Abstract
In the arboretum of Universidad Nacional de Colombia sede Medellín, Cassia fistula (Golden Shower Tree) and Leucaena leucocephala (Leucaena) exhibited early defoliation. Erysiphe quercicola and Telimena canafistulae, associated with powdery mildew and tar spot, respectively, on C. fistula, were identified using morphological and molecular techniques, both representing new records of fungi for Colombia. Similarly, Phyllactinia leucaenae was proposed as a new combination for Ovulariopsis leucaenae and was associated with powdery mildew on L. leucocephala, which to the best of our knowledge is another new record for Colombia.
Résumé
Dans l’arboretum de la branche de Medellín de l’Université nationale de Colombie, Cassia fistula (Golden Shower Tree) et Leucaena leucocephala (Leucaena) ont présenté une défoliation précoce. Erysiphe quercicola et Telimena canafistulae, associées à l’oïdium et à la tache goudronneuse, respectivement, sur C. fistula, ont été identifiées à l’aide de techniques morphologiques et moléculaires, toutes deux représentant de nouveaux enregistrements de champignons pour la Colombie. De même, Phyllactinia leucaenae a été proposé comme nouvelle combinaison pour Ovulariopsis leucaenae et était associé à l’oïdium sur L. leucocephala, qui, à notre connaissance, est un autre nouveau signalement pour la Colombie.
Introduction
Cassia fistula L. (Golden Shower Tree) and Leucaena leucocephala (Leucaena) de Wit, are frequently used as ornamental shrubs/trees in tropical urban places. They are thought to have medicinal properties and are included in environmental recuperation/protection programmes (Blair et al. Citation1990; Lenné Citation1990; Bahorun et al. Citation2005; Kanani et al. Citation2006; Valpassos et al. Citation2007; Peng et al. Citation2013; Pawar et al. Citation2017; Piñeiro-Vázquez et al. Citation2017). In addition, L. leucocephala is used for animal feed and in some places is an invasive plant species of concern for its high biological fitness (Sharratt and Olckers Citation2019).
Lenné (Citation1990) described Cassia spp. L. diseases according to their potential damage, highlighting Phyllachora bakeriana Henn., Phyllachora canafistulae F. Stevens & Dalby and Oidium sp. on C. fistula. In addition, powdery mildews caused by Phyllactinia dalbergiae Piroz. (Boa and Lenné Citation1994), Phyllactinia cassiae-fistulae U. Braun & Y.S. Paul (Braun and Paul Citation2009) and Erysiphe quercicola S. Takam. & U. Braun (Meeboon and Takamatsu Citation2020; Azevedo et al. Citation2021) were reported on C. fistula. Recently, the Phyllachorales M.E. Barr were redefined and Phyllachora canafistulae was reclassified as Telimena canafistulae (F. Stevens & Dalbey) Mardones, T. Trampe & M. Piepenbr. (Mardones et al. Citation2017).
Most diseases reported on Leucaena spp. have been found to be caused by fungi. Foliar spot caused by Camptomeris leucaenae (F. Stevens & Dalby) Syd., has been described as a limiting factor for the production of Leucaena spp. as animal feed (Lenné Citation1991). Lenné (Citation1981) reported a powdery mildew in Brazil, Colombia, Venezuela and Central America caused by Oidium sp. Similarly, Ovulariopsis sp. was recorded in Central America and Mexico on L. leucocephala (Boa and Lenné Citation1994) and Ovulariopsis leucaenae López-Pedraza, Yáñez-Morales & U. Braun was registered on leaves of Leucaena latisiliqua (L.) Gillis [= Lysiloma latisiliquum (L.) Benth] in Mexico (Braun and Yáñez-Morales Citation2009). The identity of O. leucaenae was determined using morphological features but molecular techniques were not applied.
Considering the importance of identifying the fungi associated with legumes in Colombia, the objective of the present research was to identify the microorganisms associated with powdery mildew and tar spot symptoms on C. fistula and L. leucocephala using a combination of morphological and molecular analyses.
Materials and methods
Morphological analysis
Leaves of C. fistula and L. leucocephala exhibiting symptoms and signs of powdery mildew were collected in the arboretum of Campus El Volador of Universidad Nacional de Colombia sede Medellín, Antioquia, Colombia, and in addition, tar spot signs and symptoms were observed on C. fistula. Morphological analysis was performed at Museo Micológico Universidad Nacional de Colombia sede Medellín (MMUNM) and molecular analysis was performed at the Laboratory of Fitotecnia Tropical located in the same institution. Plant material was herbalized and deposited at MMUNM.
Hand-made sections and scrapings of infected tissues were mounted on microscope slides with lacto-glycerol (100 mL of 85% lactic acid, 200 mL glycerol, 100 mL distilled water). Plant samples were cut by hand, trying to obtain slides as thin as possible in such a way that they could be observed under light microscopy. Scrapings of infected plant tissue were placed on a drop of lacto-glycerol on a microscope slide and covered with a coverslip for observation under light microscopy. Only when microorganism structures were highly hyaline to the point of being difficult to observe was lacto-glycerol with cotton blue (10 g L−1) added for staining (Liberato et al. Citation2005). Then, slides were visualized and photographed under light microscopy in a Carl Zeiss Primostar microscope coupled with an Axiocam camera (Carl Zeiss Microscopy GmbH, Jena, Germany). Thirty measurements were performed for each fungal structure observed. Morphological characteristics were analyzed to identify each fungal species using taxonomic keys available in the literature (Braun and Cook Citation2012; Mardones et al. Citation2017, Citation2020).
DNA extraction and purification
Fungal structures such as hyphae and conidia from powdery mildews were scraped from fresh infected plant tissues. Ascostromatic tissues from tar spots were cut using a sterile surgical scalpel blade. Collected tissues were individually placed in sterile Eppendorf tubes and macerated with sterile polypropylene micropistils. DNA was purified following the CTAB method. Integrity and quantity of DNA was verified by agarose gel electrophoresis (1%) and spectrophotometry (Thermo Fisher Scientific, USA).
PCR amplification and sequencing
Internal transcribed spacer regions (ITS) were amplified by the polymerase chain reaction (PCR) using primers ITS5/ITS4 following the conditions reported by White et al. (Citation1990). Amplified fragments were purified with the QIAquickTM PCR Purification Kit (QIAGEN GmbH, Hilden, Germany) following the manufacturer’s instructions and sent to the sequencing service following the company guidelines (Macrogen Inc., Seoul, Republic of Korea).
Phylogenetic analysis
The obtained sequences were cleaned and edited. Consensus sequences were constructed using the Unipro UGENE v39.0–1 software (Okonechnikov et al. Citation2012). They were compared with the GenBank database (www.ncbi.nlm.nih.gov) using the BLASTn algorithm. Additionally, sequences obtained in the present research were deposited in the same database with the accession numbers OQ225515 for T. canafistulae, OQ225513 for E. quercicola and OQ225514 for Phyllactinia leucaenae. Representative ITS sequences from Phyllachorales (Mardones et al. Citation2017, Citation2020) and tribes Phyllactinieae and Erysipheae sensu (CitationBraun and Cook Citation2012), Erysiphales, were retrieved to construct corresponding phylogenetic trees.
Sequences were aligned using the MAFFT v7.487 software with the algorithm L-INS-I (Katoh and Standley Citation2013). The best tree was determined by maximum likelihood (ML) and Bayesian inference (BI) methods. Maximum likelihood was performed using the RAxML-NG v1.0.3 software (Kozlov et al. Citation2019) with ten random starting and ten parsimony starting trees and bootstrap analysis with 1000 replicates. Bayesian inference was performed using the MrBayes v3.2.7 software (Ronquist et al. Citation2012) on XSEDE (Miller et al. Citation2010) (https://www.phylo.org/portal2), under Markov Chain Monte Carlo, with two parallel runs and four chains for 5 000 000 generations, with tree sampling every 1000 generations and discarding a burn-in fraction of 0.25. Tree selection was made following the 50% majority rule. In both methods, the nucleotide substitution model GTR and substitution rate with Gamma distribution GTR+G (Abadi et al. Citation2019) were applied.
Phylogenetic trees were visualized using the FigTree v1.4.4–1 software (Rambaut Citation2021) and edited using the Inkscape v1.1–5 software (Inkscape Project). Only bootstrap (BS) and posterior probability (PP) values equal to or higher than 70% and 0.9, respectively, were shown in the trees.
Results and discussion
Despite their importance, comprehensive knowledge about legume diseases is lacking; this is particularly true for native or naturalized non-commercial species (Sulima and Zhukov Citation2022). Here we report two new records of fungal pathogens on legume plants for Colombia: Erysiphe quercicola and Telimena canafistulae associated with powdery mildew and tar spots, respectively, on C. fistula. In addition, Phyllactinia leucaenae associated with powdery mildew on L. leucocephala was proposed as a new combination for Ovulariopsis leucaenae.
Taxonomy and dendrograms
Erysiphe quercicola S. Takamatsu & U. Braun, in Takamatsu, Braun, Limkaisang, Kom-un, Sato & Cunnington, Mycol. Res. 111(7): 819. 2007 ().
Fig. 1 Symptoms and signs of powdery mildew, Erysiphe quercicola, on leaves of Cassia fistula. A, B, Yellowing of the adaxial leaf surface. C, Abaxial part of the leaf covered with powdery mildew. D, E, Conidiophores. F, G, Detached mature conidia. G, Surface focus. Blue colour from staining with cotton blue in lacto-glycerol (E, F).
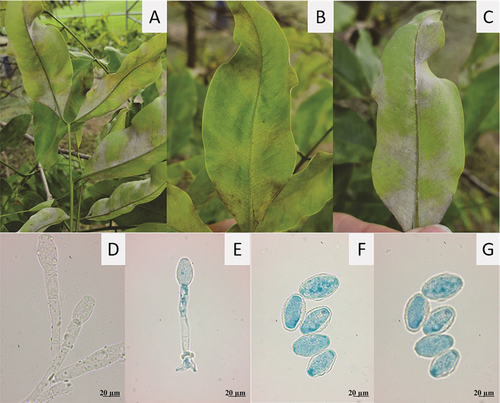
Mycelium predominantly hypophyllous, whitish, of cottony texture, in conspicuous patches, causing chlorosis on the adaxial leaf surface. Hyphae hyaline, septate, smooth, (5–) 6–7.5 µm wide, hyphal appressoria solitary, lobed. Conidiophores erect, 75–125 (–150) µm long. Foot cells cylindrical, straight to slightly curved, (30–) 37–75 (–85) µm, followed by 1–2 cells, forming single conidia. Primary conidia obovoid-ellipsoid, apex rounded, base subtruncate. Secondary conidia doliiform, ends subtruncate, (20–) 25–38 (–43) × (12–) 15–20 (–25) µm.
Material examined
On leaves of Cassia fistula L. (Fabaceae), Colombia, Antioquia, municipality of Medellin, Campus El Volador of Universidad Nacional de Colombia sede Medellín (6°15´49.5´´ N, 75°34´37.3´´ W), m.a.s.l. 1450, 12 April 2021, Samir Galvis-López & Mauricio Salazar-Yepes (MMUNM-3849).
Comments
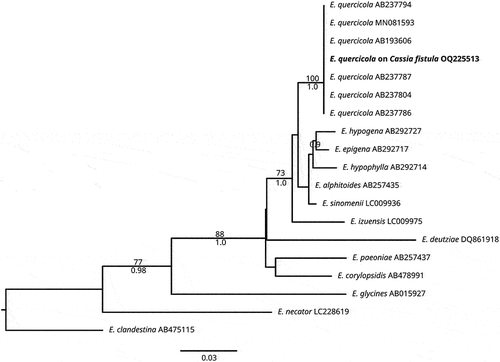
The morphology of powdery mildew on C. fistula corresponds to descriptions of Braun and Cook (Citation2012), Meeboon and Takamatsu (Citation2020) and Azevedo et al. (Citation2021). The sequence obtained in the present research (634 nucleotides in length) showed a percentage identity of 100% with the sequence from GenBank MH137256 of Erysiphe quercicola. In the first 100 sequences analyzed in a range of query coverage between 100 and 98%, several sequences of Oidium heveae, which was recently suggested as a synonym of E. quercicola, overlapped with an identity between 100 and 99.53% (Wu et al. Citation2019). Comparing our sequence in the BOLDSYSTEMS database (https://www.boldsystems.org/index.php/IDS_BlastRequest, accessed 16 January 2023), the most similar O. heveae sequence (ID: AB193589) exhibited a similarity of 99.84; and the sequence most closely related to a Erysiphe sp. with a similarity of 98.26%, corresponded to E. alphitoides (ID: AB292701). No further sequences of E. quercicola were found within the next 100 sequences, suggesting that no overlap is present for Erysiphe species using this molecular marker. A multiple alignment matrix of E. quercicola sequences comprised 734 characters and 19 sequences of the genus Erysiphe. Eryshipe clandestina (Wallr.) Link was used as the outgroup. High similarity between dendrograms was observed as indicated by the topological congruence of BI and ML trees, therefore, only the ML tree is shown with BS and PP values (). Phylogenetic analysis showed a close relationship of our sequence obtained from powdery mildew on C. fistula with other E. quercicola sequences (ML/BI = 100/1.0) (). The combination of morphological and molecular studies indicate that the fungus associated with powdery mildew corresponds to E. quercicola. To our best knowledge, this is the first report of this fungus in Colombia.
Fig. 2 Maximum likelihood tree for 19 ITS sequences of the Erysipheae showing the sequence obtained in the present research of Erysiphe quercicola on Cassia fistula (in bold). Numbers on branches represent bootstrap values of ML (above) and posterior probability of BI (below). E. clandestina was used as the outgroup. Bar indicates the number of substitutions per nucleotide position.
Phyllactinia leucaenae (López-Pedraza, Yáñez-Morales & U. Braun) Galvis-López, Morales-Osorio & Salazar, comb. nov. MB843970 ().
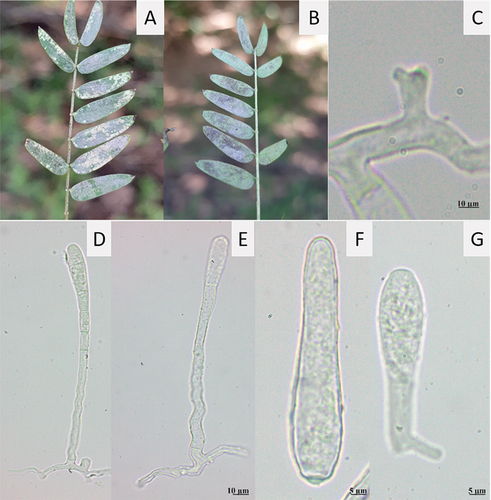
Fig. 3 Phyllactinia leucaenae on L. leucocephala. A, Symptoms on adaxial side of leaves. B, Symptoms on abaxial side of leaves. C, Appressorium. D, E, Conidiophores. F, Primary conidium. G, Secondary conidium with germ tube.
≡ Ovulariopsis leucaenae López-Pedraza, Yáñez-Morales & U. Braun, Mycotaxon 109: 154. 2009.
Mycelium hypophyllous, whitish, of powdery texture. Hyphae hyaline, irregular, septate, smooth, 6–7.5 µm wide, hyphal appressoria solitary, distinctly lobed. Conidiophores solitary, erect, (132–) 175–200 (–213) µm long, basal septum distal to 6 µm of branching point of supporting hypha. Foot cells straight to sinuous, 87–100 (–113) × 3–5 (–6) µm, followed by two cells of (12–) 15–18 (–28) µm length. Primary conidia narrowly ellipsoid-ovoid to lanceolate, apex rounded to slightly pointed, base subtruncate, (45–) 52–60 (–63) × 8–10 µm. Secondary conidia clavate to slightly obovoid, apex broadly rounded, base subtruncate, (45–) 52–60 (–63) × 8–10 µm. Conidia with sub-basal germination.
Material examined
On leaves of Leucaena leucocephala (Lam.) de Wit (Fabaceae), Colombia, Antioquia, municipality of Medellín, Campus El Volador from Universidad Nacional de Colombia sede Medellín (6°15´49.5´´ N, 75°34´37.3´´ W), m.a.s.l. 1450, 12 April 2021, Samir Galvis-López & Mauricio Salazar-Yepes (MMUNM-3847).
Comments
Based on morphological and molecular analyses, the new combination for Ovulariopsis leucaenae causing powdery mildew on L. leucocephala is proposed to be transferred to the genus Phyllactinia Lév.
The sequence obtained in the present research (646 nucleotides length) showed 91.19% identity with the sequence MH137256 from Phyllactinia mimosae. Generic affinity of this fungus with Phyllactinia mimosae Marm., Siahaan, S. Takam. & U. Braun with a well-supported relationship (ML/BI = 94/1, ) was observed. Only one other sequence of P. mimosae with the same value of identity (91.19%) was available in the database at the time of this analysis. The second most similar sequence ID (LC307201) corresponded to P. fraxini, with 86.41% identity with a query coverage of 100%. The next ten sequences of P. fraxini showed the same value of identity (86.41%), suggesting a gap of 4.78% between the two species.
Fig. 4 Maximum likelihood tree for 42 ITS sequences of the Phyllactinieae showing the position of Phyllactinia leucaenae (in bold). Numbers on branches represent the bootstrap value of ML (above) and posterior probability of BI (below). Pleochaeta shiraiana was used as the outgroup. Bar indicates the number of substitutions per nucleotide position.
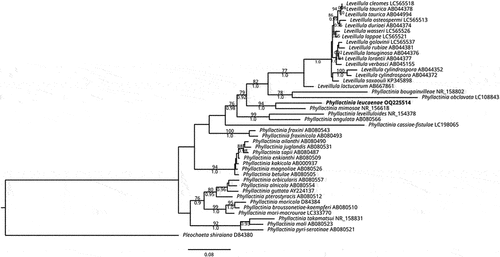
The multiple alignment matrix comprised 944 characters and 43 sequences, from which 16 belonged to Leveillula and 26 to Phyllactinia, including the sequence obtained in the present research (in bold). Pleochaeta shiraiana (Henn.) Kimbr. & Korf was used as the outgroup. High similarity between dendrograms was observed as indicated by the topologic congruence of BI and ML trees, therefore, only the ML tree is shown with BS and PP values (). The phylogenetic analysis showed a close relationship of Phyllactinia leucaenae with the sister taxon Phyllactinia mimosae (ML/BI = 94/1.0).
Phyllactinia leucaenae is similar to Phyllactinia dalbergiae Piroz. and Phyllactinia mimosae. However, the conidia of Phyllactinia leucaenae are narrowly ellipsoid-ovoid to lanceolate, apex rounded to slightly pointed, subtruncate base, (45) 52–60(63) × 8–10 µm, while Phyllactinia dalbergiae has larger, clavate to mostly soleiform conidia, 50–90 × (10) 13–24 µm and Phyllactinia mimosae presents clavate-cylindrical, shorter and narrower conidia, 40–65 × 10–18 µm. Host range and distribution: Phyllactinia leucaenae has been found on Leucaena leucocephala, in Honduras and Mexico (Braun and Cook Citation2012). To our best knowledge, this is the first report of this fungus in Colombia.
Telimena canafistulae (F. Stevens & Dalby) Mardones, T. Trampe & M. Piepenbr., in Mardones, Trampe-Jaschik, Oster, Elliott, Urbina, Schmitt & Piepenbring, Persoonia 39: 84. 2017 ().
Fig. 5 Telimena canafistulae on Cassia fistula. A, Symptoms on adaxial side of leaf. B, Section through pseudostroma. C, Section through ostiolar region. D, Asci. E, Paraphyses. F, Ascospores.
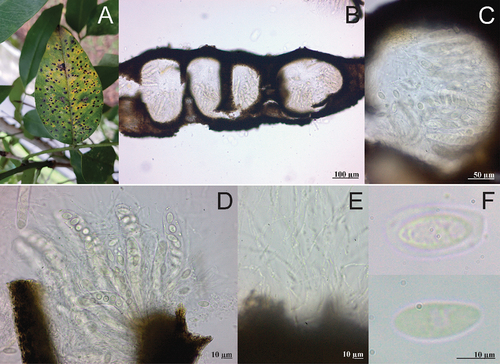
Leaf spots conspicuous, black, 1–6 mm in diameter, irregular, scattered all over the leaf blade, surrounded by a thin chlorotic zone, slightly raised above the leaf surface. Ascostromata black, occupying the mesophyll, multiloculate. Clypei amphigenous mostly developed in the adaxial side. Perithecia globose, dark brown, immersed in the stromata, (150–) 190–225 (–238) µm in diameter. Periphyses not observed. Paraphyses filiform, hyaline. Asci cylindrical to clavate, 75–100 (–105) × (12–) 15–20 µm, 8–spored. Ascospores ovoid to ellipsoid, hyaline, 12–18 (–20) × 7.5–10 (–13) µm, mainly uniseriate, surrounded by a gelatinous layer.
Material examined
On leaves of Cassia fistula L. (Fabaceae), Colombia, Antioquia, municipality of Medellin, Campus El Volador of Universidad Nacional de Colombia sede Medellín (6°15´49.5´´ N, 75°34´37.3´´ W), m.a.s.l. 1450, 12 April 2021, Samir Galvis-López & Mauricio Salazar-Yepes (MMUNM-3848).
Comments
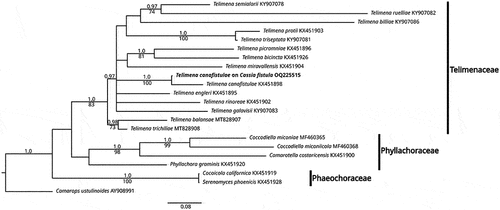
The morphology of the fungus found on C. fistula agreed with descriptions of Phyllachora made by Stevens and Dalby (Citation1919). The sequence obtained in the present research (579 nucleotides length) showed 94.97% identity with the sequence KX451898 from Telimena canafistulae. The most similar sequence corresponded to an undescribed species (Uncultured fungus clone C33 18S, Sequence ID: EU686766) with an identity of 88%. The most similar sequence (Sequence ID: KX451895) with a described species corresponded to T. engleri with 84% identity, suggesting a gap of 10.97% between these two species. The phylogenetic analysis showed that the sequence obtained in the present research grouped (PP/BS of 1.0/100) with that of T. canafistulae (KX451898) reported in the study of Phyllachorales of Mardones et al. (Citation2017) ().
The multiple alignment matrix of T. canafistulae sequences comprised 816 characters and 22 sequences, from which four belonged to Phyllachoraceae Theiss. & P. Syd., two to Phaeochoraceae K.D. Hyde, P.F. Cannon & M.E. Barr and 15 to Telimenaceae Mardones, T. Trampe & M. Piepenbr., including the sequence obtained in the present research (in bold). Camarops ustulinoides (Henn.) Nannf. was used as the outgroup. High similarity between dendrograms was observed as indicated by the topologic congruence of BI and ML trees, therefore, only the BI tree is shown with BS and PP values (). Phylogenetic analysis showed a close relationship of T. canafistulae with sequences of the Telimenaceae (BI/ML = 1.0/83) next to T. canafistulae voucher MM-13 (BI/ML = 1.0/100). Host range and distribution: on Barbieria pinnata (Pers.) Baill., Dominican Republic; on Cassia (C. fistula L., C. grandis L.f., C.javanica Vell., C. nodosa Buch.-Ham. ex Roxb., Cassia sp.), Bermuda, Brazil, Costa Rica, Cuba, Dominican Republic, EEUU, Jamaica, Panama, Puerto Rico, Trinidad and Tobago, Virgin Islands, West Indies (Farr and Rossman Citation2022) and Colombia. To our best knowledge, this is the first report of this fungus in Colombia.
Fig. 6 Bayesian inference tree for 21 ITS sequences of the three Phyllachorales families showing the position of Telimena canafistulae (in bold). Numbers on branches represent posterior probability of BI (above) and bootstrap value of ML (below). Camarops ustulinoides was used as the outgroup. Bar indicates the number of substitutions per nucleotide position.
Most studies regarding powdery mildews and other mycoflora on Fabaceae have been performed on commercial crops such as peas and beans (Kelly et al. Citation2021; Sulima and Zhukov Citation2022). The knowledge of mycoflora associated to wild Fabaceae species is limited worldwide and in Colombia is even scarcer. In the present research, three new records for Colombia with a new combination are reported, which contributes to the knowledge about fungal diversity in the world.
Acknowledgements
We are very grateful to Universidad Nacional de Colombia sede Medellín, Museo Micológico Universidad Nacional de Colombia sede Medellín (MMUNM), Laboratory of Fitotecnia Tropical and to Paola Betancur for technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abadi S, Azouri D, Pupko T, Mayrose I. 2019. Model selection may not be a mandatory step for phylogeny reconstruction. Nat Comm. 10(1):1–11. doi:10.1038/s41467-019-08822-w.
- Azevedo A, de Castro T, de Oliveira L, Firmino A, Liparini O. 2021. Erysiphe quercicola causing powdery mildew on Cassia fistula and Tamarindus indica in Brazil. Australas Plant Dis Notes. 16(1):1–4. doi:10.1007/s13314-021-00418-y.
- Bahorun T, Neergheen V, Aruoma O. 2005. Phytochemical constituents of Cassia fistula. African J Biotech. 4(13):1530–1540.
- Blair G, Catchpoole D, Horne P. 1990. Forage tree legumes: their management and contribution to the nitrogen economy of wet and humid tropical environments. Adv Agron. 44:27–54. doi:10.1016/S0065-2113(08.
- Boa E, Lenné J. 1994. Diseases of nitrogen fixing trees in developing countries: an annotated list. Chatham (UK): Natural Resources Institute.
- Braun U, Cook R. 2012. Taxonomic manual of the Erysiphales (Powdery Mildews), CBS Biodiversity Series No. 11. Utrecht (Netherlands): CBS.
- Braun U, Paul Y. 2009. The Indian Erysiphaceae revisited. Nova Hedwigia. 89:371–395. doi:10.1127/0029-5035/2009/0089-0371.
- Braun U, Yáñez-Morales M. 2009. Phyllactinia and Ovulariopsis species on legumes. Mycotaxon. 109:145–160. doi:10.5248/109.145.
- Farr DF, Rossman AY. 2022. Fungal databases, U.S. national fungus collections. ARS, USDA; [accessed 2022 Aug 8]. https://nt.ars-grin.gov/fungaldatabases/.
- Kanani J, Lukefahr S, Stanko R. 2006. Evaluation of tropical forage legumes (Medicago sativa, Dolichos lablab, Leucaena leucocephala and Desmanthus bicornutus) for growing goats. Small Rum Res. 65(1–2):1–7. doi:10.1016/j.smallrumres.2005.04.028.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kelly LA, Vaghefi N, Bransgrove K, Fechner NA, Stuart K, Pandey AK, Sharma M, Németh MZ, Liu SY, Tang SR, et al. 2021. One crop disease, how many pathogens? Podosphaera xanthii and Erysiphe vignae sp. nov. Identified as the two species that cause powdery mildew of mungbean (Vigna radiata) and black gram (V. mungo) in Australia. Phytopathology. 111(7):1193–1206. doi:10.1094/PHYTO-12-20-0554-R.
- Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi:10.1093/bioinformatics/btz305.
- Lenné J. 1981. Diseases of important tropical pasture plants in central and South America. Australas Plant Path. 10:10–12. doi:10.1071/APP9810010.
- Lenné J. 1990. Diseases of Cassia species – a review. Trop Grasslands. 24(4):311–324.
- Lenné J. 1991. Diseases of Leucaena species. Trop Pest Manag. 37(3):281–289. doi:10.1080/09670879109371600.
- Liberato JR, Barreto RW, Shivas RG. 2005. Leaf-clearing and staining techniques for the observation of conidiophores in the Phyllactinioideae (Erysiphaceae). Australas Plant Path. 34:401–404. doi:10.1071/AP05027.
- Mardones M, Trampe-Jaschik T, Oster S, Elliot M, Urbina H, Schmit I, Piepenbring M. 2017. Phylogeny of the order Phyllachorales (Ascomycota, Sordariomycetes): among and within order relationships based on five molecular loci. Persoonia. 39:74–90. doi:10.3767/persoonia.2017.39.04.
- Mardones M, Trampe-Jaschik T, Piepenbring M. 2020. Phylogenetics and taxonomy of Telimenaceae (Phyllachorales) from Central America. Mycol Prog. 19(12):1587–1599. doi:10.1007/s11557-020-01649-6.
- Meeboon J, Takamatsu S. 2020. Hosts of asexual morph of Erysiphe quercicola from Thailand. Trop Plant Path. 45(2):122–135. doi:10.1007/s40858-019-00326-8.
- Miller M, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans (USA). p. 1–8.
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 28(8):1166–1167. doi:10.1093/bioinformatics/bts091.
- Pawar A, Patil S, Killedar S. 2017. Uses of Cassia fistula Linn as a medicinal plant. Int J Adv Res Dev. 2(3):85–91.
- Peng S, Chen A, Fang H, Wu J, Liu G. 2013. Effects of vegetation restoration types on soil quality in Yuanmou dry-hot valley, China. Soil Sci Plant Nutr. 59(3):347–360. doi:10.1080/00380768.2013.785918.
- Piñeiro-Vázquez A, Jiménez-Ferrer G, Chay-Canul A, Casanova-Lugo F, Díaz-Echeverría V, Ayala-Burgos A, Solorio-Sánchez F, Aguilar-Pérez C, Ku-Vera J. 2017. Intake, digestibility, nitrogen balance and energy utilization in heifers fed low-quality forage and Leucaena leucocephala. Anim Feed Sci Techn. 228:194–201. doi:10.1016/j.anifeedsci.2017.04.009.
- Rambaut A. 2021. FigTree, a graphical viewer of phylogenetic trees. Edinburgh (UK): University of Edinburgh, Institute of Evolutionary Biology. http://tree.bio.ed.ac.uk/software/figtree/
- Ronquist F, Teslenko M, Van Der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard M, Huelsenbeck J. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Sharratt M, Olckers T. 2019. Responses of the seed-feeding beetle Acanthoscelides macrophthalmus and its recruited parasitoids to resource availability–implications for the biological control of Leucaena leucocephala in South Africa. Biol Control. 135:102–109. doi:10.1016/j.biocontrol.2019.05.013.
- Stevens F, Dalby N. 1919. Some phyllachoras from Porto Rico. Bot Gazette. 68(1):54–59. doi:10.1086/332521.
- Sulima AS, Zhukov VA. 2022. War and peas: molecular bases of resistance to powdery mildew in pea (Pisum sativum L.) and other legumes. Plants. 11:339. doi:10.3390/plants11030339.
- Valpassos M, Maltoni K, Rodrigues A, Nahas E. 2007. Recovery of soil microbiological properties in a degraded area planted with Corymbia citriodora and Leucaena leucocephala. Sci Agri. 64(1):68–72. doi:10.1590/S0103-90162007000100010.
- White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. San Diego (USA): Academic Press; p. 315–322.
- Wu H, Pan Y, Di R, He Q, Rajaofera MJN, Liu W, Zheng F, Miao W. 2019. Molecular identification of the powdery mildew fungus infecting rubber trees in China. For Pathol. 49:e12519. doi:10.1111/efp.12519.
