Abstract
New diseases in pepper plantations were discovered in La Comarca Lagunera (CL) region in September 2014, the severity of which increased by October 2016. Pepper plants exhibited mild and severe yellow leaf mosaic, deformation, stunting and chlorotic leaves. In addition, whiteflies were observed on symptomatic plants, suggesting a possible begomovirus aetiology. In this study, naturally infected pepper plants were collected during three consecutive years to identify the potential begomovirus present in pepper in CL. PCR detection using degenerate and specific primers indicated that 47 out of 49 pepper plants were infected by begomoviruses mainly in mixed infection. The complete begomovirus genomes were isolated from a representative symptomatic pepper plant and two clones for each begomovirus were fully sequenced for the corresponding year of collection (2014 to 2016). Phylogenetic analysis of complete genomes of CL begomovirus pepper isolates indicated a close homology with Tomato yellow leaf curl virus (designated TYLCV-CL) displaying 99.9–100% identity with TYLCV Sinaloa isolate, and the bipartite Pepper huasteco yellow vein virus (designated PHYVV-CL) displaying 94.5% and 84.2% identity with first PHYVV isolate from Tamaulipas for DNA A and DNA B, respectively, and 97–98% identity with PHYVV Sinaloa isolate for DNA B. In 2016, Pepper golden mosaic virus (designated PepGMV-CL) was also found that consisted of DNA-A and DNA-B genome displaying 97% and 93.5% identity with PepGMV isolate Tamaulipas. To our knowledge, this is the first report of a pepper disease associated with TYLCV in double or triple infection either with PHYVV and/or PepGMV in Mexico.
Résumé
En 2014, de nouvelles maladies ont été découvertes dans les plantations de piments de la région de La Comarca Lagunera (CL), maladies dont, en octobre 2016, la gravité s’était accrue. Les plants de piments affichaient des formes bénignes et graves de la mosaïque jaune ainsi que de la déformation, du rabougrissement et de la chlorose des feuilles. De plus, des aleurodes ont été observés sur les plants symptomatiques, suggérant une possible étiologie des maladies à bégomovirus. Dans cette étude, des plants infectés naturellement ont été collectés durant trois années consécutives afin d’identifier le possible bégomovirus infectant les piments de CL. La détection par PCR utilisant des amorces dégénérées et spécifiques a révélé que 47 des 49 plants de piments étaient infectés par des bégomovirus, principalement en infection mixte. Les génomes entiers des bégomovirus ont été isolés à partir d’un plant de piment symptomatique typique, et deux clones pour chaque bégomovirus ont été entièrement séquencés en fonction de l’année de collection (2014 à 2016). L’analyse phylogénétique des génomes entiers des isolats des bégomovirus des plants de piments de CL a indiqué une forte homologie avec le virus de la frisolée jaune de la tomate (désigné TYLCV-CL), affichant une identité de 99 à 100% avec l’isolat du TYLCV du Sinaloa et le virus biparti de la rhizomanie du piment huasteco (désigné PHYVV-CL), affichant une identité de 94.5 à 84.2% d’abord avec l’isolat du PHYVV de Tamaulipas quant à l’ADN A et l’ADN B, respectivement, puis, une identité de 97 à 98% avec l’isolat du PHYVV de Sinaloa quant à l’ADN B. En 2016, on a également découvert que le virus de la mosaïque dorée du piment (désigné PepGMV-CL) consistait en un génome d’ADN A et d’ADN B affichant une identité de 97 et 93.5% avec l’isolat du PepGMV de Tamaulipas. À notre connaissance, il s’agit de la première mention, au Mexique, d’une maladie du piment associée au TYLCV dans une infection double ou triple avec soit le PHYVV ou le PepGMV.
Introduction
Pepper (Capsicum annuum L.) is an economically important crop in Mexico and comprises 1.93% of the country's worldwide exports (FAOSTAT, Citation2013). In 2015, the national pepper production was 2.7 million metric tons valued at US$1.25 billion (SIAP, Citation2015). The Comarca Lagunera (CL) is a growing economic region in Northern Mexico comprising counties of two states, namely Durango and Coahuila. Since 2014, farmers and small producers have described the emergence of virus-like diseases affecting pepper crop yields. In 2016, the increased severity of these pepper diseases and high populations of whitefly, as observed in the affected areas, has led to a yield reduction in pepper cultivation in CL.
Members of genus Begomovirus (Family Geminiviridae) are associated with different crop diseases, causing an enormous concern for global agriculture (Leke et al., Citation2015) especially under global warming that could alter the distribution of their insect vectors. In Mexico, a pepper disease named ‘rizado amarillo’ was initially described as the co-infection of Pepper huasteco yellow vein virus (PHYVV) and Pepper golden mosaic virus (PepGMV) (Garzón-Tiznado, Citation1993). Thereafter, both viruses were reported to infect pepper in several Mexican states, such as Guanajuato, Jalisco, Oaxaca, Queretaro, San Luis Potosi, Sinaloa, Sonora, Tamaulipas and Yucatán, causing severe reductions in pepper yield production (Garzón-Tiznado, Citation1993; Torres-Pacheco et al., Citation1993; Garzón-Tiznado et al., Citation2002; Méndez-Lozano et al., Citation2003; Rodelo-Urrego et al., Citation2015). An increase in sweet pepper and tomato disease was reported in Sinaloa state in the last few years, which was found to be associated with a new isolate of PHYVV (Melendrez-Bojorquez et al., Citation2016; Moreno-Félix et al., Citation2018).
Tomato yellow leaf curl virus (TYLCV), one of the most devastating begomoviruses affecting tomatoes worldwide, was first detected in Mexico in the Yucatán peninsula in 1999 (Ascencio-Ibáñez et al., Citation1999) and then reported in Sinaloa in tomato and tomatillo crops (Gámez-Jiménez, Citation2007; Gámez-Jimenez et al., Citation2009). Subsequently, it was found singly and in mixed infections with other begomoviruses, affecting mainly tomato plants in the states of Sonora and Tamaulipas (Hernández-Zepeda et al., Citation2007; Bañuelos-Hernández et al., Citation2012).
Infection of Capsicum sp. crops by TYLCV has been reported in several countries including southern Spain (Reina et al., Citation1999), Dominican Republic (Salati et al., Citation2002), Cuba (Quiñones et al., Citation2002), Jamaica (Roye et al., Citation1999) and Mexico; mixed infection of TYLCV with a bipartite begomovirus Tomato chino La Paz virus (ToChLPV) were reported in Baja California Sur (Cardenas-Conejo et al., Citation2010). The objective of this work was to detect and identify using molecular methods the begomoviruses associated with emerging diseases in pepper in CL, Mexico.
Materials and methods
Plant sampling
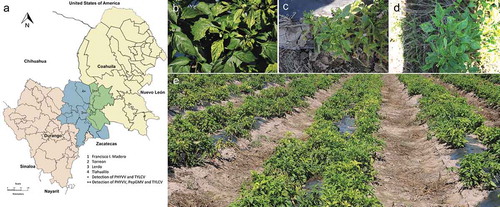
During 2014–2016, surveys for pepper diseases were carried out in six open fields in four counties of CL, according to the locations of pepper production during the year of survey (). Forty-nine mature pepper plants were collected in Torreón, Coahuila (12 in September 2014), Tlahualilo, Durango (three in May 2015), Lerdo, Durango (eight in May 2015 and 17 in October 2016) and Francisco I. Madero, Coahuila (nine in October 2016).
Fig. 1 (Colour online) Symptoms observed in pepper open fields in Comarca Lagunera (CL). (a) Map of CL which includes surveyed counties of Durango state (blue) and Coahuila state (green) of CL. (b) A pepper plant showing yellow mosaic, deforming and chlorotic leaf. (c) A stunted pepper plant with severe leaf deformation and yellowing. (d) A pepper plant with chlorotic leaf and yellow mosaic. (e) General view of pepper open field surveyed in 2016 with a 90% disease incidence caused by begomoviruses when double or triple infections were detected.
DNA isolation and PCR detection
Total genomic DNA was purified from leaves using the CTAB method (Doyle & Doyle, Citation1990). PCR was employed on the extracted DNA samples to determine the begomoviruses present using degenerate primers (Mauricio-Castillo et al., Citation2007). In addition, PCR using a set of primers specific for TYLCV, which amplify a 180 bp fragment (Rodríguez-Negrete et al., Citation2014), a set for PepGMV which amplifies a 120 bp fragment (Carrillo-Tripp et al., Citation2007) and for PHYVV, which amplifies a 161 bp fragment (Supplementary table), were performed to determine mixed viral infections.
Cloning and sequencing of viral DNA
In order to obtain the putative full-length begomovirus monomeric component (~2.7 kb fragment), total DNAs from representative pepper samples collected in 2014, 2015 and 2016 were amplified by rolling circle amplification (RCA) with Φ-29 DNA polymerase (TempliPhi, Ge Healthcare, USA) as described previously (Inoue-Nagata et al., Citation2004) or by a PCR strategy with overlapping primers for TYLCV and PHYVV DNA-A using high fidelity polymerase (iProofTM High-Fidelity DNA Polymerase, BIO-RAD®, USA). RCA amplification products were digested with BamHI, SacI, ApaI and EcoRI and cloned either into BamHI-digested pBluescript SK− vector (Agilent, USA) or SacI-, ApaI- and EcoRI-digested pGreen0029 vector (Hellens et al., Citation2000). The PCR amplified genomes of 2.7 kb were cloned into pGEM-T Easy Vector System® (Promega, USA) or NEB® PCR cloning kit (New England BioLabs, USA). Two independent clones for each viral component obtained from samples of the corresponding year were obtained and fully sequenced using the primer walking strategy. The assemblies of the sequences were obtained using the SeqMan program (DNASTAR Inc., Madison, USA), and genome comparisons were performed employing Mega 7.0 (Kumar et al., Citation2016). One sequence of each genome component per year was submitted to GenBank. Recombination analysis was performed in RDP4 with the default settings using all seven methods: RDP, GENCONV, BooScan, MaxChi, SiScan, Chimaera and 3Seq (Martin et al., Citation2015).
Results and discussion
Plant sampling and symptoms
In September 2014, a new viral disease was reported by the growers in a single pepper farm at Torreón, Coahulia and subsequently the same symptomatology was observed in May 2015, where 30% of open field pepper plants showed mild yellow mosaic, deformation and chlorotic leaves, and symptomatic plants were randomly distributed (). By October 2016, symptoms showed a dramatic increase in severity that included severe yellow mosaic, deformation and chlorotic leaves, and stunting that reduced the quality and yield to an extent that rendered farmers not able to harvest (, c, and d). The suspected viral disease incidence increased up to 90% and a high population of whitefly was observed in the surveyed pepper fields (). In order to determine the agent of the disease, a total of 49 symptomatic pepper leaf samples were collected in four counties of CL during 2014–2016, according to the pepper crop distribution in CL during the year of survey ().
Molecular detection and identification of begomoviruses in pepper samples
To investigate the presence of begomoviruses, DNA extracted from symptomatic plants were used as a template in PCR employing degenerate primers. The analysis confirmed the occurrence of begomoviruses in either single or mixed infections (data not shown). Complete begomoviruses genomes were obtained using either RCA or PCR amplification from representative pepper samples collected in 2014, 2015 and 2016. Two selected clones were fully sequenced per year, which were 99–100% identical to each other. Sequence analyses of selected genomes of the corresponding year indicated that isolates LV157-2014, LV447-2015 and LV56-2016 showed the arrangement of genes typical of the Old World monopartite begomoviruses; the sequences were submitted to GenBank (KX440610, KX440606 and MF945598) and designated as TYLCV-CL. Sequence analysis revealed that TYLCV-CL isolates showed the highest identity of 99.7–100% to TYLCV Sinaloa isolate (KU836749). In contrast, LV165/SacI-2014 and LV42-2016 isolates had a genome organization of a begomovirus DNA-A; the sequences were submitted to GenBank (KY247179 and MG582068) and designated PHYVV-CL DNA A. Isolates LV163/BamHI-2014, LV42/EcoRI-2016 had a genome organization of a begomovirus DNA-B; the sequences were submitted to GenBank (KX440614 and MG582069) and designated PHYVV DNA-B. Sequence analysis revealed that PHYVV-CL shared identity 94.5% for DNA-A and 84.2% for DNA–B to PHYVV isolate Tamaulipas DNA A and B (X70418 and X70419), respectively. Interestingly, PHYVV-CL showed 95% and 98% to PHYVV isolate Sinaloa for DNA A and DNA B (KP890827 and KP890828), respectively. Finally, isolate LV46/EcoRI-2016 had a genome organization of a begomovirus DNA-A, the sequence was submitted to GenBank (MF109819) and designated PepGMV-CL DNA A; and LV46/ApaI-2016 isolate had a genome organization of a begomovirus DNA-B, the sequence was submitted to GenBank (MF109821) and designated PepGMV-CL DNA B. Genetic analysis revealed that PepGMV-CL shared 97% and 93.5% identity with PepGMV DNA-A (U57457) and DNA B (AF499442), respectively.
Genome analysis confirms TYLCV, PHYVV and PepGMV associated with pepper disease in CL
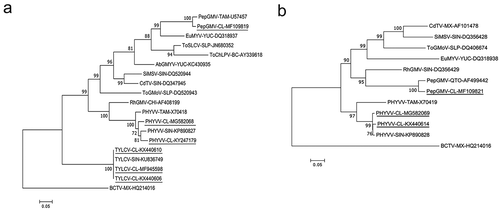
Phylogenetic analysis of the complete genomes of TYLCV-CL, PHYVV-CL and PepGMV-CL with selected begomovirus sequences available in GenBank showed the highest nucleotide sequence identity with TYLCV, PHYVV and PepGMV as described above (). The TYLCV-CL genome analysis indicates a nucleotide identity above 99% to the closest TYLCV reported in Sinaloa, Mexico, suggesting that this virus remains genetically stable in a new agro region. To date, it is not known to what extent TYLCV-CL contributes to the pepper infections since the Capsicum species has been reported as asymptomatic to single infections with TYLCV (Morilla et al., Citation2005; Polston et al., Citation2006; Kil et al., Citation2014). Further study is needed to determine the role of TYLCV-CL in single or mixed infections with PHYVV-CL and/or PePGMV-CL in pepper hybrids cultivated in Mexico. Based on genome sequence analysis of PHYVV-CL DNA-A and DNA-B, PHYVV-CL DNA-B (isolates LV165/BamHI-2014 and LV46/EcoRI-2016) had low identity (84.2%) when compared with PHYVV DNA B Tamaulipas (Garzón-Tiznado, Citation1993), whereas identity to PHYVV DNA B Sinaloa isolate (Melendrez-Bojorquez et al., Citation2016) was 98% (). It has been suggested that the DNA B component contributed significantly to symptom severity in cassava (Patil & Fauquet, Citation2015) and this may be the case for the previously described PHYVV-Sin and for PHYVV-CL DNA B obtained from CL reported in this work. A recombination analysis of PHYVV-CL DNA-B component was performed, but no recombination events were detected based on the parameters used in this study, suggesting that DNA B of PHYYV-CL isolate evolved by a different mechanism perhaps due to accumulation of point mutations.
Fig. 2 Phylogenetic trees based on multiple sequence alignment of the complete genome components. (a) DNA-A. (b) DNA-B. Phylogenetic trees were constructed using maximum likelihood method. Numbers represent bootstrap percentages values out of 1000 replicates using MEGA 7. Nodes with credibility values of 65% or high are shown. Beet curly top virus (BCTV) was used as a root. Sequences of begomoviruses isolated in this work are underlined. Acronym of virus sequences were as follows: Pepper golden mosaic virus (PepGMV), Pepper huasteco yellow vein virus (PHYVV), Tomato yellow leaf curl virus (TYLCV), Rynchosia golden mosaic virus (RhGMV), Chino del tomate virus (CdTV), Sida mosaic Sinaloa virus (SiMSV), Tomato severe leaf curl virus (ToSLCV), Tomato chino La Paz virus (ToChLPV), Abutilon golden mosaic Yucatan virus (AbGMYV), Euphorbia mosaic virus (EuMYV), Tomato golden mottle virus (ToGMV). The abbreviations after the virus acronyms represent the states of Mexico where the isolates were obtained: SIN (Sinaloa), CHI (Chiapas), YUC (Yucatan), SLP (San Luis Potosí), BC (Baja California), SON (Sonora), QTO (Querétaro), TAM (Tamaulipas), MX (No knowledge of the state of collection) and CL (La Comarca Lagunera). GenBank accession numbers are shown beside the names of each isolate.
Begomoviruses mixed infection associated with pepper diseases
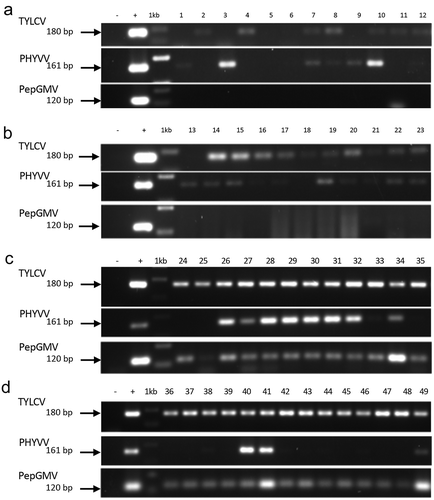
In order to individually analyze naturally infected pepper plants from CL, samples from the 2014, 2015 and 2016 crops were evaluated for the presence of TYLCV-CL, PHYVV-CL and PepGMV-CL that were previously identified. PCR specific detection confirmed the presence of begomoviruses in 47 out of 49 pepper plant samples with mixed infection being common (). In 2014, 10 out of 12 plants were positive for a single infection of TYLCV on 3 plants (; lanes 2, 4 and 11), PHYVV on 3 plants (; lanes 1, 3 and 9) or mixed infection with both viruses on 4 plants (; lanes 7, 8, 10 and 12). In 2015, mixed infections with TYLCV and PHYVV were detected on 9 out of 11 plants (; lanes 14–17 and 19–23), whereas single infections with TYLCV or PHYVV were detected only in one plant for each virus (; lanes 13 and 18). In 2016, PepGMV was also detected in addition to TYLCV and PHYVV, and it is tempting to propose that PepGMV-CL could be a possible factor contributing to observed symptom severity. Interestingly, mixed infection was also common in the 26 plants tested either with three viruses (TYLCV, PHYVV and PepGMV) in 13 plants ( and d; lanes 26–34, 38, 40–41 and 49) or with two viruses (TYLCV and PepGMV) in the remaining 13 plants (c and d; lanes 24–25, 35–39, 42–48). The constant detection of TYLCV suggested the prevalence and spread of this virus in CL. This is the first report of TYLCV in a new pepper-growing region in Mexico. It was postulated that once TYLCV is introduced in a new region, it will prevail as a source of emerging diseases (Rojas et al., Citation2005; Hoon et al., Citation2011; Lugo-Melchor et al., Citation2011; Yang et al., Citation2014). PHYVV and PepGMV have been well-documented affecting pepper crops in Mexico (Garzón-Tiznado, Citation1993; Méndez-Lozano et al., Citation2003; Holguín-Peña et al., Citation2004; Cardenas-Conejo et al., Citation2010; Ndunguru et al., Citation2015; Rodelo-Urrego et al., Citation2015; Melendrez-Bojorquez et al., Citation2016). In spite of this, CL region had no previous reports of the presence of TYLCV, PHYVV and PepGMV in pepper.
Fig. 3 Detection by PCR using specific primers for the begomoviruses TYLCV, PHYVV and PepGMV. (a) Samples collected in 2014 in CL open fields at Torreón, Coahuila. (b) Samples collected in 2015 in CL open fields at Tlahualilo, Durango. (c and d) Samples collected 2016 in CL open fields at Francisco I. Madero, Coahuila. Negative control (−), DNA of PHYVV (+) as positive control, 1Kb molecular maker (Invitrogen, USA).
Mixed infections of begomoviruses are currently associated with a more severe disease in pepper; it is well-documented that in the case of PHYVV and PepGMV, a synergistic effect increases the disease severity in pepper (Méndez-Lozano et al., Citation2003). Mixed infections offer the opportunity to begomoviruses to evolve through recombination, resulting in novel pathogenic phenotypes similar to the recombination of TYLCV and TYLCSV documented in tomato (Monci et al., Citation2002; Lefeuvre & Moriones, Citation2015). Pepper diseases are of great concern to farmers in CL since the severity of the disease has increased in the last few years. The first step to viral disease management is the identification of the virus or viruses causing the disease. In this study, molecular identification was done in pepper samples, and our findings indicated the occurrence of TYLCV with either PHYVV and/or PepGMV in mixed infection on pepper in CL, a finding reported for the first time in Mexico. The potential of viruses to evolve under mixed infections is always a risk; a follow-up in pepper-begomovirus pathosystems to check whether novel recombinant begomoviruses will develop is intriguing from an evolutionary point of view.
Supplementary_table_february_2019.docx
Download MS Word (16.4 KB)Acknowledgements
We would like to thank Instituto Politécnico Nacional (SIP 20164812) and Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico) for grant PDCPN No. 214950, for financial support of this research. JJMA and CALL were supported by a fellowship from CONACyT, Mexico and BEIFI programme of IPN.
Supplemental data
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2019.1591509
Additional information
Funding
References
- Ascencio-Ibáñez JT, Diaz-Plaza R, Méndez-Lozano J, Monsalve-Fonnegra ZI, Argüello-Astorga GR, Rivera-Bustamante RF. 1999. First report of Tomato yellow leaf curl Geminivirus in Yucatán, México. Plant Dis. 83(12):1178.
- Bañuelos-Hernández B, Mauricio-Castillo JA, Cardenas-Conejo Y, Guevara-González RG, Arguello-Astorga GR. 2012. A new strain of tomato severe leaf curl virus and a unique variant of Tomato yellow leaf curl virus from Mexico. Arch Virol. 157(9):1835–1841.
- Cardenas-Conejo Y, Argüello-Astorga GR, Poghosyan A, Hernandez-Gonzalez J, Lebsky V, Holguin-Peña J, Medina-Hernandez D, Vega-Peña S. 2010. First report of Tomato yellow leaf curl virus co-infecting pepper with Tomato chino La Paz virus in Baja California Sur, Mexico. Plant Dis. 94(10):1266.
- Carrillo-Tripp J, Lozoya-Gloria E, Rivera-Bustamante RF. 2007. Symptom remission and specific resistance of pepper plants after infection by Pepper golden mosaic virus. Phytopathology. 97:51–59.
- Doyle JJ, Doyle JL. 1990. Isolation of plant dna from fresh tissue. Focus. 12(13):39–40.
- FAOSTAT. 2013. Food and Agriculture Organization of the United Nations; [accessed 2017 Dec 11]. http://www.fao.org/statistics/en/.
- Gámez-Jiménez C 2007. Identificacion y caracterización molecular del Virus del enrollamiento de la hoja amarilla del tomate (TYLCV) y otros begomovirus asociados al cultivo de tomate en Sinaloa [Master’s thesis]. Guasave (Mexico): Instituto Politénico Nacional.
- Gámez-Jimenez C, Romero-Romero JL, Santos-Cervantes ME, Leyva-Lopez NE, Mendez-Lozano J. 2009. Tomatillo (Physalis ixocarpa) as a natural new host for Tomato yellow leaf curl virus in Sinaloa, Mexico. Plant Dis. 93(5):545.
- Garzón-Tiznado JA. 1993. Inoculation of peppers with infectious clones of a new Geminivirus by a biolistic procedure. Phytopathology. 83(5):514–521.
- Garzón-Tiznado JA, Acosta-García G, Torres-Pacheco I, González-Chavira M, Rivera-Bustamante RF, Maya-Hernández V, Guevara-González RG. 2002. Presencia de los Geminivirus, Huasteco del Chile (PHV), Texano del Chile variante Tamaulipas (TPV-T), y Chino del Tomate (VCdT), en los Estados de Guanajuato, Jalisco y San Luis Potosi, Mexico. Rev Mex Fitopatol. 20(1):45–52.
- Hellens RP, Edwards EA, Reyland NR, Bean S, Mullineaux PM. 2000. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 42:819–832.
- Hernández-Zepeda C, Idris AM, Carnevali G, Brown JK, Moreno-Valenzuela OA. 2007. Molecular characterization and phylogenetic relationships of two new bipartite begomovirus infecting malvaceous plants in Yucatan, Mexico. Virus Genes. 35(2):369–377.
- Holguín-Peña RJ, Vázquez-Juárez R, Rivera-Bustamante RF. 2004. Pepper golden mosaic virus affecting tomato crops in the Baja California Peninsula, Mexico. Plant Dis. 88:221.
- Hoon S, Oh S, Oh T, Sung J, Sei C, Hwan S, Shik Y, Kyu J, Sim S, Seo K, et al. 2011. Genetic diversity of tomato-infecting Tomato yellow leaf curl virus (TYLCV) isolates in Korea. Virus Genes. 42:117–127.
- Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T. 2004. A simple method for cloning the complete begomovirus genome using the bacteriophage Φ-29 DNA polymerase. J Virol Meth. 116:209–211.
- Kil EJ, Byun HS, Kim S, Kim J, Park J, Cho S, Yang DC, Lee KY, Choi HS, Kim JK, et al. 2014. Sweet pepper confirmed as a reservoir host for Tomato yellow leaf curl virus by both agro-inoculation and whitefly-mediated inoculation. Arch Virol. 159(9):2387–2395.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lefeuvre P, Moriones E. 2015. Recombination as a motor of host switches and virus emergence: geminiviruses as case studies. Curr Opin Virol. 10:14–19.
- Leke WN, Mignouna DB, Brown JK, Kvarnheden A. 2015. Begomovirus disease complex: emerging threat to vegetable production systems of West and Central Africa. Agric Good Secur. 4(1):1.
- Lugo-Melchor OY, Guzmán-Uriarte R, García-Estrada RS, León-Félix J. 2011. Geminivirus Transmitidos por mosca blanca (Bemisia tabaci) en tomate, en el valle agrícola de Cu, liacán, Sinaloa. Rev Mex Fitopatol. 29(2):109–118.
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. Rdp4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution. 1(1):vev003.
- Mauricio-Castillo JA, Argüello-Astorga GR, Ambriz-Granados S, Alpuche-Solís AG, Monreal-Vargas CT. 2007. First report of Tomato golden mottle virus on Lycopersicon esculentum and Solanum rostratum in Mexico. Plant Dis. 11(91):11–12.
- Melendrez-Bojorquez N, Rodríguez-Negrete EA, Magallanes-Tapia MA, Camacho-Beltran E, Armenta-Anaya C, Leyva-López NE, Mendez-Lozano J. 2016. Pepper huasteco yellow vein virus associated to sweet pepper disease in Sinaloa, Mexico. Plant Dis. 11(100):2338.
- Méndez-Lozano J, Torres-Pacheco I, Fauquet CM, Rivera-Bustamante RF. 2003. Interactions between Geminiviruses in a naturally occurring mixture: Pepper huasteco virus and Pepper golden mosaic virus. Phytopathology. 93(3):270–277.
- Monci F, Sánchez-Campos S, Navas-Castillo J, Moriones E. 2002. A natural recombinant between the Geminiviruses Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology. 303(2):317–326.
- Moreno-Félix ML, Rodríguez-Negrete EA, Meléndrez-Bojórquez N, Camacho-Beltrán E, Leyva-López NE, Méndez-Lozano J. 2018. A new isolate of Pepper huasteco yellow vein virus (PHYVV) breaks geminivirus tolerance in tomato (Solanum lycopersicum) comercial lines. Acta Hortic. 1207:35–44.
- Morilla G, Janssen D, Garcia-Andres S, Moriones E, Cuadrado IM, Bejarano ER. 2005. Pepper (Capsicum annuum) is a dead-end host for Tomato yellow leaf curl virus. Phytopathology. 95:1089–1097.
- Ndunguru J, Sseruwagi P, Tairo F, Stomeo F, Maina S, Djinkeng A, Kehoe M, Melcher U. 2015. Analyses of twelve new whole genome sequences of Cassava brown streak viruses and Ugandan cassava brown streak viruses from East Africa: diversity, supercomputing and evidence for further speciation. PLoS One. 10(10):1–18.
- Patil BL, Fauquet CM. 2015. Studies on differential behavior of cassava mosaic geminivirus DNA components, symptom recovery patterns, and their siRNA profiles. Virus Genes. 50(3):474–486.
- Polston JE, Cohen L, Sherwood TA, Ben-Joseph R, Lapidot M. 2006. Capsicum species: symptomless hosts and reservoirs of Tomato yellow leaf curl virus. Phytopathology. 96:447–452.
- Quiñones M, Fonseca D, Martinez Y, Accotto GP. 2002. First report of Tomato yellow leaf curl virus infecting pepper plants in Cuba. Plant Dis. 86:73.
- Reina J, Morilla G, Bejarano ER, Rodríguez MD, Janssen D. 1999. First report of Capsicum annuum plants infected by Tomato yellow leaf curl virus. Plant Dis. 83:1176.
- Rodelo-Urrego M, Garcia-Arenal F, Pagan I. 2015. The effect of ecosystem biodiversity on virus genetic diversity depends on virus species: a study of chiltepin-infecting begomoviruses in Mexico. Virus Evol. 1(1):vev004.
- Rodríguez-Negrete EA, Sánchez-Campos S, Cañizares MC, Navas-Castillo J, Moriones E, Bejarano ER, Grande-Pérez A. 2014. A sensitive method for the quantification of virion-sense and complementary-sense DNA strands of circular single-stranded DNA viruses. Sci Rep. 4:6438.
- Rojas A, Kvarnheden A, Marcenaro D, Valkonen JPT. 2005. Sequence characterization of Tomato leaf curl Sinaloa virus and tomato severe leaf curl virus: phylogeny of New World begomoviruses and detection of recombination. Arch Virol. 150(7):1281–1299.
- Roye ME, Wernecke ME, McLaughlin WA, Nakhla MK, Maxwell DP. 1999. Tomato dwarf leaf curl virus, a new bipartite geminivirus associated with tomatoes and peppers in Jamaica and mixed infection with Tomato yellow leaf curl virus. Plant Pathol. 48(3):370–378.
- Salati R, Nahkla MK, Rojas MR, Guzman P, Jaquez J, Maxwell DP, Gilbertson RL. 2002. Tomato yellow leaf curl virus in the Dominican Republic: characterization of an infectious clone, virus monitoring in Whiteflies, and identification of reservoir hosts. Phytopathology. 92(5):487–496.
- SIAP. 2015. Servicio de Información Agroalimentaria y Pesquera; [accessed 2017 Jun 20]. http://infosiap.siap.gob.mx/aagricola_siap_gb/icultivo/index.jsp
- Torres-Pacheco I, Garzon-Tiznado JA, Herrera-Estrella L, Rivera-Bustamante RF. 1993. Complete nucleotide sequence of Pepper huasteco virus: analysis and comparison with bipartite geminiviruses. J Gen Virol. 74(10):2225–2231.
- Yang X, Zhou M, Qian Y, Xie Y, Zhou X. 2014. Molecular variability and evolution of a natural population of Tomato yellow leaf curl virus in Shanghai, China. J Zhejiang Uni Sci B. 15(2):133–142.
