Abstract
The structure and diversity of wild grass spike-inhabiting Fusarium communities are not well understood. Fifteen common, non-cultivated grasses were surveyed across two years, regions, and land uses for spike-dwelling Fusarium spp. Eleven fungal species were identified from 857 isolates, including two, F. camptoceras and F. lactis, not recorded previously in New York state or on grass hosts. Species diversity and community structure varied by year and region. Land use and host community did not influence Fusarium communities, and no species-specific grass-Fusarium associations were detected. Fusarium communities were divided into two categories, those dominated by F. graminearum and those dominated by F. sporotrichioides. The community formation process is relevant to disease prediction and toxin monitoring in cropping systems as well as to land management practices at the intersection of agricultural and natural spaces.
Résumé
La structure et la diversité des communautés de Fusarium qui colonisent les épis de graminées sauvages ne sont pas bien comprises. Quinze espèces de graminées sauvages courantes ont été étudiées pendant deux ans dans diverses régions et sur des terres affectées à différentes utilizations afin d’y détecter des espèces de Fusarium qui colonisent leurs épis. Onze espèces fongiques ont été identifiées à partir de 857 isolats, y compris deux, F. camptoceras et F. lactis, à ce jour jamais trouvées dans l’État de New York ou sur des graminées hôtes. La diversité des espèces et la structure des communautés variaient en fonction des années et des régions. L’utilization des terres et la communauté d’hôtes n’ont pas influencé les communautés de Fusarium, et aucune association spécifique graminées-Fusarium n’a été détectée. Les communautés de Fusarium ont été divisées en deux catégories: celles dominées par F. graminearum et celles dominées par F. sporotrichioides. Le processus de formation des communautés est déterminant pour la prédiction des maladies et le suivi des toxines dans les cultures ainsi que pour la gestion des terres situées à la jonction de l’espace agricole et de l’espace naturel.
Introduction
Fusarium is a cosmopolitan genus containing many plant pathogen species with significant economic impacts (Booth Citation1971). A number of Fusarium spp. have wide host ranges that include non-cultivated grasses (Farr and Rossman Citation2019). While previous surveys have recorded the incidence of various Fusarium spp. in wild grasses (Inch and Gilbert Citation2003; Turkington et al. Citation2011; Szécsi et al. Citation2013; Lofgren et al. Citation2017) none have explicitly considered the influence of different environments or host plants on community composition. Because these fungal pathogens often damage staple crops, like wheat and maize, and contaminate grain with diverse mycotoxins (Marasas et al. Citation1984), the factors driving species diversity and structuring communities have relevance to disease prediction and monitoring in agricultural systems. Grasses growing in close proximity to crops may serve as pathogen reservoirs, contributing disease-inciting propagules and providing opportunities for survival between cropping cycles. Understanding the ecology of multi-host pathogens may also lead to land management practices that benefit natural host plants where agricultural and non-agricultural environments meet. The spillover of plant pathogens from one host to another can result in changes in host species abundance (Power and Mitchell Citation2004). Multiple grass inhabiting fusaria are capable of causing seedling blights and root rots in crops, and if Fusarium communities change as a result of proximity to agricultural production there may be an impact on natural host communities. Understanding grass-Fusarium community dynamics requires an understanding of the drivers of species diversity and community structure. In particular, the role of the environment and of the host community in shaping wild grass spike-inhabiting Fusarium communities are of interest because both are known to be important in agricultural contexts (Zhang et al. Citation2016; Bankina et al. Citation2017; Yang et al. Citation2018).
In this study, we leveraged the presence of diverse host communities found across New York State to: (i) compare Fusarium species diversity between regional and local environments; (ii) identify factors structuring these communities; (iii) and relate host communities to pathogen communities. Species diversity was expected to be highest in diverse plant communities remote from agricultural production, while proximity to agricultural production was expected to cause a decrease in species diversity and changes to community structure.
Materials and methods
Fungal cultures and species identification
The isolates used in this study were recovered in the course of a field survey recording Fusarium graminearum incidence in wild grasses (Fulcher et al. Citation2020). Briefly, non-cultivated grass spikes were collected in June 2016 and 2017 from 19 field sites in New York State (). Sampling was performed in two regions, Central and Northeastern New York, which differed in host density and level of agricultural production. Within each region, both agricultural fields and non-managed, natural spaces were sampled (). Only the Central region was sampled in 2016, while both regions were sampled in 2017. Plant tissue was taken from 1 m2 quadrats laid on transects following the margins of crop fields or along transects placed randomly in natural grass communities. Grass species richness was also recorded in quadrats at the time of sampling. The quadrats were separated by 10 m, and sites were no closer than 1 km from one another.
Fig. 1 Grass inflorescences were sampled from 19 locations over 2 years. Field sites were divided between two regions of New York State and two land-use categories: agricultural and natural
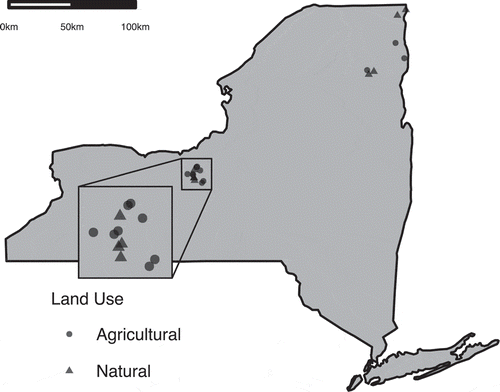
Table 1. Sampling depth over years, regions and land uses
Fusarium spp. colonies were recovered from surface-sterilized host tissue, and only a single fungal culture was saved from each grass spike. Single-spore derived isolates were stored as conidial suspensions at −80°C until further use. Fusarium graminearum was identified morphologically (Leslie and Summerell Citation2006). Taxonomic placement was confirmed for a subsample of these isolates using the molecular methods detailed below. All non-graminearum isolates were grown on potato dextrose agar for one week under 12-hr fluorescent light cycles at room temperature. Mycelia were scraped from the agar’s surface and placed into 2 mL microcentrifuge tubes with 1 g of garnet beads. Tubes were frozen at −20°C and tissue was ground using a Vortex-Genie 2 (Scientific Industries, Bohemia, NY, USA). DNA extraction proceeded using a commercial kit according to manufacturer instructions (DNeasy Plant Mini Kit, QIAGEN, Hilden, Germany). Taxonomic placement of isolates was based on partial sequences of either the translation elongation factor 1-alpha (TEF-1⍺) or RNA polymerase II subunit (RPB2) genes (Ward et al. Citation2002; O’Donnell et al. Citation2015; Lofgren et al. Citation2017). While a fragment of either gene is able to resolve Fusarium to the species complex level, the RPB2 locus was used for the majority of samples because PCR amplification was more consistent. Amplified DNA was visualized with gel electrophoresis, cleaned with a commercial silica spin column kit (QIAquick PCR Purification Kit, QIAGEN, Hilden, Germany; Monarch PCR & DNA Cleanup Kit, New England Biolabs, Ipswich, MA, USA), and submitted to the Cornell Biotechnology Resource Centre for sanger sequencing (ABI 3730xl, Applied Biosystems, Foster City, CA, USA). Sequences were managed in Geneious Prime version 2019.0.4 (Biomatters, Auckland, New Zealand), trimmed for quality, and compared to those from known specimens deposited in curated culture collections (e.g. CBS or NRRL) and type cultures accessioned in NCBI Genbank using BLAST (Altschul et al. Citation1990). Species or species complex identification was considered positive when sequence homology was ≥ 98% to a single taxonomic assignment.
Statistical analyses
All statistical analyses were performed in RStudio version 1.1.453 (RStudio Development Team, Citation2016). Individual Fusarium species incidence was analysed with a multinomial regression model using the ‘nnet’ package (Venables and Ripley Citation2002). A full model was built using year, region, land use, site, and grass host as predictors. Automated, stepwise model selection was performed to minimize Akaike information criterion (AIC) (Hastie and Pregibon Citation1992; Venables and Ripley Citation2002). The final model, with the lowest AIC, was subjected to analysis of variance to identify the variables that most effectively explained the data, and these were included in further analyses. The probability of each species occurring given region and land use was estimated with 95% confidence intervals using the ‘emmeans’ package (Lenth Citation2019).
Alpha diversity of Fusarium spp. was measured using Hill numbers, or effective species numbers (Gotelli et al. Citation2014). Rarefaction and estimation of effective species numbers (Hill numbers) at orders 0, 1, and 2 was performed using the ‘iNEXT’ package (Hsieh et al. Citation2016). Following Shapiro-Wilk tests for normal distributions (Royston Citation1992), mean Fusarium species numbers were contrasted between years, regions, and land uses with linear models and analysis of variance. Beta diversity was measured and analysed using the ‘vegan’ package (Oksanen et al. Citation2018). Community dissimilarity, using Bray-Curtis distances, was contrasted by year, region, and land use with a permutational multivariate analysis of variance. Dispersion within each group was assessed with analysis of variance to ensure any PERMANOVA significance was the result of differences in mean not dispersion (Anderson Citation2006). Community dissimilarity was also visualized in nonmetric multi-dimensional scaling plots using k = 2 dimensions.
Fusarium community structure was described with a principle components analysis of species abundance data and a Mantel test (using 10 000 random permutations) comparing community distance and physical distance matrices. The diversity and structure of grass communities were also examined using the above methods. The relationship between grass and Fusarium species was assessed by correlating their effective species numbers with a linear model and their community dissimilarities with a partial-Mantel test accounting for spatial autocorrelation. Preferential associations between grasses and Fusarium species were assessed with a permutation test using 10 000 simulations to generate a X2 null distribution.
Results
Species identification
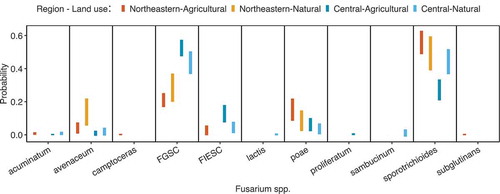
There were 51 samples discarded after DNA extraction, PCR amplification or sequencing failed. In total, 857 Fusarium isolates were identified as belonging to 11 species (). Partial gene sequences were deposited to NCBI GenBank (Accession nos.: MN013432-MN013439; MN183343-MN183753). Year (X2 = 167, P < 0.001), region (X2 = 240, P < 0.001), and land use (X2 = 29, P = 0.001) were retained in a multinomial model of species incidence with ρ2 = 0.180, indicating a good model fit (McFadden Citation1977). Sample site and grass host identity were discarded during the model selection step because they lacked power to explain Fusarium species occurrence. The differences in the probability of each species occurring in grass spikes given region and land use is shown in .
Table 2. Fusarium isolates recovered from grass species
Alpha and beta diversity of Fusarium communities
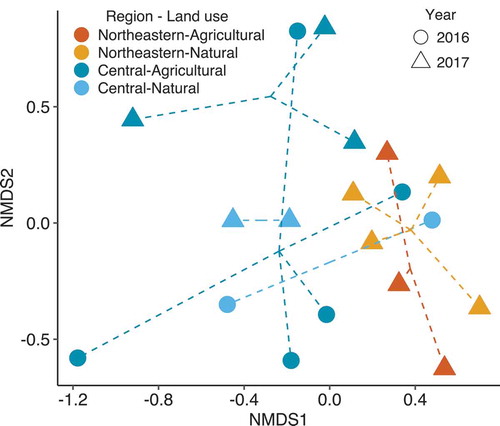
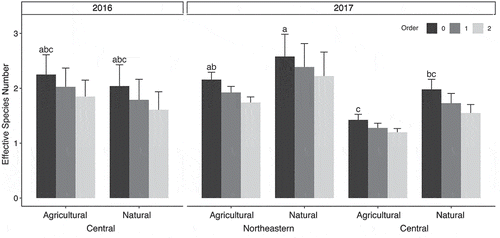
Effective species number (qD) for order 0 ranged from 1.34 to 3.00, with a mean of 2.12. Increasing to orders 1 and 2 resulted in slightly lower estimates of qD (). Effective species’ numbers were assumed normal following Shapiro-Wilk tests (P ≥ 0.307). Year (F1,18 ≥ 6.092, P ≤ 0.026) and region (F1,18 ≥ 11.499, P ≤ 0.004), but not land use (F1,18 ≤ 2.397, P ≥ 0.142), had significant effects on effective species number at all three orders (R2 = 0.400–0.441). Natural sites in Northeastern New York had the highest species diversity, while agricultural sites in Central New York had the lowest. Community dissimilarity was associated with differences between year (R2 = 0.167, F1,18 = 4.200, P = 0.004) and region (R2 = 0.182, F1,18 = 4.575, P = 0.003), but not land use (R2 = 0.540, F1,18 = 1.368, P = 0.236) according to PERMANOVA. Dispersion around group means did not vary for year (P = 0.415), region (P = 0.884) or land use (P = 0.180). An nMDS plot shows moderate differentiation for year and region ().
Fig. 3 Effective species, or Hill numbers (qD), were obtained by rarifying species abundance and compared across year, region, and land use. Fusarium species diversity differed significantly between years and regions but not between land uses. Filled bars represent mean values and error bars indicate standard deviation. Pairwise comparisons were conducted to identify differences in mean effective species number at each order of q, and Tukey’s Honest significant differences were calculated at alpha = 0.05. Bars of the same shade with the same letter were not significantly different
Fusarium community structure
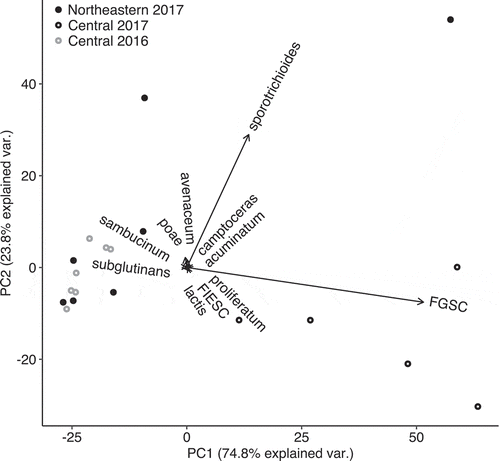
The principle components analysis showed nearly all variation in Fusarium populations was attributed to the relative abundance of F. graminearum and sporotrichioides, the two most frequently recovered species (). The communities found in Central New York during 2016 grouped together with the communities found in Northeastern New York during 2017. No spatial correlation in community dissimilarity was detected with a Mantel test (r = 0.106, P = 0.117).
Fig. 5 Principal components analysis of Fusarium species abundance at 19 sites. Community differences were attributable to large variation in the occurrence of two Fusarium species, sporotrichioides and graminearum. Central New York sites in 2016 experienced a drought, which led to communities more similar to those in low host density Northeastern New York during 2017 than to nearby communities in 2017 during a year of average rainfall
Relating Fusarium and grass communities
The effective species estimates for grass and Fusarium communities were not correlated (r = −0.13, P = 0.599 for qD of order 0). Community dissimilarity was not correlated according to a partial Mantel test (r = 0.56, P = 0.281). The X2 test failed to find differential association between Fusarium species and grass species (P = 0.061).
Discussion
This is the first study to catalogue Fusarium species diversity in non-cultivated grasses in New York, including those found in the Adirondack Mountains’ wilderness and a 4,000 hectare national wildlife refuge. The geographic or host ranges of several uncommon Fusarium spp. were expanded by this study. Our findings also show communities are likely structured by annual and regional factors, rather than the local environment or host community. This contrasts with findings for soil-borne Fusarium communities, which vary significantly between hosts (LeBlanc et al. Citation2017), and does not support our hypothesis of positive correlation between fungal pathogen diversity and grass diversity, which has been shown for foliar fungal pathogens (Rottstock et al. Citation2014). Our results may help predict pathogen population diversity and, by extension, toxin potential in different environments, specifically those with varying levels of rainfall and host density, which is potentially useful for monitoring disease and toxin content in crops. The findings also suggest altered land management practices at the intersection of crop and natural host communities, like adding buffer strips or restricting crop production, would have little influence on pathogen populations in nearby grasses.
The most common Fusarium species, such as F. graminearum and sporotrichioides, were well distributed across hosts, evidenced by a lack of any particular host-Fusarium associations. Most of the Fusarium species recorded here have previously been reported from numerous hosts, particularly plants in the true grass family, Poaceae (Farr and Rossman Citation2019). Culture-based approaches to fungal community surveys are limited in their depth and are likely to underestimate diversity. The results are inherently biased towards the most abundant species and species with high growth rates under the conditions imposed by isolation. However, even with the current approach, saving only a single isolate from each plant sample, singleton isolates were recovered that represent the first instances of certain species occurring in New York State or in association with a grass host. Fusarium camptoceras is most often found in association with rotting tropical fruits (Li-Sha et al. Citation2013; Abd Murad et al. Citation2017), but was once recorded from shattered wild rice (Zizania palustris) in Minnesota (Nyvall et al. Citation1999). This is the first recorded occurrence in New York and from smooth brome (Bromus inermis). Fusarium lactis causes internal fruit rots in sweet pepper and fig (Ploetz Citation2003; Van Poucke et al. Citation2012). This isolate is the first from any grass host (Phalaris arundinacea), and the first found in New York State.
Annual and regional environments were more important to Fusarium species diversity than local environments or grass communities. Despite not being informative at the community level, land use explained significant variation in the incidence of individual Fusarium spp. according to multinomial regression. The categorization of sites into two land uses, natural and agricultural, captured specific differences of interest related to disturbance and management practices, like residue removal, tillage, and crop rotation, but failed to account for many other differentiating factors. No attempt was made to quantify landscape features including non-host plant ground cover, elevation or land use in the immediate vicinity. It is likely that quantitative differences between field sites environments lead to changes in community that the present approach was unable to detect. Variation expected between year and region, for example rainfall and host density, are clearly important factors and may also vary on a field to field basis. Indeed, work focusing on F. graminearum has shown rainfall and host density are able to explain variation in pathogen incidence in grasses (Fulcher et al. Citation2020), and these factors were already recognized as driving crop disease epidemics (Leplat et al. Citation2013; Manstretta et al. Citation2016). The occurrence of various Fusarium species in crop hosts has also been related to regional differences in host community composition (Xu and Nicholson Citation2009), and the regions sampled in this study did vary in both host density and the ratio of agricultural to non-agricultural hosts. Within Fusarium communities, two species were dominant. Alongside F. graminearum, a number of less common pathogens or saprophytes were found. Environments more favourable to F. sporotrichioides also had a greater incidence of other crop pathogens, like F. avenaceum and poae. These differences in composition could be linked to individual life-histories. Because F. graminearum is homothallic, and the only species observed to reproduce sexually under natural conditions, it is more readily spread via airborne ascospores than other species capable only of producing asexual conidia. Conidia are splash or wind dispersed over short distances, while ascospores are capable of kilometre-scale movement (Paul et al. Citation2004; Maldonado-Ramirez et al. Citation2005; Keller et al. Citation2014).
It was surprising to find no spatial correlation in community dissimilarity matrices after seeing strong differentiation of communities based on regional location. The Mantel test used to assess spatial structure has been the subject of criticism in part because of its low power (Legendre et al. Citation2015). The Mantel test was unable to account for annual variation, which might mask the spatial variation in a rough analysis but is easily incorporated into linear models.
To summarize, this study recorded significant regional and annual variation in Fusarium communities and species diversity that was not influenced by local land use or individual host species. Two distinct communities, corresponding to a high and low incidence of the major agricultural pathogen F. graminearum, were detected and contained different species capable of producing distinct mycotoxin cocktails. Understanding when these different communities form will aid crop disease management and toxin monitoring efforts. The potential impact of these different communities on grasses in natural spaces should be further investigated, especially at the level of individual plant-fungus interactions.
Acknowledgements
The authors thank Extension Specialists K. Severson, M. Stanyard and K. O’Neil, C. Nobles and the Uihlein Seed Potato Farm, M. Davis and Willsboro Research Farm, L. Ziemba and the Montezuma National Wildlife Refuge, and the small grains producers who assisted with sampling.
Additional information
Funding
References
- Abd Murad NB, Mohamed Nor NMI, Shohaimi S, Mohd Zainudin NAI. 2017. Genetic diversity and pathogenicity of Fusarium species associated with fruit rot disease in banana across Peninsular Malaysia. J Appl Microbiol. 123:1533–1546. doi:10.1111/jam.13582.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. doi:10.1016/S0022-2836(05)80360-2.
- Anderson MJ. 2006. Distance-based tests for homogeneity of multivariate dispersion. Biometrics. 62:245–253. doi:10.1111/j.1541-0420.2005.00440.x.
- Bankina B, Bimšteine G, Neusa-Luca I, Roga A, Fridmanis D. 2017. What influences the composition of fungi in wheat grains? Acta Agrobot. 70:1–8. doi:10.5586/aa.1726.
- Booth C. 1971. The genus Fusarium. Wallingford (UK): Commonwealth Mycological Institute, Kew.
- Farr DF, Rossman AY. 2019. Fungal databases, U.S. National Fungus collections, ARS, USDA. [ accessed 2019 Apr 8]. https://nt.ars-grin.gov/fungaldatabases/.
- Fulcher MR, Winans JB, Quan M, Bergstrom GC. 2020. The incidence of Fusarium graminearum in wild grasses is associated with rainfall and cumulative host density in New York. Plant Disease. in press. doi:10.1094/PDIS-02-20-0286-RE
- Gotelli NJ, Hsieh TC, Sander EL, Colwell RK. 2014. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr. 84:45–67. doi:10.1890/13-0133.1.
- Hastie TJ, Pregibon D. 1992. Generalized linear models. In: Chambers JM, Hastie TJ, editors. Statistical models in S. London (UK): Chapman & Hall/CRC; p. 195–248.
- Hsieh TC, Ma KH, Chao A. 2016. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol. 7:1451–1456. doi:10.1111/2041-210X.12613.
- Inch S, Gilbert J. 2003. The incidence of Fusarium species recovered from inflorescences of wild grasses in southern Manitoba. Can J Plant Pathol. 25:379–383. doi:10.1080/07060660309507093.
- Keller MD, Bergstrom GC, Shields EJ. 2014. The aerobiology of Fusarium graminearum. Aerobiologia. 30:123–136. doi:10.1007/s10453-013-9321-3.
- LeBlanc N, Kinkel L, Kistler HC. 2017. Plant diversity and plant identity influence Fusarium communities in soil. Mycologia. 109:128–139. doi:10.1080/00275514.2017.1281697.
- Legendre P, Fortin MJ, Borcard D. 2015. Should the Mantel test be used in spatial analysis? Methods Ecol Evol. 6:1239–1247. doi:10.1111/2041-210X.12425.
- Lenth R 2019. Estimated Marginal means, aka least-squares means. R package version 1.3.4.
- Leplat J, Friberg H, Abid M, Steinberg C. 2013. Survival of Fusarium graminearum, the causal agent of Fusarium head blight. A review. Agron Sustain Dev. 33:97–111. doi:10.1007/s13593-012-0098-5.
- Leslie JF, Summerell BA. 2006. The Fusarium laboratory manual. Oxford: Blackwell Publishing.
- Li-Sha Z, Zhi-Hui Z, Shun L, Zhuo-Jun X, Li MH, Ping-Gen X, Zi-De J. 2013. The Fusarium species isolated from banana and their phylogenetic relationships. Mycosystema. 3232:617–632.
- Lofgren LA, LeBlanc NR, Certano AK, Nachtigall J, LaBine KM, Riddle J, Broz K, Dong Y, Bethan B, Kafer CW, et al. 2017. Fusarium graminearum: pathogen or endophyte of North American grasses? New Phytol. 217:1203–1212. doi:10.1111/nph.14894.
- Maldonado-Ramirez SL, Schmale DG, Shields EJ, Bergstrom GC. 2005. The relative abundance of viable spores of Gibberella zeae in the planetary boundary layer suggests the role of long-distance transport in regional epidemics of Fusarium head blight. Agric For Meteorol. 132:20–27. doi:10.1016/j.agrformet.2005.06.007.
- Manstretta V, Morcia C, Terzi V, Rossi V. 2016. Germination of Fusarium graminearum ascospores and wheat infection are affected by dry periods and by temperature and humidity during dry periods. Phytopathology. 106:262–269. doi:10.1094/PHYTO-05-15-0118-R.
- Marasas WFO, Nelson PE, Tousser TA. 1984. Toxigenic Fusarium species: identification and mycotoxicology. University Park: The Pennsylvania State University Press.
- McFadden D 1977. Quantitative methods for analysing travel behacious of individuals: some recent developments. Cowles Foundation Discussion Paper No. 474. New Haven: Yale University.
- Nyvall RF, Percich JA, Mirocha CJ. 1999. Fusarium head blight of cultivated and natural wild rice (Zizania palustris) in Minnesota caused by Fusarium graminearum and associated Fusarium spp. Plant Dis. 83:159–164. doi:10.1094/PDIS.1999.83.2.159.
- O’Donnell K, Ward TJ, Robert VARG, Pedro WC, Geiser DM, Seogchan K. 2015. DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica. 43:583–595. doi:10.1007/s12600-015-0484-z.
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, et al. 2018. Vegan: community ecology package. R package version 2.5.
- Paul PA, Lipps PE, Madden LV. 2004. Rain splash dispersal of Gibberella zeae within wheat canopies in Ohio. Phytopathology. 94:1342–1349. doi:10.1094/PHYTO.2004.94.12.1342.
- Ploetz RC. 2003. Diseases of tropical fruit crops. Wallingford (UK): CABI Publishers.
- Power AG, Mitchell CE. 2004. Pathogen spillover in disease epidemics. Am Nat. 164:S79–S89. doi:10.1086/424610.
- Rottstock T, Joshi J, Kummer V, Fischer M, Rottstock T, Joshi J, Kummer V, Fischer M. 2014. Higher plant diversity promotes higher diversity of fungal pathogens, while it decreases pathogen infection per plant. Ecology. 95:1907–1917. doi:10.1890/13-2317.1.
- Royston P. 1992. Approximating the Shapiro-Wilk W-test for non-normality. Stat Comput. 2:117–119. doi:10.1007/BF01891203.
- RStudio Development Team. Version 1.1.453. 2016. RStudio.
- Szécsi Á, Magyar D, Tóth S, Szőke C. 2013. Poaceae:a rich source of endophytic fusaria. Acta Phytopathol Entomol Hungarica. 48:19–32. doi:10.1556/APhyt.48.2013.1.2.
- Turkington TK, Clear RM, Demeke T, Lange R, Xi K, Kumar K. 2011. Isolation of Fusarium graminearum from cereal, grass and corn residues from Alberta, 2001–2003. Can J Plant Pathol. 33:179–186. doi:10.1080/07060661.2011.560189.
- Van Poucke K, Monbaliu S, Munaut F, Heungens K, De Saeger S, Van Hove F. 2012. Genetic diversity and mycotoxin production of Fusarium lactis species complex isolates from sweet pepper. Int J Food Microbiol. 153:28–37. doi:10.1016/j.ijfoodmicro.2011.10.011.
- Venables WN, Ripley BD. 2002. Modern applied statistics with S. 4th ed. New York: Springer.
- Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci U S A. 99:9278–9283. doi:10.1073/pnas.142307199.
- Xu X, Nicholson P. 2009. Community ecology of fungal pathogens causing wheat head blight. Annu Rev Phytopathol. 47:83–103. doi:10.1146/annurev-phyto-080508-081737.
- Yang M, Zhang H, Kong X, Van Der Lee T, Waalwijk C, van Diepeningen A, Xu J, Chen W, Feng J. 2018. Host and cropping system shape the Fusarium population: 3ADON-producers are ubiquitous in wheat whereas NIV-producers are more prevalent in rice. Toxins. 10:115. doi:10.3390/toxins10030115.
- Zhang H, Brankovics B, Luo W, Xu JS, Guo C, Guo JG, Jin SL, Chen WQ, Feng J, Van Diepeningen AD, et al. 2016. Crops are a main driver for species diversity and the toxigenic potential of Fusarium isolates in maize ears in China. World Mycotoxin J. 9:701–715. doi:10.3920/WMJ2015.2004.