Abstract
Leaf yellowing and reddening symptoms on Vitis vinifera vines were observed in different grapevine nurseries in the province of Kurdistan in the west of Iran during 2016–2017. Total DNA was extracted from 27 symptomatic and four symptomless plants and tested for the presence of phytoplasma DNA by nested polymerase chain reaction (PCR) using P1/P7 and R16F2n/R16R2 primer pairs. PCR amplicons of about 1.8 and 1.2 kb in size were obtained from all symptomatic V. vinifera but not from symptomless plants. Restriction fragment length polymorphism (RFLP) profiles of R16F2n/R2 amplicons using AluI, HhaI, HpaII, RsaI, TaqI, KpnI, HaeIII and Sau3AI restriction enzymes were identical to each other and to those obtained from 16SrI phytoplasmas, indicating that the phytoplasmas associated with grapevine yellowing and reddening were members of the 16SrI group. A R16F2n/R16R2 16S rDNA sequence of Kurdistan V. vinifera yellowing and reddening strain (KVvY) was submitted to GenBank as accession number MH638316. The Blast analysis of KVvY showed 100% sequence identity with those of the ‘Candidatus Phytoplasma asteris’-related strains. Phylogenetic analysis confirmed that the phytoplasma associated with KVvY clustered within the 16SrI phytoplasma clade closer to the Onion yellows mild strain (OY-M), a 16SrI-B phytoplasma. Restriction analysis using iPhyClassifier confirmed that virtual RFLP patterns of the KVvY were identical (similarity coefficient 1.00) to the reference pattern of 16Sr group I, subgroup B (GenBank accession AP006628).
Résumé
En 2016-2017, des symptômes du jaunissement et du rougissement des feuilles chez Vitis vinifera ont été observés dans diverses pépinières de vignes de la province du Kurdistan, dans l’ouest de l’Iran. L’ADN total a été extrait de 27 plants symptomatiques et de 4 plants asymptomatiques, et a été testé par réaction en chaîne de la polymérase nichée (PCR) à l’aide des paires d’amorces P1/P7 et R16F2n/R16R2 pour y détecter l’ADN de phytoplasmes. Les amplicons résultant de la PCR, d’environ 1.8 et 1.2 kb, ont été obtenus de tous les plants symptomatiques de V. vinifera, mais pas des plants asymptomatiques. Les profils du polymorphisme de longueur des fragments de restriction (RFLP) des amplicons R16F2n/R2, obtenus avec les enzymes de restriction AluI, HhaI, HpaII, RsaI, TaqI, KpnI, HaeIII et Sau3AI, étaient identiques entre eux ainsi qu’à ceux obtenus des phytoplasmes 16SrI, indiquant que les phytoplasmes associés au jaunissement et au rougissement de la vigne appartenaient au groupe 16SrI. Une séquence 16S de l’ADNr de R16F2n/R16R2 de la souche du jaunissement et du rougissement de V. vinifera du Kurdistan (KVvY) a été soumise à la GenBank en tant qu’obtention numéro MH638316. L’analyse de type BLAST de KVvY a révélé une identité de séquence de 100% avec celle des souches associées à ‘Candidatus Phytoplasma asteris’. L’analyse phylogénétique a confirmé que les phytoplasmes associés à KVvY se regroupaient au sein du clade du phytoplasme 16SrI ressemblant le plus à la souche faible du jaunissement de l’oignon (OY-M), un phytoplasme du groupe 16SrI-B. Une analyse de restriction basée sur iPhyClassifier a confirmé que les schémas virtuels du RFLP de KVvY étaient identiques (coefficient de conformité de 1.00) au schéma de référence du sous-groupe B du groupe I de 16Sr (GenBank, obtention AP006628).
Introduction
Phytoplasmas, members of the class Mollicutes, are wall-less, phloem-limited bacterial plant pathogens that infect a wide range of plant species worldwide (Bertaccini et al., Citation2014). They are transmitted mainly by leafhoppers and psyllids and cause yellowing, discolouration, witches’ broom, dwarfing, virescence and phyllody disease symptoms in different plant species. Among the plant species harbouring phytoplasmas, grapevine has been associated with several phytoplasma diseases worldwide. Bois Noir (16SrXII-A), flavescence doree (16SrV-C and 16SrV-D), Australian grapevine yellows and North American grapevine yellows are known as grapevine yellows phytoplasmas (Constable & Bertaccini, Citation2017). Also grapevine yellows and reddening disease associated with Ca. Phytoplasma pruni (16SrIII-A), Ca. Phytoplasma asteries (16SrI-B), Ca. Phytoplasma fraxini (16SrVII-A), Ca. Phytoplasma solani (16SrXII-A), Ca. Phytoplasma australiense (16srXII-B), Ca. Phytoplasma ulmi (16SrV-A), 16SrI-C, 16SrV-C, 16SrV-D,16SrVI, 16SrIX and 16SrX-B were reported from Australia, Lebanon, Turkey, Chile, Tunisia, Georgia, Armenia, Jordan, Italy, Serbia, South Africa and USA (Prince et al., Citation1993; Choueiri et al., Citation2002; M’hirsi et al., Citation2004; Botti & Bertaccini, Citation2006; Gajardo et al., Citation2009; Canik et al., Citation2011; Salem et al., Citation2013; Roggiabb et al., Citation2014; Ertunc et al., Citation2015; Davis et al., Citation2015; Quaglino et al., Citation2016; Zambon et al., Citation2018).
Fig. 1 (Colour online) Symptoms of (A) leaf curling (B) reddening (C) yellowing (D) little leaves in Vitis vinifera infected plant in Kurdistan (Iran).

Fig. 2 Polyacrylamide gel showing RFLP profiles of 16S rDNA amplified by nested PCR using P1/P7 followed by R16F2n/R16R2 primer pairs from KVvY (GenBank accession number MH638316). PCR products were digested by AluI, HhaI, HpaII, RsaI, TaqI, KpnI, HaeIII, Sau3AI restriction enzymes. M, 100 bp DNA ladder.
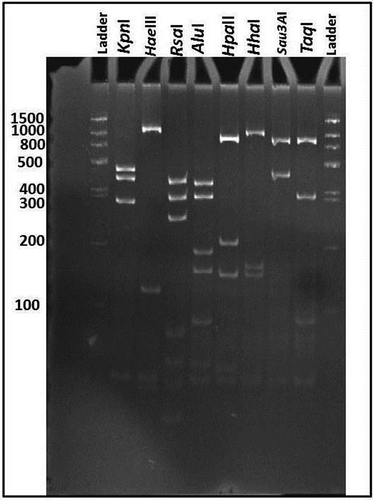
Fig. 3 Neighbour-joining phylogenetic tree based on the 1.2 kb 16S rDNA sequence of KVvY (GenBank accession number MH638316) and those of selected phytoplasma strains from GenBank. A. laidlawii was used as an outgroup. GenBank accession numbers are shown on the right of phytoplasma strains. Numbers on branches are bootstrap values of 1000 replicates.
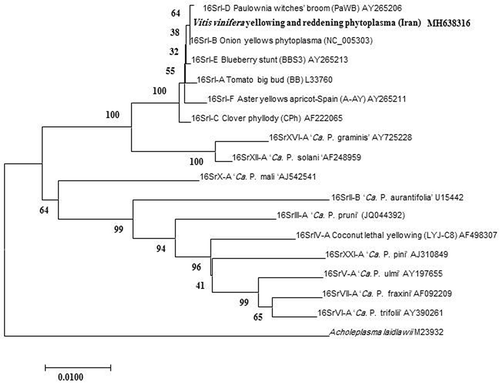
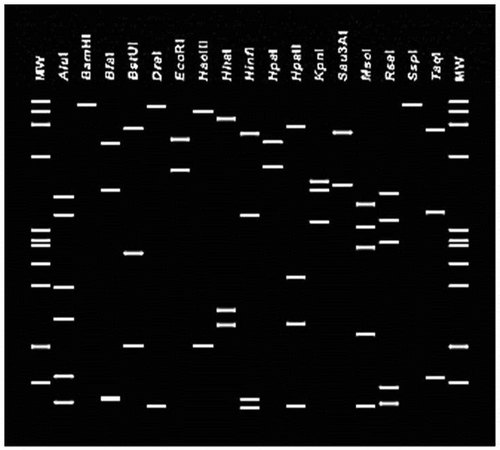
Fig. 4 Virtual RFLP profiles from the R16F2n/R16R2 sequence of the VivY phytoplasma (GenBank accession number MH638316) using 17 restriction endonucleases with iPhyClassifier (Zhao et al., Citation2009).
Grapevine production areas, especially for rainfed cultivars, are very important in west areas of Iran and the grapes are used mostly for fresh fruit or export. Phytoplasma symptoms were observed in vineyards in the central and north east regions of Iran which were associated with phytoplasmas groups 16SrVII-A, 16SrXII-A, Ca. Phytoplasma phoenicium (16SrIX-B) and Ca. Phytoplasma aurantifolia (16SrII-B) (Karimi et al., Citation2009; Salehi et al., Citation2014, Citation2016a; Mirchenari et al., Citation2015; Ghayeb Zamharir et al., Citation2017). The aim of this work was to identifiy and characterize phytoplasmas present in grapevine nurseries located in the Kurdistan province (west of Iran) to improve our understanding of the disease and develop sustainable disease management for new grapevine orchards.
Materials and methods
Plant sampling and infection incidence
During 2016–17, sampling at two times was carried out in grapevine nurseries located in the Kurdistan province in the west part of Iran. Grapevine nurseries were sampled on a diagonal transect across each. Plants expressing phytoplasma disease-like symptoms such as leaf curling, leaf yellowing and necrotic reddening of leaves, as well as death of shoot tips, were collected. Young leaf and petiole samples were taken and transferred on ice to the lab and used immediately for DNA extraction. A total of 27 symptomatic and four asymptomatic samples were collected and subjected to nested-PCR assays.
DNA extraction and nested PCR amplification
Total DNA was extracted from 0.2g of midrib tissue of symptomatic and symptomless grapevine plants according to the procedure described by Healey et al. (Citation2014). Total DNA extracted from a periwinkle plant infected with alfalfa witches’ broom phytoplasma strain (16SrXII-A) (Esmailzadeh Hosseini et al., Citation2016) was used as a positive control and a DNA sample from a grapevine plant grown in the greenhouse was used as a negative control. The DNA quality was estimated by agarose gel electrophoresis and 100 ng of nucleic acid was used for PCR reactions. For nested-PCR, the universal primer pair P1/P7 amplifying 16SrRNA gene, 16S-23S spacer region (SR) and the 5ʹ end of 23S rRNA gene (Deng & Hiruki, Citation1991; Schneider et al., Citation1995) and the R16F2n/R2 primers amplifying ~1.2 kbp inside the 16SrRNA gene (Gundersen & Lee, Citation1996) were used. The PCR reaction was performed in a Emeraldamp MaxHS master mix (Takara, Japan). The reaction cycled 35 times in a Bio-Rad (USA) thermal cycler with the following parameters: denaturation for 30 s at 94°C (2 min in the first round), annealing for 30 s at 55°C and primer extension for 1 min at 72°C (10 min in the final cycle). PCR conditions for the nested PCR were the same except that the annealing temperature was 58°C. Following PCR, 5 µL of each reaction mixture was electrophoresed in a 1% (w/v) agarose gel containing 0.3 µg mL−1 ethidium bromide in 0.5× TBE buffer (22.5 mM TRIS–borate, 1 mM EDTA, pH 8.0) to verify amplification of target DNAs.
Restriction fragment length polymorphism (RFLP) analysis
RFLP analysis of 1.2 kb nested PCR products were used for identification of the phytoplasma associated with the grapevine disease. Restriction fragment length polymorphism (RFLP) analysis of R16F2n/R16R2 amplicons using AluI, HhaI, HpaII, RsaI, TaqI, KpnI, HaeIII and Sau3AI restriction enzymes was performed according to the manufacturer’s instructions (Fermentas, Vilnius, Lithuania). The restriction products were then separated by electrophoresis through an 8% polyacrylamide gel, stained by ethidium bromide and visualized with a UV transilluminator. The resulting RFLP patterns were compared with those previously published for 16S rDNA of other phytoplasmas (Lee et al., Citation1998).
Sequencing and phylogenetic analyses
R16F2n/R16R2 primed PCR product (1.2 kb) from DNA extracted from infected plants was directly sequenced in both directions and the assembled sequence was compared with deposited sequences in the GenBank database using BLAST (National Center for Biotechnology Information, Bethesda, MD, USA) and aligned with Bioedit tools. The 1250 bp of 16S rDNA sequences of 18 phytoplasmas including a strain of KVvY phytoplasma disease were aligned using MEGA7 software (Kumar et al., Citation2016) and a phylogenetic tree was constructed using the neighbour-joining method. Acholeplasma laidlawii was used as an outgroup to root the tree. Bootstrapping was performed 1000 times to estimate the stability and support for the branches.
Virtual RFLP analysis
Virtual RFLP analysis using iPhyClassifier (Zhao et al., Citation2009) was used to determine the subgroup affiliation of KVvY phytoplasma. Each aligned DNA fragment was digested in silico with 17 distinct restriction enzymes: AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, MboI (Sau3AI), MseI, RsaI, SspI and TaqI.
Results
Disease symptoms and incidence
During two sets of sampling times in grapevine nurseries in Kurdistan province, various symptoms including yellowing, reddening and inward leaf curling and malformed leaf shape were observed in different grapevine nurseries (). In red fruit varieties, the early season symptoms consisted mostly of leaf yellowing and the late season symptoms consisted mostly of leaf reddening. The incidence of the symptomatic plants in the visited nurseries ranged from 5–10%.
PCR-RFLP analyses
PCR amplicons of ~1.8 kb were observed in all of the samples from the 27 symptomatic plants. No amplification was obtained from samples collected from symptomless plants and the negative control. The subsequent nested PCR using P1/P7 and R16f2n/R2 primer pairs yielded a 1.25 kb product in all 27 symptomatic samples. The RFLP patterns of the 1.25 kb 16S rDNA amplicons of all 27 symptomatic KVvY phytoplasma samples from the surveyed regions were identical to each other and similar to those published for members of the Aster yellows (16SrI) phytoplasma group (Lee et al., Citation1998) ().
Sequencing, virtual RFLP and phylogenetic analysis
The 1.2 kb DNA fragments amplified from the KVvY phytoplasma samples were directly sequenced and showed 100% sequence identity to each other. One of the sequences was named KVvY phytoplasma and submitted to GenBank as accession number MH638316. The BLAST analysis of the KVvY consensus sequence showed 100% sequence identity with those of the ‘Candidatus Phytoplasma asteris’-related strains. Phylogenetic analysis (MEGA software version 7.0) confirmed that the phytoplasma associated with KVvY clustered within the 16SrI phytoplasma clade and is closely associated with the Onion yellows mild strain (OY-M) a 16SrI-B phytoplasma (). Restriction analysis using iPhyClassifier confirmed that virtual RFLP patterns of the KVvY are identical (similarity coefficient 1.00) to the reference pattern of 16Sr group I, subgroup B (GenBank accession number: AP006628) ().
Discussion
The Aster yellows phytoplasmas group (16SrI) is the most prevalent phytoplasmas group, as it can infect a broad range of host plants, including monocotyledonous and dicotyledonous species of herbaceous and woody plants. The 16SrI group contains numerous subgroups that are distributed in grapevine cultivation regions worldwide. Numerous plants such as Eucalyptus camaldulensis, Brassica napus, Spinacia oleracea, Eruca sativa, Medicago sativaand Lactuca serriola and several ornamental plants with symptoms of little leaf, phyllody, yellowing, purpling, leaf rolling and virescence were identified as infected by Aster yellows phytoplasma strains in Iran (Babaie et al., Citation2007; Asghari Tazehkand et al., Citation2010; Rashidi et al., Citation2010; Salehi et al., Citation2011; Esmailzadeh Hosseini et al., Citation2015a, Citation2015b; Salehi et al., Citation2016b).
The 16SrI-B phytoplasma subgroup has been reported to occur in grapevines in Italy, Slovenia, Croatia, Chile, Tunisia and Turkey (Alma et al., Citation1996; Saric et al., Citation1997; M’hirsi et al., Citation2004; Gajardo et al., Citation2009; Jezič & Karoglan Kontić, Citation2013; Landi et al., Citation2013; Ertunc et al., Citation2015). This is the most common phytoplasma associated with grapevine yellows and it was reported in South Africa (Engelbrecht et al., Citation2010) and in grapevines grown in Canada, along with the subgroups 16SrI-A and 16SrI-C (Olivier et al. Citation2009). 16SrI-B is mostly transmitted by the leafhoppers Euscelis incisus and Macrosteles quadripunctulatus (Alma et al., Citation2001), which can live on different plant host species and transmit phytoplasmas from reservoir hosts to grapevine (Krüger et al., Citation2015).
Symptoms caused by phytoplasmas are similar to each other but also dependent on the varietal resistance, the age of plants during infection, contamination with other pathogens especially viruses, and effects of abiotic stress. Different phytoplasmas groups or subgroups with different insect or plant hosts can produce similar symptoms in grapevine (Bertaccini et al., Citation1998). Identification and characterization of phytoplasmas groups can help in understanding the disease epidemiology and planning for disease management. Symptomless phytoplasmas infection in grapevine is common and presence of phytoplasmas in one nursery can show severe and high incidence of infection (Bertaccini et al., Citation1998).
Phytoplasmas infection in grapevine nurseries probably originated from propagation materials or through transmission by insect vectors. As grapevine is propagated by stem cuttings, movement of propagation materials can distribute and spread phytoplasmas infection in grapevines grown in other geographic areas. Characterization of phytoplasmas groups in grapevine nurseries can determine transportation limits for quarantine of stem cuttings. Cultivation of grapevine varieties resistant to phytoplasmas can help to manage the disease, although to date, no variety resistant to phytoplasmas causing yellowing has been identified (Laimer et al., Citation2009).
In this study, we detected for the first time the presence of phytoplasmas group 16SrI-B in grapevine nurseries grown in the west region of Iran. This phytoplasmas subgroup was also reported in Turkey, a neighbouring country of Iran (Ertunc et al., Citation2015). Further studies are necessary to identify the main insect vectors, their biology and their origin of infection in order to determine the geographic distribution of the phytoplasma and to develop sustainable disease management.
Acknowledgements
This work was supported by the Chaharmahal and Bakhtiari Agricultural Organization, Shahrekord, Iran. We thanks Mrs M. Zandian for assistance in this research.
References
- Alma A, Davis RE, Vibio M, Danielli A, Bosco D, Arzone A, Bertaccini A. 1996. Mixed infection of grapevines in Northern Italy by phytoplasmas including 16S rRNA RFLP subgroup 16SrI-B strains previously unreported in this host. Plant Dis. 80:418–421.
- Alma A, Palermo S, Boccardo G, Conti M. 2001. Transmission of chrysanthemum yellows, a subgroup 16SrI-B phytoplasma, to grapevine by four leafhopper species. J Plant Pathol. 83:181–187.
- Asghari Tazehkand S, Hosseini Pour A, Heydarnejad J, Massumi H, Azadvar M. 2010. Identification of phytoplasmas associated with cultivated and ornamental plants in Kerman province, Iran. J Phytopathol. 158:713–720.
- Babaie G, Khatabi B, Bayat H, Rastgou M, Hosseini A, Salekdeh GH. 2007. Detection and characterization of phytoplasma infecting ornamental and weed plants in Iran. J Phytopathol. 155:368–372.
- Bertaccini A, Borgo M, Martini M. 1998. Continous grapevine yellows epidemic in Veneto. L’Inf.tore Agrario. 54:85–90.
- Bertaccini A, Duduk B, Paltrinieri S, Contaldo N. 2014. Phytoplasmas and phytoplasma diseases: a severe threat to agriculture. Am J Plant Sci. 5:1763–1788.
- Botti S, Bertaccini A. 2006. First report of phytoplasmas in grapevine in South Africa. Plant Dis. 90:1360.
- Canik D, Ertunc F, Paltrinieri S, Contaldo N, Bertaccini A. 2011. Identification of different phytoplasmas infecting grapevine in Turkey. Bull Insectology. 64:225–226.
- Choueiri E, Jreijiri F, El Zammar S. 2002. First report of grapevine “Bois Noir” disease and a new phytoplasma infecting solanaceous plants in Lebanon. Plant Dis. 86:697.
- Constable F, Bertaccini A. 2017. Worldwide Distribution and Identification of Grapevine Yellows Diseases. In: Dermastia M, Bertaccini A, Constable F, Mehle N, editors. Grapevine Yellows Diseases and Their Phytoplasma Agents. Switzerland: Springer Briefs in Agriculture. Springer International Publishing AG; p. 17–46.
- Davis RE, Dally EL, Zhao Y, Lee IM, Wei W, Wolf TK, Beanland LeDoux DG, Johnson DA, Fiola JA, Walter Peterson H, et al. 2015. Unraveling the etiology of North American Grapevine Yellows (NAGY): novel NAGY phytoplasma sequevars related to `Candidatus Phytoplasma pruni’. Plant Dis. 99:1087–1097.
- Deng S, Hiruki C. 1991. Amplification of 16S rRNA genes from culturable and non-culturable mollicutes. J Microbiol Meth. 14:53–61.
- Engelbrecht M, Joubert J, Burger JT. 2010. First report of aster yellows phytoplasma in grapevines in South Africa. Plant Dis. 94:373.
- Ertunc F, Orel D, Bayram S, Paltrinieri S, Bertaccini A, Topkaya S, Soylemezoglu G. 2015. Occurrence and identification of grapevine phytoplasmas in main viticultural regions of Turkey. Phytoparasitica. 43:303–310.
- Esmailzadeh Hosseini SA, Khodakaramian G, Salehi M, Bertaccini A. 2016. Molecular identification and phylogenetic analysis of phytoplasmas associated with alfalfa witches’ broom diseases in the western areas of Iran. Phytopathogenic Mollicutes. 6:16–22.
- Esmailzadeh Hosseini SA, Salehi M, Khodakaramian G, Mirchenari SM, Bertaccini A. 2015b. An up to date status of alfalfa witches’ broom disease in Iran. Phytopathogenic Mollicutes. 5:9–18.
- Esmailzadeh Hosseini SA, Salehi M, Salehi E. 2015a. First report of a 16SrI-B subgroup related phytoplasma associated with Eruca sativa phyllody in Iran. New Dis Reports. 32:22.
- Gajardo A, Fiore N, Prodan S, Paltrinieri S, Botti S, Pino AM, Zamorano A, Montealegre J, Bertaccini A. 2009. Phytoplasmas associated with grapevine yellows disease in Chile. Plant Dis. 93:789–796.
- Ghayeb Zamharir M, Paltrinieri S, Hajivand S, Taheri M, Bertaccini A. 2017. Molecular dentification of diverse ‘Candidatus Phytoplasma’species associated with grapevine decline in Iran. J Phytopathol. 165:407–413.
- Gundersen DE, Lee I-M. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer sets. Phytopathol Mediterr. 35:144–151.
- Healey A, Furtado A, Cooper T, Henry RJ. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 10:21. doi:10.1186/1746-4811-10-21
- Jezič M, Karoglan Kontić J. 2013. Grapevine yellows affecting the Croatian indigenous grapevine cultivar Grk. Acta Bot Croat. 72:287–294.
- Karimi M, Contaldo N, Mahmoudi B, Duduk B, Bertaccini A. 2009. Identification of stolbur-related phytoplasmas in grapevine showing decline symptoms in Iran. Le Progrès Agricole et Viticole HS. 31:208–209.
- Krüger K, Pietersen G, Smit N, Carstens R. 2015. Epidemiology of aster yellows phytoplasma: alternate host plants and the vector Mgenia fuscovaria (Hemiptera: cicadellidae) in South Africa. In: Proceedings of the 18th congress of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine (ICVG); Ankara, Turkey. p. 126–127.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evolution. 33:1870–1874.
- Laimer M, Lemaire O, Herrbach E, Goldschmidt V, Minafra A, Bianco PA, Wetzel T. 2009. Resistance to viruses, phytoplasmas and their vectors in the grapevine in Europe: a review. J Plant Pathol. 91:7‐23.
- Landi L, Isidoro N, Riolo P. 2013. Natural phytoplasma infection of four phloem-feeding Auchenorrhyncha across vineyard agroecosystems in Central-Eastern Italy. J Econ Entomol. 106:604–613.
- Lee I-M, Gundersen-Rindal DE, Davis RE, Bartoszyk IM. 1998. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int J Syst Evol Microbiol. 48:1153–1169.
- M’hirsi S, Acheche H, Fattouch S, Boccardo G, Marrakchi M, Marzouki N. 2004. First report of phytoplasmas in the aster yellows group infecting grapevine in Tunisia. Plant Pathol. 53:521.
- Mirchenari SM, Massah A, Zirak L. 2015. ‘Bois noir’: new phytoplasma disease of grapevine in Iran. J Plant Prot Res. 55:88–93.
- Olivier CY, Lowery DT, Stobbs LW. 2009. First report of aster yellows phytoplasmas Candidatus Phytoplasma asteris in Canadian grapevines. Plant Dis. 93:669.
- Prince JP, Davis RE, Wolf TK, Lee IM, Mogen BD, Dally EL, Bertaccini A, Credi R, Barba M. 1993. Molecular detection of diverse mycoplasmlike organisms (MLOs) associated with grapevine yellows and their classification with aster yellow, X-disease, and elm yellows phytoplasmas. Phytopathology. 83:1130–1137.
- Quaglino F, Maghradze D, Casati P, Chkhaidze N, Lobjanidze M, Ravasio A, Passera A, Venturini G, Failla O, Bianco PA. 2016. Identification and characterization of new ‘Candidatus Phytoplasma solani’ strains associated with Bois Noir Disease in Vitis vinifera L. cultivars showing a range of symptom severity in Georgia, the Caucasus Region. Plant Dis. 100:904–915.
- Rashidi M, Ghosta Y, Bahar M. 2010. Molecular identification of a phytoplasma associated with Russian olive witches’ broom in Iran. Euro J Plant Pathol. 127:157–159.
- Roggiabb C, Caciaglia P, Galettoa L, Pacificoa D, Verattia F, Boscob D, Marzachi C. 2014. Flavescence doree phytoplasma titre in field-infected Barbera and Nebbiolo grapevines. Plant Pathol. 63:31–41.
- Salehi E, Salehi M, Taghavi SM, Izadpanah K. 2016a. First report of a 16SrIX group (Pigeon pea witches’ broom) phytoplasma associated with grapevine yellows in Iran. J Plant Pathol. 98:376.
- Salehi E, Taghavi SM, Salehi M, Izadpanah K. 2014. Partial biological and molecular characterization of phytoplasmas associated with grapevine yellows in Fars and Lorestan provinces of Iran. Iranian J Plant Pathol. 50:55–64.
- Salehi M, Esmailzadeh Hosseini SA, Salehi E. 2016b. First report of a ‘Candidatus Phytoplasma asteris’ related phytoplasma associated with Eucalyptus little leaf disease in Iran. J Plant Pathol. 98:175.
- Salehi M, Izadpanah K, Siampour M. 2011. Occurrence, molecular characterization and vector transmission of a phytoplasma associated with rapeseed phyllody in Iran. J Phytopathol. 159:100–105.
- Salem NM, Quaglino F, Abdeen A, Casati P, Bulgari D, Alma A, Bianco PA. 2013. First report of ‘Candidatus Phytoplasma solani’ associated with grapevine Bois noir disease in Jordan. p. 23–24. COST Action FAO 807 Final Meeting. Integrated Management of Phytoplasma Epidemics in Different Crop Systems; Lisbon, Portugal. p. 85.
- Saric A, Skoric D, Bertaccini A. 1997. Molecular detection of phytoplasmas infecting grapevines in Slovenia and Croatia. In: 12th ICVG meeting; Lisbon. p. 77–78.
- Schneider B, Seemüller E, Smart CD, Kirkpatrick BC. 1995. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas. In: Razin S, Tully JG, editors. Molecular and diagnostic procedures in Mycoplasmology. San Diego (CA): Academic Press; p. 369–380.
- Zambon Y, Canel A, Bertaccini A, Contaldo N. 2018. Molecular diversity of phytoplasmas associated with grapevine yellows disease in North-Eastern Italy. Phytopathology. 108:206–214.
- Zhao Y, Wei W, Lee I-M, Shao J, Suo X, Davis RE. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int J Syst Evol Microbiol. 59:2582–2593.
