Abstract
Powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt) is a major disease of wheat (Triticum aestivum) in Ontario, which can cause 20% yield loss. The development of resistant commercial wheat cultivars is the most economical means of controlling this disease, but only if the resistance genes used are incompatible with the virulence phenotypes present in the pathogen population. The virulence structure of Bgt in Ontario was examined in 2018 and 2019. Of the 42 single colony isolates collected in Ontario greenhouses and commercial fields, 40 virulence phenotypes, assigned VP1 to VP40, were identified on a set of 24 single-gene differential genotypes. Of the 24 resistance genes possessed by the differential genotypes, eight genes including Pm1a, Pm1b, Pm1c, Pm12, Pm16, Pm21, Pm37, and MlAG12 were effective against all of the Bgt isolates. Four genes including Pm3d, Pm29, Pm34, and NCAG13 were mostly effective with resistance reactions to more than 80% of the isolates. There were no significant differences in numbers of virulence genes per isolate between the two years, or between the greenhouse and field origins. The virulence frequencies of Bgt isolates for these effective and mostly effective genes were also not significantly affected by the year of collection and their origins, suggesting that the Bgt population is relatively stable. The effective genes identified in this study may be deployed singly or used for gene pyramiding in wheat breeding programs for developing powdery mildew-resistant cultivars in Ontario.
Résumé
Le blanc, causé par Blumeria graminis f. sp. tritici (Bgt), est une grave maladie du blé (Triticum aestivum) en Ontario qui peut engendrer des pertes de rendement de 20%. Le développement de cultivars de blé résistants est le moyen le plus économique de lutter contre cette maladie, mais seulement si les gènes de résistance utilisés sont incompatibles avec les phénotypes de virulence trouvés dans la population d’agents pathogènes. La structure de virulence de Bgt en Ontario a été étudiée en 2018 et 2019. Des 42 isolats à colonie unique collectés dans les serres et les champs cultivés commercialement en Ontario, 40 phénotypes de résistance, désignés de VP1 à VP40, ont été identifiés sur une série de 24 génotypes différentiels monogéniques. Des 24 gènes de résistance possédés par les différents génotypes, 8 gènes, y compris Pm1a, Pm1b, Pm1c, Pm12, Pm16, Pm21, Pm37 et MlAG12, étaient efficaces contre tous les isolats de Bgt. Quatre gènes, y compris Pm3d, Pm29, Pm34 et NCAG13, étaient généralement efficaces, résistant à plus de 80% des isolats. Il n’y a pas eu de différences significatives quant au nombre de gènes de virulence par isolat d’une année à l’autre ou quant à la provenance, soit les serres ou les champs. Les fréquences de virulence des isolats de Bgt pour ces gènes efficaces et généralement efficaces n’ont pas été sensiblement influencées non plus par l’année de collecte ni leur provenance, ce qui suggère que la population de Bgt est relativement stable. Les gènes efficaces identifiés dans le cadre de cette étude peuvent être déployés seuls ou utilisés pour accumulation pyramidale dans les programmes d’amélioration du blé afin de développer des cultivars résistants au blanc en Ontario.
Introduction
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is a destructive fungal disease of wheat (Triticum aestivum) worldwide (Cowger et al. Citation2012; Basandrai and Basandrai Citation2017). In the United States, yield losses of 13% to 34% due to powdery mildew have been reported in winter wheat (Johnson et al. Citation1979; Leath and Bowen Citation1989; Griffey et al. Citation1993), and in Canada yield losses of up to 20% have been described (Conner et al. Citation2003). In Ontario, where both winter wheat and spring wheat are grown, powdery mildew used to be considered an important disease of winter wheat only (Bailey and MacNeill Citation1983; Menzies and MacNeill Citation1986). The disease appears soon after the plants emerge in the fall and continues to affect the foliage throughout the spring and early summer of the subsequent year. Bgt infects wheat by either ascospores from overwintered-cleistothecia or conidia from diseased plants in Ontario or from neighbouring U.S. states. The spores of Bgt are spread by wind to growing plants in Ontario. Recent surveys on wheat diseases in Ontario have found that powdery mildew has become increasingly important in spring wheat (Xue and Chen Citation2016, Citation2018, Citation2019, Citation2020; Xue et al. Citation2017).
The control of powdery mildew on wheat in Ontario has been based on the use of resistant cultivars, crop rotation, and tillage in combination with fungicide applications when the environment was conducive to the disease (Brown et al. Citation2017). If prolonged wet, humid weather is forecast during the heading and flowering stages of wheat growth, foliar fungicides are recommended. However, fungicide application increases production costs to producers, in addition to the negative impact on the environment. The use of powdery mildew-resistant cultivars is the most effective disease control method and imperative for preventing pathogen outbreaks and increasing the profitability of wheat production in Ontario.
There are more than 90 powdery mildew-resistance genes, designated Pm1 to Pm68, and approximately 50 more newly identified genes with temporary designations reported in wheat and its relatives (Basandrai and Basandrai Citation2017; Li et al. Citation2019; He et al. Citation2021). However, wheat cultivars with known Pm genes are not available or the Pm genotype is not known in Ontario. Several cultivars including Benefit, OAC Flight, Priesley, Venture, Wentworth, WB425, 25R46, and 25R34 have shown some level of resistance to powdery mildew based on field observations, but there is no information on the type of resistance and possible Pm genes contained in these cultivars. This knowledge gap is largely due to the lack of information on the virulence structure of the Ontario Bgt population and the effectiveness of the available Pm genes in Ontario.
The increased occurrence and severity of powdery mildew in both winter and spring wheat in Ontario may be due to the defeat of certain Pm genes that have been widely deployed in commercial wheat production. Bgt is known for its highly pathogenic variability and new virulent strains may develop in response to the release of resistant cultivars (Cowger et al. Citation2012; Basandrai and Basandrai Citation2017). The new strains of Bgt with greater virulence complexity have not only shortened the effective life-span of these cultivars, but also have virulence against resistance genes that are not currently present in wheat cultivars or breeding lines, rendering these genes ineffective even before use in a breeding program (Niewoehner and Leath Citation1998; Imani et al. Citation2002). There have been no studies to examine the pathogenic variability of Bgt populations in Ontario or in any wheat-growing province of Canada for the past more than 30 years. The objectives of this study were to determine the virulence spectra of the Bgt population in Ontario and effectiveness of the available powdery mildew-resistance genes. This information is needed by wheat breeding programs and producers to deploy and pyramid Pm genes that are effective against the current Bgt population in Ontario.
Materials and methods
Collection and development of single-colony isolates
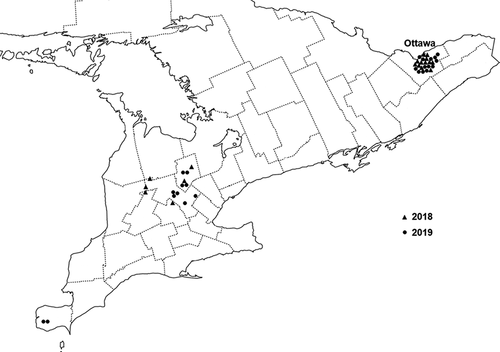
A total of 42 Bgt isolates were obtained from symptomatic wheat leaves in Ontario between 2018 and 2019. Of the 42 Bgt isolates, 12 were collected from infected plants in greenhouses at the Ottawa Research and Development Centre (ORDC) during December to April each year; and the remaining 30 isolates were collected from commercial fields and research plots of both spring and winter wheat across Ontario between May and July each year. Powdery mildew-resistance genes in the host crop of the sampled fields and research plots were not known and not expected to play a large role in the selection of Bgt isolates, given that most cultivars currently grown in Ontario have no effective Pm genes. The 30 field sites were chosen at random in densely cultivated regions in central and eastern Ontario where these crops are grown (). At each site, three symptomatic wheat plants were selected at random, and leaves with Bgt colonies were cut into 5-cm segments, and gently placed in a petri dish containing water agar (5% agar) amended with 50 mg L−1 of benzimidazole to delay leaf senescence (Imani et al. Citation2002). Within 24 h of the sample collection, the agar plates containing the infected leaf segments were transported in a cooler to the Crop Pathology Laboratory at ORDC, where they were placed in an incubator at 15°C with 12 h of artificial light (cool-white fluorescent lamps providing light at 40 ± 5 µmol m–2 s–1) for 2 d to promote sporulation.
Fig. 1 Location of origin of 42 isolates of Blumeria graminis f. sp. tritici collected in Ontario, Canada, in 2018 and 2019
To develop single-colony isolates, a 5-cm long piece was cut from the central part of a healthy, fully expanded primary leaf of 12-day-old Chancellor wheat, a susceptible cultivar that carries no effective resistance genes to powdery mildew (Srnic et al. Citation2005). The leaf fragments were rinsed with autoclaved sterilized water and placed with the adaxial surface up on plates containing water agar amended with benzimidazole as described above. These Chancellor wheat leaf fragments were then inoculated with Bgt conidia from a single isolated colony of each collected sample using a cooled sterile dissection needle. The inoculated leaf fragments were incubated in a chamber under the same conditions as described above for infection and disease development. One single-colony isolate was obtained from each sampling site and was transferred twice to ensure purity. These single-colony isolates were then maintained by transferring cultures to fresh leaves every 12 d until they were tested for virulence on a set of 24 selected single-gene powdery mildew differential genotypes.
Selection and growth of differential genotypes
To develop the powdery mildew differential set that is capable of detecting the virulence phenotypes of Bgt in Ontario, a small amount of seed of 67 wheat cultivars and isogenic lines, each with a reported resistance gene to powdery mildew, were obtained from the United States and China in 2017. Of these genotypes, 61 were accessions in the powdery mildew differential nursery maintained by the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) in Raleigh, NC, USA and the remaining six were provided by the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China. Seeds of the 67 wheat genotypes were increased in a growth room at ORDC in 2017 and stored at 4°C during the course of this study.
To prepare fresh leaves for inoculation with each Bgt isolate, the 67 wheat genotypes, along with Chancellor as the susceptible control, were grown in a growth room operated at 20°C with a 16-h photoperiod at a light intensity of 360 mol.m−2s−1 for 12 d. Seeds were surface-sterilized by soaking in 2% sodium hypochlorite for 5 min and washed thoroughly in autoclaved distilled water before each planting. Five seeds of each genotype were sown in 10-cm-diameter plastic pots containing autoclaved soil:peat:perlite mixture (1:1:1). After planting, the pots were covered with a clear plastic plant-propagation dome to avoid contamination by airborne pathogens. After emergence, plants were watered twice weekly from the bottom of the pots. The potting mixture contained sufficient nutrients to support healthy plant growth for the course of the experiments.
In a preliminary study on the development of the differential set, all 67 wheat genotypes were inoculated with six isolates of Bgt collected in 2018. Twenty-four wheat genotypes, excluding Pm1d and Pm1e, that showed resistant reactions to at least one of the six isolates were selected as the differential set, and the remaining genotypes were dropped from further testing (). Pm1d and Pm1e were not selected because they showed the same resistant response as Pm1a, Pm1b and Pm1c.
Table 1. List of 24 differential wheat genotypes and their resistance genes used to characterize the virulence structure of Blumeria graminis f. sp. tritici populations in Ontario, Canada, in 2018 and 2019
Inoculation and disease assessment
The virulence phenotypes of the 42 single-colony isolates on the 24 single-gene differential wheat genotypes were determined using the detached leaf segment method described above. For each isolate and differential genotype combination, inoculation tests were conducted on two replicate Petri dishes, each containing two 2.5-cm long leaf segments that were placed end-to-end for each differential genotype. One leaf segment of Chancellor was placed along each of the four sides within each Petri dish as susceptible controls (). To inoculate leaf segments with a Bgt isolate, a settling tower made from a 0.75-L plastic bottle (10 cm in diameter at the bottom, 13 cm high) with a 5-cm opening at the top was used to distribute conidia over the individual Petri dishes containing leaf segments of differential lines and the susceptible control. The settling tower was modified by Imani et al. (Citation2002). Once inoculated, the Petri dishes were placed in an incubator at 15°C with a 12-h photoperiod (40 mol.m−2s−1) for infection and disease development.
Fig. 2 (Colour online) Powdery mildew symptoms developed on leaf segments of wheat genotypes containing Pm1a (1), Pm1c (3), Pm1d (4), Pm1e (5), Pm2 (6), or no effective Pm (c) genes, 12 d after inoculation with a Blumeria graminis f. sp. tritici isolate from Virulence Phenotype 33
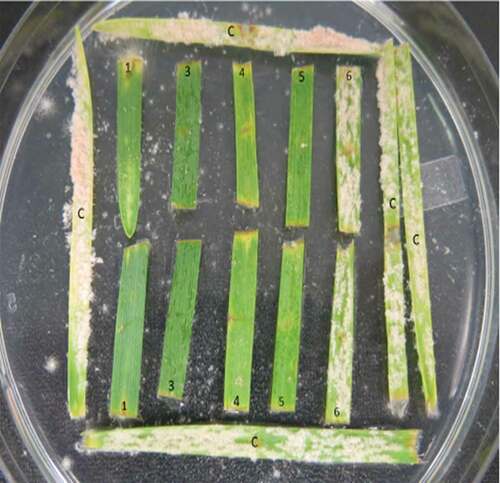
Leaf segments were assessed for infection type (IT) at 12 d after inoculation using the modified 0–9 scale of Parks et al. (Citation2008), where 0 = no visible symptoms; 1 = small flecks only; 2 = chlorotic lesions; 3 = chlorotic and necrotic lesions; 4 = chlorotic lesions with mycelium, conidia barely detectable; 5 = a few small to medium colonies with conidia, <10% of leaf area with symptoms (LAS); 6 = medium colonies with conidia, <20% of LAS; 7 = large colonies with conidia, 20–50% of LAS; 8 = large colonies with conidia, 50–75% of LAS; and 9 = large colonies with conidia, >75% of LAS. Infection types of 0, 1, 2, 3, and 4 were considered resistant reactions, and 5, 6, 7, 8 and 9 were considered susceptible reactions.
Statistical analyses
The data on numbers of virulence genes and frequencies of virulence of the Bgt isolates to the 24 single-gene differential wheat genotypes were subjected to the Student t-test to determine if there were significant differences between the two years of collection and between the greenhouse and field origins of these isolates. If the t-tests were significant (P < 0.05), means were separated by Fisher’s Least Significant Difference (LSD0.05). The analysis was conducted using the general linear model in SAS version 9.3 (SAS Institute Inc., Cary, NC, USA).
Results
There were 40 virulence phenotypes, assigned VP1 to VP40, identified from the 42 Bgt isolates analysed in 2018 and 2019 (). Of these virulence phenotypes, 17 were from 18 isolates collected in 2018 and 23 from 24 isolates collected in 2019. All virulence phenotypes were each detected in a single year and represented by a single isolate, except for VP20 and VP33, which comprised two isolates in 2019 and two isolates in 2018, respectively.
Table 2. Virulence phenotypes (VP) identified from 42 Blumeria graminis f. sp. tritici isolates collected from greenhouses (G) and commercial fields and research plots (F&P) in Ontario, Canada, in 2018 and 2019
The virulence complexities of the 40 Bgt phenotypes are presented in . The numbers of virulence genes per isolate ranged from 3 to 12 in 2018 and 2 to 11 in 2019. The mean number of virulence genes per isolate was 7.3 in 2018 and 6.2 in 2019, which was not significantly different (P = 0.121). Over the two years, the mean number of virulence genes possessed by the 12 isolates collected from greenhouses was 7.3 and that possessed by the 30 isolates from commercial fields was 6.4; that difference was also not significant (P = 0.261).
Virulence frequencies of Bgt isolates within the 24 single-gene differential genotypes are presented in . The frequencies of virulence to Pm4b, Pm25, and Pm32 were significantly less in 2019 as compared with 2018 (P < 0.05). The frequencies of virulence to Pm2, Pm4b, Pm30 and Pm32 were significantly less in commercial fields as compared with greenhouses; whereas, the opposite was observed for Pm3a, Pm5a, Pm34, and Pm35. Overall, eight genes (Pm1a, Pm1b, Pm1c, Pm12, Pm16, Pm21, Pm37, and MlAG12) were effective against all 42 Bgt isolates. Four genes (Pm3d, Pm29, Pm34, and NCAG13) were mostly effective, with resistant reactions to more than 80% of the isolates. The remaining 12 resistance genes were ineffective, having susceptible reactions to 23.8–100% of the Bgt isolates.
Table 3. Frequency of virulence of Blumeria graminis f. sp. tritici isolates collected from greenhouses and commercial fields in Ontario, Canada, in 2018 and 2019, on 24 single-gene powdery mildew differential lines
Discussion
The presence of 17 virulence phenotypes among 18 isolates collected in 2018 and 23 virulence phenotypes among 24 isolates collected in 2019 demonstrates a high degree of pathogenic variability of Bgt in Ontario. The results of this study are generally in agreement with the findings of Menzies et al. (Citation1989) who reported the existence of more than 30 phenotypes in the Bgt population in southern Ontario each year in 1985, 1986 and 1987. A similar high level of virulence phenotype diversity was observed in a large sample of the Bgt population in the US states that border Ontario (Cowger et al. Citation2016, Citation2018).
The present study used 24 single-gene differential genotypes, while only eight single-gene differentials had been used in the 1980s to determine the pathogenic variability of Bgt in Ontario (Bailey and MacNeill Citation1983; Menzies and MacNeill Citation1986; Menzies et al. Citation1989). As a result, eight effective genes and four mostly effective genes were identified in the present study (). Except for Pm1a, the effective and mostly effective Pm genes identified in this study were not included in the previous studies of Bgt population in Ontario or were not available at the time. It is worth noting that Pm1a, coded as Pm1 at the time (Parks et al. Citation2008; Hao et al. Citation2015), was also one of the three most effective genes in Ontario in the 1980s (Menzies and MacNeill Citation1986). The other two effective Pm genes in the 1980s were Pm3a and Pm3b. Gene Pm3a was ineffective in the present study (). Gene Pm3b was not included in the differential set due to its susceptible reactions to the first six Bgt isolates in the preliminary study. Pm3a and Pm3b were also considered defeated genes in the southern and eastern United States (Niewoehner and Leath Citation1998; Parks et al. Citation2008; Cowger et al. Citation2018).
Although shifts in virulence in Bgt populations which are characterized by the defeat of newly introduced resistance genes are commonly observed, some Pm genes have been shown to be more durable than others (Parks et al. Citation2008; Cowger et al. Citation2009, Citation2018). Of the eight effective genes identified for Ontario in the present study, Pm1a, Pm1b, and Pm1c were among the few Pm genes available during the 1980s and 1990s (Hsam et al. Citation1998; Szunics and Szunics Citation1999). Pm1a remained effective throughout the United States, despite sporadic regional deployment, until at least 2014 (Cowger et al. Citation2018), although it is ineffective in Western Australia, China, and Egypt (Zeng et al. Citation2014; Golzar et al. Citation2016; Abdelrhim et al. Citation2018). Pm1c, also known as Pm18 according to Wu et al. (Citation2019), was defeated in the United States (Li et al. Citation2016) but has remained effective in Nordic countries (Hysing et al. Citation2007) and in the northeast and northwest provinces of China (Wu et al. Citation2019). Genes Pm12, Pm16, Pm21, Pm37, and MlAG12, although not being widely deployed, have not been reported on their losses of resistance to virulent Bgt populations (Hysing et al. Citation2007; Li et al. Citation2016; Abdelrhim et al. Citation2018; Wu et al. Citation2019).
Breeding for powdery mildew-resistant wheat cultivars in Ontario and eastern Canada has started at ORDC in recent years, using Pm21 as a key source of resistance based on the findings of several studies demonstrating that Pm21 can provide robust and long-lasting resistance to powdery mildew (Chen et al. Citation1995; Huang et al. Citation1997; Czembor et al. Citation2014; Abdelrhim et al. Citation2018; Wu et al. Citation2019). Advanced elite lines with Pm21 have been selected from crossing programs using a Pm21-containing wheat line obtained from the Wheat Genetic Resources Centre of Kansas State University in the United States. New wheat cultivars containing Pm21 for powdery mildew resistance will hopefully be released in Ontario and Canada within 4–5 years. Results of the present study confirmed that Pm21 is among the eight effective genes for Ontario during 2018–2019. The identification of Pm21 and the additional seven effective genes and four mostly effective genes in the present study provides new information on selecting powdery mildew-resistance genes to be deployed or used for gene pyramiding in breeding programs in Ontario in the future. At the same time, research is underway at ORDC to screen the current cultivars and breeding lines in Ontario for possible effective Pm genes and for partial and adult-plant resistances to the natural Bgt populations under field conditions. It will be important to identify agronomically desirable germplasm that possesses quantitative powdery mildew resistance. Deploying Pm21 in such genotypes, whether alone or in combination with other Pm genes, will be desirable in order to maximize the durability of the major-gene resistance (Brun et al. Citation2010; Cowger et al. Citation2012, Citation2018) and mitigate the impact of a resistance breakdown should it occur.
Acknowledgements
We thank Emily Meyers, Mihaela Stanescu, Karla Delacerdacollazo, Yasamin Al-Rewashdy, Kyle Simon, Magean Ng, Ricardo Spada, Sarah Watt, John Matsoukas, and Erin Francispillai for technical assistance.
Additional information
Funding
References
- Abdelrhim A, Abd-Alla HM, Abdou E-S, Ismail ME, Cowger C. 2018. Virulence of Egyptian Blumeria graminis f. sp. tritici population and response of Egyptian wheat cultivars. Plant Dis. 102:391–397. doi:https://doi.org/10.1094/PDIS-07-17-0975-RE
- Bailey K, MacNeill BH. 1983. Virulence of Erysiphe graminis f. sp. tritici in southern Ontario in relation to vertical genes for resistance in winter wheat. Can J Plant Pathol. 5:148–153. doi:https://doi.org/10.1080/07060668309501616
- Basandrai AK, Basandrai D. 2017. Powdery mildew of wheat and its management. In: Singh DP, editor. Management of wheat and barley diseases. Waretown (NJ): Apple Academic Press; p. 133–182.
- Briggle LW, Sears ER. 1966. Linkage of resistance to Erysiphe graminis f. sp. tritici (Pm3) and hairy glume (Hg) on chromosome 1A of wheat. Crop Sci. 6:559–561. doi:https://doi.org/10.2135/cropsci1966.0011183X000600060017x
- Brown C, Follings J, Moran M, Rosser B. 2017. Agronomy guide for field crops. Toronto (ON): Publication 811, Ontario Ministry of Agriculture, Food and Rural Affairs.
- Brun H, Chèvre A-M, Fitt BDL, Powers S, Besnard A-L, Ermel M, Huteau V, Marquer B, Eber F, Renard M, et al. 2010. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 185:285–299. doi:https://doi.org/10.1111/j.1469-8137.2009.03049.x
- Chen PD, Qi LL, Zhou B, Zhang SZ, Liu DJ. 1995. Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet. 91:1125–1128. doi:https://doi.org/10.1007/BF00223930
- Conner RL, Kuzyk AD, Su H. 2003. Impact of powdery mildew on the yield of soft white spring wheat cultivars. Can J Plant Sci. 83:725–728. doi:https://doi.org/10.4141/P03-043
- Cowger C, Mehra L, Arellano C, Meyers E, Murphy JP. 2018. Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern United States. Phytopathology. 108:402–411. doi:https://doi.org/10.1094/PHYTO-06-17-0211-R
- Cowger C, Miranda L, Griffey C, Hall M, Murphy P, Maxwell J. 2012. Wheat powdery mildew. In: Sharma I, editor. Disease resistance in wheat. London (UK): CABI; p. 84–19.
- Cowger C, Parks R, Kosman E. 2016. Structure and migration in U.S. Blumeria graminis f. sp. tritici populations. Phytopathology. 106:295–304. doi:https://doi.org/10.1094/PHYTO-03-15-0066-R
- Cowger C, Parks R, Marshall D. 2009. Appearance of powdery mildew of wheat caused by Blumeria graminis f. sp. tritici on Pm17-bearing cultivars in North Carolina. Plant Dis. 93:1219. doi:https://doi.org/10.1094/PDIS-93-11-1219B
- Czembor HJ, Domeradzka O, Czembor JH, Mańkowski DR. 2014. Virulence structure of the powdery mildew (Blumeria graminis) population occurring on Triticale (Triticosecale) in Poland. J Phytopathol. 162:499–512. doi:https://doi.org/10.1111/jph.12225
- Golzar H, Shankar M, D’Antuono M. 2016. Responses of commercial wheat varieties and differential lines to western Australian powdery mildew (Blumeria graminis f. sp. tritici) populations. Austr Plant Pathol. 45:347–355. doi:https://doi.org/10.1007/s13313-016-0420-9
- Griffey CA, Das MK, Stromberg EL. 1993. Effectiveness of adult plant resistance in reducing loss to powdery mildew in winter wheat. Plant Dis. 77:618–622. doi:https://doi.org/10.1094/PD-77-0618
- Hao Y, Parks R, Cowger C, Chen Z, Wang Y, Bland D, Murphy JP, Guedira M, Brown-Guedira G, Johnson JW. 2015. Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theor Appl Genet. 128:465–476. doi:https://doi.org/10.1007/s00122-014-2445-1
- He H, Liu R, Ma P, Du H, Zhang H, Wu Q, Yang L, Gong S, Liu T, Huo N, et al. 2021. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor Appl Genet. 134:53–62. doi:https://doi.org/10.1007/s00122-020-03681-2
- He R, Chang Z, Yang Z, Yuan Z, Zhan H, Zhang X, Liu J. 2009. Inheritance and mapping of powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor Appl Genet. 118:1173–1180. doi:https://doi.org/10.1007/s00122-009-0971-z
- Heun M, Friebe B. 1990. Introgression of powdery mildew resistance from rye into wheat. Phytopathology. 80:242–245. doi:https://doi.org/10.1094/Phyto-80-242
- Heun M, Friebe B, Bushuk W. 1990. Chromosomal location of the powdery mildew resistance gene of Amigo wheat. Phytopathology. 80:1129–1133. doi:https://doi.org/10.1094/Phyto-80-1129
- Hsam SLK, Huang XQ, Ernst F, Hartl L, Zeller FJ. 1998. Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 5. Alleles at the Pm1 locus. Theor Appl Genet. 96:1129–1134. doi:https://doi.org/10.1007/s001220050848
- Hsam SLK, Lapochkina IF, Zeller FJ. 2003. Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 8. Gene Pm32 in a wheat-Aegilops speltoides translocation line. Euphytica. 133:367–370. doi:https://doi.org/10.1023/A:1025738513638
- Huang XQ, Hsam SLK, Zeller FJ. 1997. Identification of powdery mildew resistance genes in common wheat (Triticum aestivum L. em Thell.). IX. Cultivars, land races and breeding lines grown in China. Plant Breed. 116:233–238. doi:https://doi.org/10.1111/j.1439-0523.1997.tb00988.x
- Hysing S, Merker A, Liljeroth E, Koebner RMD, Zeller FJ, Hsam SLK. 2007. Powdery mildew resistance in 155 Nordic bread wheat cultivars and landraces. Hereditas. 144:102–119. doi:https://doi.org/10.1111/j.2007.0018-0661.01991.x
- Imani Y, Ouassou A, Griffey CA. 2002. Virulence of Blumeria graminis f. sp. tritici populations in Morocco. Plant Dis. 86:383–388. doi:https://doi.org/10.1094/PDIS.2002.86.4.383
- Johnson JW, Baenziger PS, Yamazaki WT, Smith RT. 1979. Effects of powdery mildew on yield and quality of isogenic lines of ‘Chancellor’ wheat. Crop Sci. 19:349–352. doi:https://doi.org/10.2135/cropsci1979.0011183X001900030018x
- Leath S, Bowen KL. 1989. Effects of powdery mildew, triadimenol seed treatment, and triadimefon foliar sprays on yield of winter wheat in North Carolina. Phytopathology. 79:152–155. doi:https://doi.org/10.1094/Phyto-79-152
- Li G, Cowger C, Wang X, Carver BF, Xu X. 2019. Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theor Appl Genet. 132:2625–2632. doi:https://doi.org/10.1007/s00122-019-03377-2
- Li G, Xu X, Bai G, Carver B, Hunger R, Bonman JM. 2016. Identification of novel powdery mildew resistance sources in wheat. Crop Sci. 56:1817–1830. doi:https://doi.org/10.2135/cropsci2015.09.0551
- Liu Z, Sun Q, Ni Z, Nevo E, Yang T. 2002. Molecular characterization of a novel powdery mildew resistance gene Pm30 in wheat originating from wild emmer. Euphytica. 123:21–29. doi:https://doi.org/10.1023/A:1014471113511
- Lukaszewski AJ, Cowger C. 2017. Re-engineering of the Pm21 transfer from Haynaldia villosa to bread wheat by induced homoeologous recombination. Crop Sci. 57:2590–2594. doi:https://doi.org/10.2135/cropsci2017.03.0192
- Maxwell JJ, Lyerly JH, Cowger C, Marshall D, Brown-Guedira G, Murphy JP. 2009. MlAG12: a Triticum timopheevii-derived powdery mildew resistance gene in common wheat on chromosome 7AL. Theor Appl Genet. 119:1489–1495. doi:https://doi.org/10.1007/s00122-009-1150-y
- McIntosh RA, Baker EP. 1970. Cytogenetical studies in wheat. IV. Chromosome location and linkage studies involving the Pm2 locus for powdery mildew resistance. Euphytica. 19:71–77. doi:https://doi.org/10.1007/BF01904668
- Menzies JG, MacNeill BH. 1986. Virulence of Erysiphe graminis f. sp. tritici in southern Ontario in 1983, 1984, and 1985. Can J Plant Pathol. 8:338–341. doi:https://doi.org/10.1080/07060668609501810
- Menzies JG, MacNeill BH, Peng G. 1989. Virulence spectrum of Erysiphe graminis f. sp. tritici in southern Ontario in 1986 and 1987. Can J Plant Pathol. 11:148–152. doi:https://doi.org/10.1080/07060668909501130
- Miranda LM, Murphy JP, Marshall D, Cowger C, Leath S. 2007. Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L.). Theor Appl Genet. 114:1451–1456. doi:https://doi.org/10.1007/s00122-007-0530-4
- Miranda LM, Murphy JP, Marshall D, Leath S. 2006. Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor Appl Genet. 113:1497–1504. doi:https://doi.org/10.1007/s00122-006-0397-9
- Murphy JP, Navarro RA, Marshall D, Cowger C, Cox TS, Kolmer JA, Leath S, Gaines CS. 2007. Registration of NC06BGTAG12 and NC06BGTAG13 powdery mildew–resistant wheat germplasm. J Plant Regist. 1:75–77. doi:https://doi.org/10.3198/jpr2006.04.0244crg
- Niewoehner AS, Leath S. 1998. Virulence of Blumeria graminis f. sp. tritici on winter wheat in the eastern United States. Plant Dis. 82:64–68. doi:https://doi.org/10.1094/PDIS.1998.82.1.64
- Parks R, Carbone I, Murphy JP, Marshall D, Cowger C. 2008. Virulence structure of the eastern U.S. wheat powdery mildew population. Plant Dis. 92:1074–1082. doi:https://doi.org/10.1094/PDIS-92-7-1074
- Perugini LD, Murphy JP, Marshall D, Brown-Guedira G. 2008. Pm37, a new broadly effective powdery mildew resistance gene from Triticum timopheevii. Theor Appl Genet. 116:417–425. doi:https://doi.org/10.1007/s00122-007-0679-x
- Reader SM, Miller TE. 1991. The introduction into bread wheat of a major gene for resistance to powdery mildew from wild emmer wheat. Euphytica. 53:57–60. doi:https://doi.org/10.1007/BF00032033
- Sears ER, Briggle LW. 1969. Mapping the gene Pm1 for resistance to Erysiphe graminis f. sp. tritici on chromosome 7A of wheat. Crop Sci. 9:96–97. doi:https://doi.org/10.2135/cropsci1969.0011183X000900010033x
- Shi AN, Leath S, Murphy JP. 1998. A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology. 88:144–147. doi:https://doi.org/10.1094/PHYTO.1998.88.2.144
- Singh D, Park RF, McIntosh RA. 2001. Postulation of leaf (brown) rust resistance genes in 70 wheat cultivars grown in the United Kingdom. Euphytica. 120:205–218. doi:https://doi.org/10.1023/A:1017578217829
- Srnic G, Murphy JP, Lyerly JH, Leath S, Marshall DS. 2005. Inheritance and chromosomal assignment of powdery mildew resistance genes in two winter wheat germplasm lines. Crop Sci. 45:1578–1586. doi:https://doi.org/10.2135/cropsci2004.0530
- Szunics L, Szunics LU. 1999. Wheat powdery mildew resistance genes and their application in practice. Acta Agro Hung. 47:69–89.
- Wu XX, Xu XF, Ma DX, Chen RZ, Li TY, Cao YY. 2019. Virulence structure and its genetic diversity analyses of Blumeria graminis f. sp. tritici isolates in China. BMC Evol Biol. 9:183. doi:https://doi.org/10.1186/s12862-019-1511-3
- Xue AG, Chen Y. 2016. Diseases of spring wheat in central and eastern Ontario in 2015. Can Plant Dis Surv. 96:134–135.
- Xue AG, Chen Y. 2018. Diseases of spring wheat in central and eastern Ontario in 2017. Can Plant Dis Surv. 98:148–149.
- Xue AG, Chen Y. 2019. Diseases of spring wheat in central and eastern Ontario in 2018. Can J Plant Pathol. 41(Suppl.):115–116.
- Xue AG, Chen Y. 2020. Diseases of spring wheat in Ontario in 2019. Can J Plant Pathol. 42(Suppl.):92–93.
- Xue AG, Chen Y, Al-Rewashdy Y. 2017. Diseases of spring wheat in central and eastern Ontario in 2016. Can Plant Dis Surv. 97:148–149.
- Yi YJ, Liu HY, Huang XQ, An LZ, Wang F, Wang XL. 2008. Development of molecular markers linked to the wheat powdery mildew resistance gene Pm4b and marker validation for molecular breeding. Plant Breed. 127:116–120. doi:https://doi.org/10.1111/j.1439-0523.2007.01443.x
- Zeller FJ, Kong L, Hartl L, Mohler V, Hsam SLK. 2002. Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.) 7. Gene Pm29 in line Pova. Euphytica. 123:187–194. doi:https://doi.org/10.1023/A:1014944619304
- Zeller FJ, Lutz J, Stephan U. 1993. Chromosome location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L.) 1. Mlk and other alleles at the Pm3 locus. Euphytica. 68:223–229. doi:https://doi.org/10.1007/BF00029876
- Zeng FS, Yang LJ, Gong SJ, Shi WQ, Zhang XJ, Wang H, Xiang LB, Xue MF, Yu DZ. 2014. Virulence and diversity of Blumeria graminis f. sp. tritici populations in China. J Integr Agric. 13:2424–2437. doi:https://doi.org/10.1016/S2095-3119(13)60669-3
