Abstract
Begomoviruses (Geminiviridae) cause severe diseases in several crops, often causing significant losses due to reduced yield and quality. These viruses are easily transmitted by the whitefly (Bemisia tabaci) vector. Begomovirus-like symptoms were observed in wild poinsettia (Euphorbia cyathophora) and cultivated papaya (Carica papaya) plants in the Nizwa, Al-Dakhliya and Al-Batinah regions of Oman, where papaya is grown extensively. The aim of this study was to identify the suspected viral causal agent associated with yellow mosaic and leaf curl symptoms in the poinsettia and papaya plants. Symptomatic leaf samples were collected and subjected to DNA isolation followed by PCR amplification with begomovirus-coat protein specic primers. An amplicon of betasatellites was also amplified using betasatellite-specific primers. The amplicons were cloned and sequenced bi-directionally. The full-length begomoviral genome was determined to be 2766 and 2752 bp in length for the wild poinsettia and papaya isolates, respectively. The respective betasatellites were found to be 1354 and 1370 bp. Sequence analysis indicated that the papaya isolate, herein tomato yellow leaf curl virus (TYLCV)-Pap-Oman, shared 95.0% nucleotide identity with the wild poinsettia isolate, herein TYLCV-Poin-Oman, followed by 93.5% identity with TYLCV-Iran (GU076448). Phylogenetic analysis grouped TYLCV-Pap-Oman and TYLCV-Poin-Oman with TYLCV isolates previously reported from Iran, Saudi Arabia and Sudan, whereas the two betasatellites grouped them with TYLCB isolates previously reported from Oman. Based on the nucleotide sequence identity and phylogenetic tree analysis, the Begomovirus identified from papaya and wild poinsettia is considered an isolate of TYLCV from plant host species previously reported in the Arabian Peninsula.
Résumé
Les bégomovirus (Geminiviridae) causent de graves maladies à plusieurs cultures, entraînant souvent d’importantes pertes à cause de la baisse de rendement et de la dégradation de la qualité. Ces virus sont facilement transmis par la mouche blanche (Bemisia tabaci). Des symptômes semblables à ceux provoqués par les bégomovirus ont été observés chez le poinsettia sauvage (Euphorbia cyathophora) et la papaye cultivée (Carica papaya) dans les régions de Nizwa, d’Al-Dakhliya et d’Al-Batinah d’Oman où la papaye est cultivée intensivement. Le but de cette étude était d’identifier l’agent causal viral soupçonné, associé à la mosaïque jaune et aux symptômes de la frisolée du poinsettia et de la papaye. Des échantillons de feuilles symptomatiques ont été collectés et soumis à l’isolement de l’ADN suivi de l’amplification par PCR avec des amorces spécifiques de la protéine de l’enveloppe du bégomovirus. Un amplicon des betasatellites a également été amplifié avec des amorces spécifiques des betasatellites. Les amplicons ont été clonés et séquencés bidirectionnellement. La longueur totale du génome du bégomovirus mesurait de 2 766 à 2 752 bp chez les isolats de poinsettia sauvage et de papaye, respectivement. Les betasatellites correspondants mesuraient de 1 354 à 1 370 bp. L’analyse séquentielle a indiqué que l’isolat de la papaye, ci-après virus de la frisolée jaune de la tomate (TYLCV)-Pap-Oman, partageait 95,0% d’identité en nucléotides avec l’isolat du poinsettia sauvage, ci-après TYLCV-Poin-Oman, suivi d’une identité de 93,5% avec le TYLCV-Iran (GU076448). L’analyse phylogénétique a regroupé TYLCV-Pap-Oman et TYLCV-Poin-Oman avec les isolats du TYLCV rapportés auparavant en Iran, en Arabie saoudite et au Soudan, tandis que les deux betasatellites les ont regroupés avec les isolats du TYLCB rapportés auparavant à Oman. En se basant sur l’identité de séquence des nucléotides et l’analyse de l’arbre phylogénétique, le bégomovirus identifié chez la papaye et le poinsettia sauvage est considéré comme un isolat du TYLCV provenant de l’espèce de plantes hôtes rapportées auparavant dans la péninsule arabique.
Introduction
The whitefly transmitted begomoviruses (genus Begomovirus, family Geminiviridae) cause severe disease with yellow mosaic and leaf curl symptoms in many plant species worldwide (Varma et al. Citation2011). In the Arabian Peninsula, begomoviruses are among the most damaging viral pathogens of many cultivated and economically important crops, such as Amaranthus, chilli, cucurbits, mint, papaya and tomato, causing significant yield losses ranging from 10% to 100% at the end of the season (Sohrab Citation2017, Citation2020a, Citation2020b; Sohrab and Daur Citation2018). In Oman, ~130 ha is planted to papaya production, with an annual yield estimated at 20 tons of fruit, making it among the most important fruit crops. Papaya production is significantly affected by viral infection (Khan et al. Citation2012). In addition to cultivated vegetable and fruit species, begomoviruses affect some wild hosts including the wild poinsettia, E. heterophylla L. (Euphorbiacea) (Barreto et al. Citation2013). Wild poinsettia is native to North and South America and is naturalized in many locations worldwide, including Oman, where it becomes an invasive weed (Cerdeira et al. Citation2011; Chacalis Citation2015).
Begomoviruses have a circular, single-stranded DNA (ssDNA) with either a monopartite (DNA-A) or bipartite genome (DNA- A and DNA- B, each approximately 2.7kb), encoding a genome size of ~5.2 kb. Each genomic component is encapsulated in a geminate icosahedral particle. Currently, the family Geminiviridae has been divided into nine genera: Becurtovirus, Begomovirus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus Mastrevirus, Topocuvirus, and Turncurtovirus (Muhire et al. Citation2014; Brown et al. Citation2015; Zerbini et al. Citation2017). Begomviruses are transmitted only by the whitey Bemisia tabaci (Genn.), which is considered to be a species complex comprised cryptic species (De Moya et al. Citation2019). Monopartite begomovirus DNA-A components frequently associate with at least one or more satellites, known as betasatellites (~1.4 kb), or alphasatellites (~1.3 kb) (previously referred to as DNA 1) containing a single ORF (Rep). Both satellites require the ‘helper’ begomovirus for vector transmission and infection of the plant host. However, while betasatellites rely on the helper begomovirus for replication and systemic spread in the host plant, alphasatellites encode their own replicase, or Rep, which is reminiscent of nanoviruses, but requires the helper virus for systemic infection (Gnanasekaran et al. Citation2019).
The Arabian Peninsula and Southeastern Asia are considered the centres of diversification for at least five monopartite tomato-infecting begomoviruses. In Iran, at least five tomato yellow leaf curl virus (TYLCV) strains have now been described (Kenyon et al. Citation2014), and one or more have emerged as economically important viral pathogens in vegetables and tropical fruit crops such as papaya. At least one TYLCV strain was described from Iran, which was earlier reported to cause disease in papaya plants in the Al-Batinah region, and in Saudi Arabia (Sohrab Citation2020a) and Sudan (Idris and Brown Citation2005).
Here, we discuss the molecular diagnosis, full-length genome sequence analysis, and phylogenetic relationship of begomovirus associated with leaf curl and yellow mosaic disease of wild poinsettia and papaya plants from the Al-Dakhliya region of Oman. Based on the literature, published information and data generated in this study, this is the first report of the natural infection of TYLCV in poinsettia and papaya plants from Oman.
Materials and methods
Plant sample collection
During the field survey in Al-Dakhliya, in Southern Oman, and in Al-Batinah, in northern Oman, in August and September 2017, poinsettia plants with severe yellow vein mosaic symptoms () and papaya plants exhibiting severe leaf curling, green vein banding, thickening, and chlorosis were found (). The incidence of disease in papaya trees was very high. Approximately 70%-80% of trees showed leaf curl symptoms in the field. The characteristic begomovirus-like symptoms and presence of the whitefly vector, B. tabaci, were suggestive of begomovirus aetiology. Symptomatic leaf samples were collected from 10 randomly selected poinsettia and papaya plants and transported to the lab under controlled conditions for further processing.
DNA isolation and begomovirus-satellite detection
Total DNA was isolated from 10 symptomatic poinsettia and 10 papaya leaves using the method of Rouhibakhsh et al. (Citation2008). The purified DNA was used as a template for PCR amplification of begomoviral genome(s) using the nearly universal forward and reverse Begomovirus AVcore and ACcore primers, F-5ʹ-GCCHATRTAYAGRAAGCCMAGRAT-3ʹ and R-5ʹ-GGRTTDGARGCATGHGTACANGCC-3ʹ (Brown et al. Citation2015).
The betasatellites were amplied by PCR using theuniversal betasatellite primers, β01 F-5ʹ-AGCCTTAGCTACGCCGGAGC-3ʹ and β02 R-5ʹ- GCTGCGTAGCGTAGAGGTTT-3ʹ (Briddon et al. Citation2002). The PCR products were gel purified, ligated and cloned into the pGEM®-T Easy plasmid vector (Promega Corporation), and transformed into E.coli competent cells.
Cloning and sequencing of full-length begomoviral genomes and betasatellites
Rolling circle amplication (RCA), using a TempliPhi 100 Amplication Kit (GE Healthcare Life Sciences, Piscataway, NJ), was performed following the manufacturer’s instructions. Only one sample from each plant was used to perform RCA and the RCA product was linearized with BamHI endonuclease. The ~2.7-kb full-genome products were cloned into a pGreenII binary vector. The sequence of each clone was determined with bidirectional, automated Sanger dideoxy chain termination sequencing (Eton Biosciences, San Diego, CA). The DNA sequences were assembled, aligned, and edited using Lasergene software (DNASTAR, Madison, and WI). The sequences generated from both isolates (papaya and poinsettia) were first used to search for homology with BLAST, and the top 100 hits were selected for analysis based on their similarity with other viral isolates. Sequence analysis of the putative full-length begomoviral genome and betasatellite molecules were performed using the Sequence Demarcation Tool v. 1.2 (SDTv1.2) (Muhire et al. Citation2014).
Results and discussion
The results of the PCR amplification experiments indicated partial amplification of the begomoviral coat protein gene and full-length betasatellites fragments from the tested samples. The resultant betasatellites sequences from poinsettia and papaya plants were 1354 bp and 1370 bp, respectively. The generated sequences were initially searched using BLASTn and compared with the top 100 hits. The BLAST result showed 89.0–95.8% similarity for TYLCV-Poin-MH745750, 90.3–93.8% -TYLCV-beta-Poin, 93.0–97.5-TYLCV-Pap, and 88.1–92.8-TYLCV-Pap-beta with 0.0 e-values. Based on the sequence identity, the isolates were tentatively identified and designated as TYLCV-like isolates reported previously from Oman. However, this virus and associated betasatellites have not been reported infecting papaya or wild poinsettia in Oman.
The full-length viral genome was found to be 2766 bp and 2752 bp for the isolates from poinsettia and papaya, respectively. All the generated sequences have been deposited into GenBank with the accession numbers MH745750, MH745751 (full-length) and MH732725, MH732726 (betasatellites) for poinsettia and papaya, respectively, and tentatively designated as TYLCV-Poinsettia-Oman and TYCLV-Pap-Oman. presents the features of sequences generated in this study. The sequences of TYCLV-Pap-Oman isolate shared highest identity (95.0%) with TYLCV-Poin-Oman, followed by 93.5% shared identity with the TYLCV-Iran isolate previously reported from tomato (GU076448). The lowest nucleotide sequence identity (79.6%) was observed with the isolate TYLCV-Egypt (). The betasatellites sequence of the TYLCB-Pap-Oman isolate showed the highest nucleotide sequence identity (90.9%) with TYLCV-Poinsettia-Oman, while the lowest identity (43.3%) was observed with three isolates of ToLCYB reported from Yemen ().
Table 1. Features of begomovirus genomic components isolated, cloned and sequenced, from Euphorbia cyathophora and Carica papaya in Oman
Table 2. Sequence identity matrix of tomato yellow leaf curl betasatellite -Pap-Oman (MH732726) with selected betasatellites
Table 3. Sequence identity matrix of tomato yellow leaf curl virus-Pap-Oman (MH745751) with full genome of selected begomoviruses
The phylogenetic relationship with selected begomoviruses was analysed using full-length and betasatellites sequences. The phylogenetic tree contained the multiple clusters, and the TYLCV-Pap-Oman and TYLCV-Poinsettia formed a cluster with TYLCV isolates reported from Saudi Arabia, Iran, and Sudan (). The phylogenetic relationship analysis of betasatellite sequences showed close clustering with TYLCB isolates reported from Oman (). Based on the sequence identity and phylogenetic relationship analysis, the identied Begomovirus from both papaya and poinsettia was suspected to be an isolate of TYLCV reported earlier from the Arabian Peninsula.
Fig. 3 Phylogenetic tree based on complete genome sequences showing the relationships between tomato yellow leaf curl virus (TYLCV) isolates and revealing clades/sister.
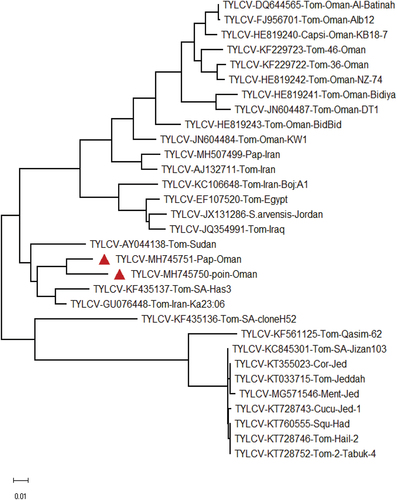
Fig. 4 Phylogenetic relationships of tomato yellow leaf curl virus betasatellite (TYLCB) with selected viruses based on betasatellite genomes.

Considering the results of this study, it is expected that the incidence of begomovirus infection may increase in other cultivated and wild crops. Cultivated crops and weed species in Oman are reported to be frequently infected with Begomovirus (Khan et al. Citation2014). The genetic diversity of full-length as well as betasatellite genomes compared with selected begomoviruses reported from the Arabian Peninsula also provide evidence for the emergence and spread of begomoviral disease to many crops/plants (Idris et al. Citation2012; Sohrab et al. Citation2016; Sohrab Citation2017; Sohrab and Daur Citation2018; Sohrab Citation2020a, Citation2020b). Several factors are relevant for the emergence and spread of the new begomovirus strains to economically important crops in the Arabian Peninsula. These factors include sequence variation caused by frequent recombination and mutation, vectors, climatic changes, and changes in the cropping system (Idris et al. Citation2012, Citation2014; Sohrab and Daur Citation2018; Sohrab Citation2020a, Citation2020b).
In the Al-Batinah region of Oman, leaf curl symptoms in papaya have been common since 1993 or earlier (Khan et al. Citation2014), and the Cotton leaf curl Gezira virus was found to be associated with the disease (Khan et al. Citation2012). Although the association of TYLCV with symptomatic papaya in Oman has not been previously reported, an isolate of TYLCV-IL has also been detected in papaya plants in Texas, USA (Alabi et al. Citation2017). Globally, many viruses infecting poinsettia and papaya species have been identied and published. As Oman is an international market-hub reachable by air and sea, the virus and/or the whitefly vector may have been introduced on infected/infested plants, and/or it may represent a previously unidentified, endemic isolate of TYLCV.
The information generated in this study showed the genetic diversity of begomoviruses infecting poinsettia and papaya in Oman. The sequence analysis results, using full-length and betasatellite genomes of isolates TYCLV-Pap-Oman and TYLCV-Poi-Oman, showed the highest identity with TYLCV-Iran isolate (GU076448). The phylogenetic relationship analysis showed variable clusters, and TYLCV-Pap and TYLCV-Poinsettia showed close clustering with TYLCV isolates reported from Saudi Arabia, Iran, and Sudan. The possibility of infection with other viruses on both plants cannot be overruled, and there may be infection with other viruses that should be examined and determined in Oman. The results of this study provided preliminary information regarding the infection by TYLCV of papaya and wild poinsettia plants in the Sultanate of Oman.
We conclude that the identified begomovirus causing disease in poinsettia and papaya could be a strain of tomato yellow leaf curl virus. The presence of other viruses in these crops requires further examination and detailed research work.
Acknowledgements
The authors are thankful to the Department of Biological Sciences and Chemistry, College of Arts and Sciences, University of Nizwa, for the support in completion of this work. The authors would like to thank and gratefully acknowledge the research facility provided by the School of Plant Sciences, University of Arizona, Tucson, USA.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alabi OJ, Al Rwahnih M, Jifon JL, Sétamou M, Brown JK, Gregg L, Park JW. 2017. A mixed infection of lettuce chlorosis virus, papaya ringspot virus and tomato yellow leaf curl virus-IL detected in a Texas papaya orchard affected by a virus-like disease outbreak. Plant Dis. 101(7):1094–1102. doi:https://doi.org/10.1094/PDIS-01-17-0118-RE.
- Barreto SS, Hallwass M, Aquino OM, Inoue-Nagata AK. 2013. A study of weeds as potential inoculum sources for a tomato-infecting begomovirus in central Brazil. Phytopath 103(5):436–444. doi:https://doi.org/10.1094/PHYTO-07-12-0174-R.
- Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. 2002. Universal primers for the PCR-mediated amplification of DNA β. Mol Biot. 20(3):315–318. doi:https://doi.org/10.1385/MB:20:3:315.
- Brown JK, Zerbini FM, Castillo JN, Moriones E, Sobrinho RR, Silva JCF, Olive EF, Briddon RW, Zepeda CHN, Idris A, et al. 2015. Revision of begomovirus taxonomy based on pair wise sequence comparisons. Arch Virol. 60(6):1593–1619. doi:https://doi.org/10.1007/s00705-015-2398-y.
- Cerdeira AL, Gazziero DL, Duke SO, Matallo MB. 2011. Agricultural impacts of glyphosate-resistant soybean cultivation in South America. J Agric Food Chem. 59(11):5799–5807. doi:https://doi.org/10.1021/jf102652y.
- Chacalis D. 2015. Wild poinsettia (Euphorbia heterophylla): an emerging weed in cotton and processing tomato in Greece. Hellenic Plant Prot J. 8(1):27–32. doi:https://doi.org/10.1515/hppj-2015-0005.
- De Moya RS, Brown JK, Sweet AD, Walden KKO, Paredes Montero JR, Waterhouse RM, Johnson KP. 2019. Nuclear orthologs derived from whole genome sequencing indicate cryptic diversity in the Bemisia tabaci (Insecta: aleyrodidae) complex of whiteflies. Diversity. 11(9):151. doi:https://doi.org/10.3390/d11090151.
- Gnanasekaran P, KishoreKumar R, Bhattacharyya D, Vinoth Kumar R, Chakraborty S. 2019. Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol Plant Path. 20(7):1019–1033. doi:https://doi.org/10.1111/mpp.12800.
- Idris AM, Abdullah NM, Brown JK. 2012. Leaf curl diseases of two Solanaceous species in Southwest Arabia are caused by a monopartite begomovirus evolutionarily most closely related to a species from the Nile Basin and unique suite of betasatellites. Virus Res. 169(1):296–300. doi:https://doi.org/10.1016/j.virusres.2012.07.014.
- Idris AM, Al-Saleh M, Piatek M, Al-Shahwan I, Ali S, Brown JK. 2014. Viral metagenomics: analysis of begomoviruses by illumina high-throughput sequencing. Viruses. 6(3):1219–1236. doi:https://doi.org/10.3390/v6031219.
- Idris AM, Brown JK. 2005. Evidence for interspecific recombination for three monopartite begomoviral genomes associated with the tomato leaf curl disease from central Sudan. Arch Virol. 150(5):1003–1012. doi:https://doi.org/10.1007/s00705-004-0484-7.
- Kenyon L, Tsai WS, Shih SL, Lee LM. 2014. Emergence and diversity of begomoviruses infecting Solanaceous crops in East and Southeast Asia. Virus Res. 186:104–113.
- Khan AJ, Akhtar S, Al-Shihi AA, Al-Hinai FM, Briddon RW. 2012. Identification of cotton leaf curl Gezira virus in Papaya in Oman. Dis Notes. 96:1704–1712.
- Khan AJ, Mansoor S, Briddon RW. 2014. Oman: a case for a sink of begomoviruses of geographically diverse origins. Trends in Plant Sci. 19(2):2–14. doi:https://doi.org/10.1016/j.tplants.2013.11.004.
- Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classication tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 9(9):e108277. doi:https://doi.org/10.1371/journal.pone.0108277.
- Rouhibakhsh A, Priya J, Periasamy M, Haq QMI, Malathi VG. 2008. An improved DNA isolation method and PCR protocol for efficient detection of multicomponents of begomovirus in legumes. Virol Method. 147(1):37–42. doi:https://doi.org/10.1016/j.jviromet.2007.08.004.
- Sohrab SS. 2020a. Genetic diversity of begomoviruses infecting tomato in Saudi Arabia. Saudi Journ of Biol Sci. 27(1):222–228. doi:https://doi.org/10.1016/j.sjbs.2019.08.015.
- Sohrab SS. 2020b. Molecular diagnosis of begomovirus associated with Chili leaf curl disease in Jeddah, Saudi Arabia. Saudi Journal of Biol Sci. 27(11):3060–3064. doi:https://doi.org/10.1016/j.sjbs.2020.09.009.
- Sohrab SS, Daur I. 2018. Identification of a monopartite begomovirus associated with yellow vein mosaic of Mentha longifolia in Saudi Arabia. 3Biotech. 8:92. doi:https://doi.org/10.1007/s13205-018-1120-7
- Sohrab SS. 2017. Current status of begomoviruses infecting cultivated crops and weeds in Saudi Arabia. In: Saxena S, Tiwari A, editors. Begomoviruses: occurrence and management in Asia and Africa. Singapore: Springer; p. 219–228. doi:https://doi.org/10.1007/978-981-10-5984-1_13.
- Sohrab SS, Yasir M, Kafrawy SA, Magdi A, Mousa MA, Bakhshwain AA. 2016. Phylogenetic relationships, recombination analysis and genetic variability of tomato yellow leaf curl virus infecting tomato in Jeddah, Saudi Arabia. Plant Omics J. 9(1):90–98.
- Varma A, Mandal B, Singh MK. 2011. Global emergence and spread of whitey (Bemisia tabaci) transmitted Geminiviruses. In: Thompson WMO, editor. The whitey, Bemisia tabaci (Homoptera: aleyrodidae) interaction with geminivirus-infected host plants. Berlin: Springer; p. 205–292.
- Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A. 2017. ICTV report consortium. ICTV virus taxonomy prole: geminiviridae. J Gen Virol. 98(2):131–133. doi:https://doi.org/10.1099/jgv.0.000738.


