. Schlechtendalia. 3:48–54.
- Braun U, Cook RTA. 2012. Taxonomic Manual of the Erysiphales (Powdery Mildews). Utrecht (The Netherlands): CBS-KNAW Fungal Biodiversity Centre; p. 707.
- Braun U, Cook RTA, Inman AJ, Shin HD. 2002. The Taxonomy of the Powdery Mildew Fungi. In: Bélanger RR, Bushnell WR, Dik AJ, Carver TLW, editors. The Powdery Mildews: a comprehensive treatise. St. Paul Minnesota (MN): American Phytopathological Society; p. 13–55.
- Cook RTA, Braun U. 2009. Conidial germination patterns in powdery mildews. Mycol Res. 113:616–636.
- Cook RTA, Inman AJ, Billings C. 1997. Identification and classification of powdery mildew anamorphs using light and scanning electron microscopy and host range data. Mycol Res. 101:975–1002.
- Crous PW, Phillips AJL, Baxter AP. 2000. Phytopathogenic Fungi from South Africa. Stellenbosch (South Africa): University of Stellenbosch, Department of Plant Pathology Press.
- Eliade E. 1990. Monografia Erysiphacee lor dein Romania. Acta Bot Hort Bucurest. 1989–1990:105–574.
- Farr DF, Rossman AY 2018. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. [accessed Mar 18]. https://nt.ars-grin.gov/fungaldatabases/.
- French AM. 1989. California plant disease host index. Sacramento (CA): California Department of Food and Agriculture.
- Glawe DA. 2006. First report of powdery mildew of Eschscholzia californica (California poppy) caused by Erysiphe cruciferarum in North America. Plant Health Progr. doi:10.1094/PHP-2006-1213-01-BR
- Gleason ML, Daughtrey ML, Chase AR, Moorman GW, Mueller DS. 2009. Diseases of Herbaceous Perennials. St. Paul (MN): APS Press.
- Lim TK. 2014. Edible Medicinal and Non-Medicinal Plants: volume 8, Flowers. Dordrecht: Springer.
- Mori Y, Sato Y, Takamatsu S. 2000. Evolutionary analysis of the powdery mildew fungi (Erysiphales) using nucleotide sequences of the nuclear ribosomal DNA. Mycologia. 92:74–93.
- Nomura Y. 1997. Taxonomical study of Erysiphaceae of Japan. Tokyo (Japan): Yokendo Ltd.
- Pastirčáková K, Jankovics T, Komáromi J, Pintye A, Pastirčák M. 2016. Genetic diversity and host range of powdery mildews on Papaveraceae. Mycol Prog. 15:36.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes v. 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 61:539–542.
- Sarbhoy AK, Lal G, Varshney JL. 1971. Fungi of India (1967-71). New Delhi (India): Navyug Traders.
- Schmidt A, Scholler M. 2011. Studies in Erysiphales anamorphs (4): species on Hydrangeaceae and Papaveraceae. Mycotaxon. 115:287–301.
- Scholler M, Schmidt A, Siahaan SAS, Takamatsu S, Braun U. 2016. A taxonomic and phylogenetic study of the Golovinomyces biocellatus complex (Erysiphales, Ascomycota) using asexual state morphology and rDNA sequence data. Mycol Prog. 15:56.
- Staden R, Beal KF, Bonfield JK. 1998. The Staden package, 1998. In: Misener S, Krawetz SA, editors. Bioinformatics methods and protocols. New York: Humana; p. 115–130.
- Takamatsu S, Kano Y. 2001. PCR primers useful for nucleotide sequencing of rDNA of the powdery mildew fungi. Mycoscience. 42:135–139.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX Windows interface: flexible strategies for multiple alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistic. 27:171–180.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; p. 315–322.
- Yáñez-Morales MJ, Braun U, Minnis AM, Tovar-Pedraza JM. 2009. Some new records and new species of powdery mildew fungi from Mexico. Schlechtendalia. 19:47–61.

