Abstract
Changes in pathogenicity of populations of Leptosphaeria maculans, the cause of blackleg disease of canola and other Brassica spp., have been observed in western Canada. These changes in the population are believed to have resulted from the use of specific resistance (Rlm) genes in Brassica spp., many of which have only recently been identified. We determined the frequency of 10 avirulence alleles for 96 isolates of L. maculans collected between 1997 and 2005 from Alberta, Saskatchewan and Manitoba, Canada from infested canola stubble in field plots or farmers’ fields. Cotyledon interaction phenotypes were scored after inoculation of each isolate on a differential set of Brassica varieties or lines carrying the known resistance genes LepR3 and Rlm1 to Rlm10, except for Rlm8. The avirulence (AvrLm) alleles present in each isolate were inferred from the data. Due to the presence of confounding resistance genes in the host differentials, AvrLm1 and AvrLm2 could not be determined in nine of the isolates, AvrLm6 in two isolates and AvrLepR3 in 49. Based on the number of isolates in which the avirulence genes were confirmed, AvrLm1 was detected in 46.0% and AvrLm9 in 60.4% of the isolates in this collection. A high proportion of isolates carried AvrLm2 (96.6%) and AvrLepR3 (97.9%), and all isolates carried AvrLm6 and AvrLm10. The proportion of L. maculans isolates carrying AvrLm3, AvrLm4, AvrLm5 and AvrLm7 was much lower and varied between 10.4 and 29.2%. In total, 16 races of L. maculans were identified based on 10 avirulence alleles, with seven races accounting for 89.7% of the isolates. Information on the avirulence frequency in the pathogen population will be useful for development of resistance management strategies to control blackleg disease in B. napus oilseed rape in western Canada.
Résumé: Des changements dans la pathogénicité des populations de Leptosphaeria maculans, l'agent responsable de la maladie de la Nécrosse du collect chez le canola ainsi que chez d'autres Brassica spp., ont été observés dans l'Ouest canadien. On croit que ces changements résultent de l'utilisation de gènes spécifiques de résistance (Rlm) chez Brassica spp., dont plusieurs n'ont été que récemment identifiés. Nous avons déterminé la fréquence de 10 allèles d'avirulence pour 96 isolats de L. maculans collectés, de 1997 à 2005, en Alberta, en Saskatchewan et au Manitoba, sur du chaume de canola infecté provenant de parcelles expérimentales et de champs cultivés. Les phénotypes d'interaction ont été notés sur cotyiédons après inoculation de chaque isolat sur un groupe différentiel de variétés de Brassica ou de lignées portant les gènes de résistance connus LepR3 et Rlm1 à Rlm10, sauf Rlm8. Les allèles d'avirulence (AvrLm) présents dans chaque isolat ont été déduits des données. Étant donnée la présence simultanèe de gènes de résistance dans certains hôtes différentiels, AvrLm1 et AveLm2 n'ont pu être confirmés dans neuf des isolats, AvrLm6, dans deux et AvrLepR3, dans 49. En se basant sur le nombre d'isolats dans lesquels la présence de gènes d'avirulence a été confirmée, AvrLm1 a été détecté dans 46,0 % des isolats et AvrLm9, dans 60,4 % de cette collection. Une forte proportion d'isolats portait AvrLm2 (96,6 %) et AvrLepR3 (97,9 %), et tous les isolats portaient AvrLm6 et AvrLm10. La proportion des isolats de L. maculans portant AvrLm3, AvrLm4, AvrLm5 et AvrLm7 était beaucoup plus faible et variait de 10,4 à 29,2 %. En tout, 16 races de L. maculans ont été identifiées à partir de 10 allèles d'avirulence, sept de ces races comprenant 89,7 % des isolats. L'information relative à la fréquence d'avirulence dans les populations d'agents pathogènes servira à élaborer des stratégies de gestion de la résistance permettant de lutter contre la maladie de la Nécrosse du collet chez B. napus dans l'Ouest canadien.
Introduction
Changes in virulence in the population of Leptosphaeria maculans Ces. & de Not., the cause of blackleg disease or phoma stem canker of Brassica spp., have been observed in western Canada (Chen & Fernando, Citation2006; Kutcher et al., Citation2007). This has been attributed to the adaptation of the pathogen to varieties of canola with high levels of genetic resistance to the disease (Kutcher et al., Citation2007), similar to that which occurred in France (Rouxel et al., Citation2003a) and in Australia (Li et al., Citation2003; Sprague et al., Citation2006). Resistance to the pathogen is thought to be of two types: quantitative or qualitative (Delourme et al., Citation2006). Quantitative resistance is believed to be race-nonspecific, is inherited polygenically and expressed at the adult plant stage as reduced development of necrotic tissue at the stem base compared with susceptible Brassica napus L. varieties. Qualitative resistance is controlled by race-specific resistance (Rlm) genes and a resistant response is presumed to result from recognition of corresponding pathogen avirulence (AvrLm) genes in a gene-for-gene manner (Ansan-Melayah et al., Citation1998). In the L. maculans–Brassica interaction, race-specific resistance is usually effective at the site of infection on leaves and as early as the cotyledon stage. At present, all current B. napus varieties in Canada are considered to have some degree of resistance to L. maculans although the basis for this resistance is generally unknown (Rimmer, Citation2006).
Specific interactions of L. maculans isolates with B. napus varieties have been used to classify isolates into pathogenicity groups (PGs). This classification of L. maculans is based on the interaction phenotypes (IP) of each isolate on only two B. napus varieties, ‘Quinta’ [Rlm1 (Ansan-Melayah et al., Citation1995) and either Rlm3 (Kutcher & Yu, unpublished data) or Rlm4 (Balesdent et al., Citation2001) depending on the seed lot] and ‘Glacier’ [Rlm2 (Ansan-Melayah et al., Citation1998) and Rlm3 (Balesdent et al., Citation2002)]. Isolates are classified as: PG2 – resistant reaction on both varieties; PG3 – ‘Quinta’ resistant, ‘Glacier’ susceptible; PGT – ‘Quinta’ susceptible, ‘Glacier’ resistant; or PG4 – both varieties susceptible (Mengistu et al., Citation1991). Isolates of L. maculans from Alberta, Saskatchewan and Manitoba in western Canada and from North Dakota, USA have recently been classified into various PGs (Chen & Fernando, Citation2006; Kutcher et al., Citation2007). The PG classification system has been useful as a tool to recognize changes in pathogenicity of the pathogen population since the early 1990s in western Canada. However, the information obtained from the PG classification system is limited by the fact that the system is based only on the three or possibly four resistance genes discussed, while as many as 14 resistance genes have been reported (Rimmer, Citation2007). Assuming that all 14 resistance genes are different from each other, gene-for-gene interactions indicate that 214 or 16 384 races of the pathogen are at least theoretically possible. Balesdent et al. (Citation2005) reported that as many as nine distinct races could be classified into a single PG. This indicates that classification of isolates into races based on their avirulence allele frequencies and patterns, corresponding to known resistance genes in the host, provides a much greater understanding of the host–pathogen interaction than the PG system.
Race-specific resistance genes were initially observed to be very effective in controlling blackleg of oilseed rape in Europe and Australia (Howlett, Citation2004). However, populations of L. maculans adapted rapidly under selection from large-scale cultivation of varieties with specific resistance genes. For example, Rlm1 in France (Rouxel et al., Citation2003a) and LepR3 in Australia (Li et al., Citation2003; Sprague et al., Citation2006) became ineffective after a few years of widespread use. Similar results were previously obtained experimentally for Rlm6 (Brun et al., Citation2000). Australian populations of L. maculans in particular have been reported to have a high level of variability for pathogenicity compared with western Canadian isolates (Kutcher et al., 1993) and to be extremely diverse for avirulence alleles compared to Canadian and European isolates (Balesdent et al., Citation2005).
Information on the frequency of individual avirulence alleles present in the pathogen population can provide insights into the corresponding resistance genes present in commercially grown varieties, including an indication of their potential durability for blackleg control. This will be of benefit for the development of strategies to manage blackleg disease of canola through cultural controls and breeding efforts. This paper examines the frequency of 10 individual avirulence genes among isolates of L. maculans from Alberta, Saskatchewan and Manitoba collected between 1997 and 2005 using the terminology and standard set of plant differentials proposed by Balesdent et al. (Citation2005).
Materials and methods
Fungal material
Ninety-six isolates of L. maculans were isolated from unidentified varieties of mature canola plants from farmer's fields and small experimental plots from the provinces of Alberta, Saskatchewan and Manitoba in western Canada during the years 1997 to 2005. The isolates were a subsample from the laboratory collection with contributions from co-operators, which were collected during two periods: 1997–2000 and 2003–2005. In Alberta, eight of the 17 isolates (47.1%) were collected during the first period and the balance from the latter period. In Saskatchewan, 20 of 61 isolates (32.8%) were from the 1997–2000 period, while in Manitoba, the proportion was 15 of 18 isolates (83.3%) collected during 1997–2000. Isolations were made from basal stem cankers and test cultures were prepared from single pycnidiospores. Pycnidiospores of isolates were stored at –20 ˚C on 6.5 mm sterile filter paper discs in sterile microfuge tubes.
Inoculum preparation
Cultures of each isolate were recovered by placing an infested filter paper disc on malt extract agar to which 1% streptomycin sulphate was added and the resulting colony grown for 21 days at room temperature under ambient light in the laboratory. Isolates were subcultured onto V8 juice agar and inoculum solutions of each isolate consisting of 1 × 107 pycnidiospores mL−1 were prepared as described by Kutcher et al. (Citation2007).
Host material
A set of differential genotypes (varieties or lines) with previously characterized resistance genes in B. napus or Brassica juncea L. genetic backgrounds were chosen for the study: ‘Westar’ – no resistance genes (Delourme et al., Citation2004), ‘MT29’ – Rlm1,9 (Delourme et al., Citation2008), ‘Samouraï’ – Rlm2,9 (Rouxel et al., Citation2003b), ‘22-1-1’ – Rlm3 (Delourme et al., Citation2004), ‘Falcon’ – Rlm4 (Rouxel et al., Citation2003b), ‘150-2-1’ – Rlm5 (Stachowiak et al., Citation2006), FalconMX – Rlm4,6 and DarmorMX – Rlm6,9 (Chèvre et al., Citation1997; Balesdent et al., Citation2006), ‘23-2-1’ – Rlm7 (Delourme et al., Citation2004), ‘Darmor’ – Rlm9 (Delourme et al., Citation2004), ‘Westar 74’ – Rlm10 (Chèvre et al., Citation1996; Chèvre, personal communication) and ‘Surpass 400’ – Rlm1, LepR3 (Van de Wouw et al., Citation2008; Yu et al., Citation2008). Since ‘MT29’ and ‘Samouraï’ each contain Rlm9 as well as Rlm1 and Rlm2, respectively, these lines could not be used to discriminate the presence of AvrLm1 and AvrLm2 in those isolates with AvrLm9. In these cases, isolates were retested using B. napus varieties which had Rlm1 or Rlm2 but which also contained other resistance genes: ‘Cooper’ – Rlm1,4, ‘Grizzly’ – Rlm1,3, ‘Glacier’ – Rlm2,3 and ‘Verona’ – Rlm2,4 (Balesdent et al., Citation2002; M.H. Balesdent, personal communication). These additional differential lines allowed for discrimination of AvrLm1 and AvrLm2 in isolates which harboured AvrLm9, when AvrLm3 and/or AvrLm4 were not present. The avirulence allele AvrLepR3, corresponding to resistance gene LepR3, could not be determined in isolates that carried AvrLm1. Similarly, AvrLm6 could not be characterized in isolates that contained both AvrLm4 and AvrLm9.
Cotyledon evaluation
Seeds of each differential Brassica genotype were placed in Petri dishes on water-dampened filter paper to promote germination. Between 48 to 72 h later, the resulting plantlets were transplanted into flats of a soil-sand-peat mix in two replicates of two plants each and maintained in the greenhouse for 10 days. Shortly after transplanting, a plastic-coated wire mesh with a 1 cm2 pattern, cut to fit the surface area of each tray of plants, was laid on the soil surface. As the plants grew, the mesh was raised to support the plants by placing strips of styrofoam under the mesh.
Cotyledons of the differential lines were inoculated with the test isolates 10 days after transplanting. Both lobes of each cotyledon were wounded with the sharp point of a set of tweezers that had been modified to puncture the cotyledon surface by bending and sharpening one arm of the tweezers. A 10 μL droplet of inoculum was placed onto each of the two wounds on each cotyledon (four wounds per plant in total). In order to maintain high humidity, trays containing the inoculated plants were covered with a fitted hard plastic cover and placed into the growth chamber. The trays were incubated in the dark by covering with black plastic for 24 h. After incubation a photoperiod of 14 h light and 10 h dark at 22 and 16 ˚C, respectively, was maintained in the growth chamber. Emerging leaves were removed from all seedling plants weekly to delay senescence of cotyledons. Both cotyledons of each plant were evaluated for interaction phenotype (IP) 14 and 21 days post inoculation (dpi) on a scale of 0 (no symptoms) to 11 (severe) based on lesion size and necrosis or chlorosis (Brun et al., Citation2001; Leflon et al., Citation2007). The mean score of the resulting 16 lesions for each isolate-Brassica genotype was used to classify the IP into one of two categories: Avr = avirulent or resistant (IP 0 to 4.9) or avr = virulent or susceptible (IP 5 or greater) for the IP at 14 dpi, or, Avr = IP 0 to 6.9 and avr = IP 7 or greater for IP at 21 dpi. Cotyledon IP for all isolate–Brassica genotype combinations were almost always able to be identified as virulent or avirulent from the 14 dpi assessment and were confirmed by the 21 dpi assessment. In the rare cases where a difference in IP occurred between 14 and 21 dpi, the 21 dpi IP was accepted.
Results
Avirulence alleles corresponding to resistance genes Rlm3, Rlm4, Rlm5, Rlm7, Rlm9 and Rlm10 were determined in all 96 L. maculans isolates characterized in this study. The avirulence alleles AvrLm1 and AvrLm2 were confirmed in 87 of the 96 isolates, but could not be determined in nine isolates due to the presence of other confounding resistance genes in the host differentials. Similarly, AvrLm6 was confirmed in 94 isolates, but not in two of the 96 isolates. Since ‘Surpass 400’ is believed to carry both Rlm1 and LepR3 (Van de Wouw et al., Citation2008), the presence or absence of AvrLepR3 could be confirmed in only 47 of the 96 isolates.
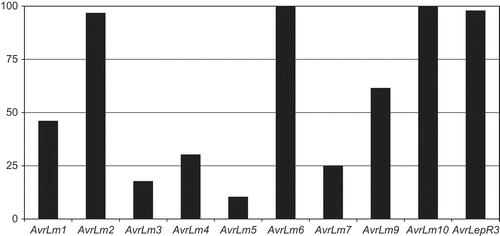
All isolates that were characterized for AvrLm6 (94) and AvrLm10 (96) carried these avirulence alleles (). The AvrLepR3 allele was found in 97.9% of the 47 isolates for which it was analyzed, and the AvrLm1 and AvrLm2 alleles were carried by 46.0 and 96.6%, respectively, of the 87 isolates for which the presence of these genes could be determined. The AvrLm9 allele was detected in 60.4% of the isolates. The remaining avirulence alleles were carried by a lower proportion of the isolates: AvrLm3 – 17.7%, AvrLm4 – 29.2%, AvrLm5 – 10.4% and AvrLm7 – 25.0%.
Fig. 1. Percentage of Leptosphaeria maculans isolates carrying each avirulence gene. Data represent 47 isolates for AvrLepR3, 87 isolates for AvrLm1 and AvrLm2, 94 isolates for AvrLm6 and 96 isolates for all other avirulence alleles. AvrLm8 was not assessed.
Of the 87 isolates for which all avirulence alleles except AvrLm6 and AvrLepR3 could be determined, 16 races (combinations of Avr genes in one isolate) of the pathogen were identified based on analysis of isolate IP on the 10 host resistance genes examined in this study (). Ninety per cent of the isolates belonged to one of seven races, each of which was composed of between three and 20 isolates. The remaining nine races were represented by single isolates. Three races, Av1-2-6-(8)-9-10-(LepR3), Av1-2-6-(8)-10-(LepR3) and Av2-6-(8)-9-10-(LepR3) represented 52 isolates (59.8%) and differed from each other only due to variation for AvrLm1 or AvrLm9. Following the nomenclature of Balesdent et al. (Citation2005), only the avirulence alleles carried by each race are listed and the alleles of the genes in parentheses were not or could not be determined in these races. One race, represented by a single isolate, carried the maximum number of avirulence alleles possible in this study, i.e. avirulence alleles corresponding to eight and possibly all 10 resistance genes. The races with the least number of avirulence alleles: Av1-2-6-(8)-10-(LepR3) and Av-2-6-(8)-10-LepR3, included 13 and three isolates, respectively. The other 14 races carried between five and seven avirulence alleles.
Table 1. Race identification of 87 isolates of Leptosphaeria maculans collected in western Canada during 1997–2000 and 2003–2005.1
The frequency of each avirulence allele was examined by province in western Canada and by collection period (). The avirulence allele AvrLm3 was not detected in Manitoba and was less frequent in Saskatchewan than Alberta, while AvrLm2 and AvrLm5 were less frequent in Manitoba than the other provinces. Frequencies of the remaining avirulence alleles were similar among the three provinces. Between the isolate collection periods, 1997–2000 and 2003–2005, the frequency of all avirulence alleles were similar except for AvrLm7, which was less frequent in the earlier period than in the most recent.
Table 2. Percentage of isolates carrying an avirulence allele (Avr) at each avirulence locus grouped by province and collection period
Discussion
Characterization of L. maculans isolates on the host differential set in this study confirmed the avirulence alleles for 10 genes, which has the potential to differentiate 210 or 1024 races. The fact that only 16 races were detected indicates that variation on many resistance genes is low, or that races with many virulence alleles may be less fit and consequently more difficult to detect. Moreover, linkage of avirulence genes (AvrLm1-2-6 and AvrLm3-4-7-9) as reported by Balesdent et al. (Citation2002), and our inability to determine all 10 avirulence alleles in some isolates, may have masked the identification of other races.
Frequencies in avirulence alleles were highly variable among the avirulence genes characterized, ranging from 10.4% to 100% (). The avirulence alleles AvrLm6 and AvrLm10 were carried by 100% of the isolates in which they were discriminated. This indicated that the corresponding resistance genes, Rlm6 and Rlm10, conditioned resistance against all the isolates tested in this collection. Both of these specific resistance genes have been introgressed into B. napus from other Brassica spp., Rlm6 from B. juncea (Chèvre et al., Citation1997) and Rlm10 from B. nigra (Chèvre et al., Citation1996; Chèvre, personal communication). Therefore, the fact that all isolates were avirulent on these resistance genes is not surprising since these genes have been used only experimentally and are not believed to be present in commercial varieties of canola.
The avirulence allele AvrLepR3 was present in all but one of the isolates examined and AvrLm2 in all but three of the isolates in which its presence could be determined. This is somewhat surprising because the corresponding resistance genes (LepR3 and Rlm2) have been available to Canadian plant breeders for some time and therefore might be expected to be carried by some current Canadian B. napus varieties. Subsequently, a greater degree of adaptation of the pathogen to these genes was expected. The fact that an isolate virulent on LepR3 was obtained from western Canada, indicates that this resistance gene may be overcome relatively rapidly if widely deployed. It is noteworthy that the three isolates in this study that did not carry AvrLm2, and were virulent to Rlm2, were collected from southern Manitoba, where isolates classified as PG3 and PG4 have been obtained (Chen & Fernando, Citation2006; Kutcher et al., Citation2007). PG3 and PG4 isolates are virulent on ‘Glacier’ (Rlm2 and Rlm3), which indicates they do not carry AvrLm2. The fact that we observed isolates virulent on Rlm2 and Rlm3, indicates that varieties solely dependent on these genes are at risk if these races increase in frequency.
Less than half of the isolates in this collection carried AvrLm1. In a previous study of isolates collected in western Canada, it was found that 13% (Kutcher et al., Citation2007) were classified as PGT. PGT isolates overcome the resistance carried by ‘Quinta’, which is known to be due to Rlm1 (Ansan-Melayah et al., Citation1995), and therefore suggests that 87% of the isolates in the previous study carried AvrLm1, which is much greater than the 46% observed in this study. However, in addition to Rlm1, some seed lots of ‘Quinta’ are believed to carry Rlm3 (Kutcher & Yu, unpublished data), and other seed lots Rlm4 (Balesdent et al., Citation2001). It is possible that some of the isolates avirulent on ‘Quinta’ and classified as PG2 and PG3 in the previous studies were actually avrLm1 and AvrLm3 or AvrLm4. Isolates carrying either AvrLm3 or AvrLm4, but not AvrLm1, would result in a resistant reaction on ‘Quinta’ seed stocks that carry Rlm3 or Rlm4. The differences in seed lots and variation in the resistance genes harboured by ‘Quinta’ may explain the lower frequency of AvrLm1 in this study, in which ‘MT29’, ‘Cooper’ and ‘Grizzly’ were used to determine the presence of AvrLm1, compared to ‘Quinta’ in the previous study.
In western Canada in the 1980s, all L. maculans isolates collected by Kutcher et al. (Citation1993) were observed to be PG2. This indicated they were avirulent on ‘Quinta’ and therefore possibly avirulent on Rlm1, although as discussed above an avirulent reaction on ‘Quinta’ could occur for isolates carrying AvrLm3 or AvrLm4 but not AvrLm1, due to the presence of either Rlm3 or Rlm4 in the particular seed lot of ‘Quinta’ used. The fact that AvrLm1 was observed at a moderate level (46.0%) in the present study suggests that the pathogen population has adapted to Rlm1, which is an indication that this resistance gene may be present in some Canadian B. napus varieties. However, the sources of resistance carried by Canadian canola varieties are unknown for the most part (Rimmer, Citation2006) and information on the area cropped to each of the hundreds of varieties marketed in each province of the Canadian prairies is not available. It is interesting to note that the frequency of AvrLm1 was lower in Manitoba than either Saskatchewan or Alberta (). It must also be noted however, that there were large differences in the number of isolates characterized from each province, and from each collection period of each province, that may also account for the variability of the results observed among provinces.
The avirulence alleles AvrLm3 and AvrLm4 were observed at relatively low frequency in this study. AvrLm3 may not be present in the Manitoba pathogen population but occurs in populations of the pathogen in Saskatchewan and Alberta at relatively low levels (). This suggests the pathogen population has adapted to the corresponding resistance gene, Rlm3, rendering it relatively ineffective, particularly in Manitoba. This can be explained if Rlm3 was present in Canadian B. napus varieties for some time. It may be that a higher proportion of the cropping area in Manitoba was seeded with varieties that harboured Rlm3 than in Alberta and Saskatchewan. The resistance gene Rlm3 was present in a number of older open-pollinated Canadian varieties and Rlm4 in some current Australian varieties (Kutcher et al., Citation2008). The proportions of AvrLm3 and AvrLm4 in our set of isolates from western Canada show a similar, though less pronounced, trend to that observed in the pathogen population in France and other oilseed rape growing countries of northern Europe (Balesdent et al., Citation2006; Stachowiak et al., Citation2006), where these avirulence alleles were reported at very low levels.
The observation that only a minority of the isolates carried AvrLm5 and AvrLm7 is surprising since the corresponding resistance genes are not believed to be in use in Canadian canola varieties. Similar situations have been observed by both French and Australian researchers who detected isolates of L. maculans with the ability to overcome specific resistance genes prior to exposure of the pathogen population to commercial varieties carrying these sources of resistance (Li et al., Citation2005; Balesdent et al., Citation2006). The Rlm5 gene is found in B. juncea and is not likely present in any B. napus lines. However, it is possible that virulent isolates of L. maculans were selected through the production of B. juncea condiment mustard crops or canola-quality B. juncea even though blackleg is generally not a problem for mustard growers and the production of canola-quality B. juncea is recent and very limited. This species is usually grown in the warmer, drier region of the southern prairies, the brown soil zone, and few isolates examined in this study were collected from this region. The proportion of isolates carrying AvrLm5 appeared to be lower in Manitoba than in other provinces (), which again is surprising since little B. juncea mustard or canola-quality B. juncea is produced in Manitoba.
The proportions of the isolates carrying AvrLm7 were similar among provinces, although it was observed that the proportion was greater in the 2003–2005 collection period than in 1997–2000 (). This is curious since there is no evidence that Rlm7 is present in either older or current Canadian B. napus varieties, although Rlm7 has been suggested to be present in B. rapa germplasm (Leflon et al., Citation2007), and previously B. rapa varieties were commonly grown in western Canada. It is also possible that some Brassica or closely related weed species may harbour this gene. The proportions of both AvrLm5 and AvrLm7 are very high in both France and Europe (Balesdent et al., Citation2006; Stachowiak et al., Citation2006), and Rlm7 is carried by a number of current French oilseed rape varieties. Similar to the isolates in this study, Australian isolates of L. maculans were shown to vary for the presence of AvrLm7, with a high proportion able to overcome the corresponding resistance gene, Rlm7 (Balesdent et al., Citation2005).
In this study, AvrLm9 was detected in 60.4% of the isolates (), indicating that Rlm9 may be beneficial as a source of resistance in Canadian canola varieties, depending on how and where this resistance is employed. European populations of L. maculans have been reported to lack AvrLm9 (Balesdent et al., Citation2006; Stachowiak et al., Citation2006). Rlm9 is common in European winter-type oilseed rape varieties and selection pressure to overcome this resistance gene has been proposed as the reason why AvrLm9 is not present in the European pathogen population (Balesdent et al., Citation2005).
This study provides an analysis of the avirulence gene frequency among isolates from the population of L. maculans in western Canada and an indication of the races present. Generally, the results of this study agree with those observed by Balesdent et al. (Citation2005), who found the absence of AvrLm7 (and AvrLm8), but the presence of AvrLm1, AvrLm2, AvrLm3 and AvrLm9 in isolates from Saskatchewan. Differences in frequencies of avirulence alleles observed between Canadian and European pathogen populations may be due to differences in the resistance genes used between the continents and to the length of time the various resistance genes have been carried by B. napus varieties. While a larger sample size of isolates from western Canada would be desirable for determining allele frequencies of these avirulence genes, Balesdent et al. (Citation2006) suggested that analyses of collections of intermediate sample size (100–200 isolates) allow a first description of the races or race structure of the pathogen population in a country. In France, smaller population surveys have been shown to agree well with a large-scale survey. In the present study, it must be remembered that the canola varieties from which the isolates were collected were not identified and may have carried race-specific resistance genes against L. maculans, although there is little knowledge of the resistance genes carried by past or current canola varieties in Canada. Therefore, it is possible that the proportions of isolates carrying some of the avirulence alleles in this study are lower than in the pathogen population as a whole in western Canada since isolates were obtained from visible stem lesions, which normally develop only as a result of the pathogen's ability to overcome any specific resistance gene(s) of the variety from which it was collected. On the other hand, it is possible that some varieties carry only quantitative resistance and do not harbour specific resistance genes. Nevertheless, this study provides significant knowledge of avirulence allele variation of the pathogen population of L. maculans in western Canada.
It appears that the western Canadian L. maculans population is different from that of Europe. The study indicated that some resistance genes, including some not likely to be currently present in Canadian canola varieties, are unlikely to be effective against the population of L. maculans in western Canada. However, other resistance genes, including currently unexploited resistance genes, may be useful sources of resistance. Development of effective genetic resistance management strategies will require complementary studies to determine the frequency of specific resistance genes present in current Canadian canola varieties.
This research was based on a rather limited number of isolates collected from unidentified varieties of B. napus. Consequently, a more precise study is being undertaken to obtain isolates from a variety of B. napus known to have no specific resistance genes to be sown at various locations to obtain an unbiased and extensive characterization of the western Canadian L. maculans population. Knowledge of avirulence gene frequency in the pathogen population is important to maintain durable resistance. From this knowledge, coupled with information on specific gene resistance in canola varieties, strategies can be developed to mitigate resistance breakdown, which has been shown to occur experimentally (Brun et al., Citation2000) and in practice (Li et al., 2003; Rouxel et al., Citation2003b; Sprague et al., Citation2006). The information provided by this study can be used to promote cultural blackleg control measures and develop effective strategies for the deployment and rotation of specific-resistance genes and the breeding of varieties with both qualitative and quantitative resistance.
Acknowledgements
The authors thank Dan Cross, Colleen Kirkham, Magali Ermel and Bruno Marquer for technical assistance, to co-operators who provided fungal isolates and to Dr T. K. Turkington for a review of the manuscript. The senior author wishes to acknowledge the support of the Organisation for Economic Co-operation and Development's Co-operative Research Programme: Biological Resource Management for Sustainable Agriculture Systems in the funding of this research. Additional financial support for this work was provided by the Saskatchewan Canola Development Commission (SCDC) and the Alberta Canola Producers Association (ACPC) through the Canola Agronomic Research Program of the Canola Council of Canada and the Matching Investment Initiative program of Agriculture and Agri-Food Canada. “Merci beaucoup” à l'Institut National de la Recherche Agronomique (INRA), Rennes for hosting the senior author while on work transfer and to Agriculture and Agri-Food Canada for supporting the work transfer.
References
- Ansan-Melayah , D. , Balesdent , M.-H. , Buée , M. and Rouxel , T. 1995 . Genetic characterization of AvrLm1, the first avirulence gene of Leptosphaeria maculans . Phytopathology , 85 : 1525 – 1529 .
- Ansan-Melayah , D. , Balesdent , M.-H. , Delourme , R. , Pilet , M.L. , Tanguy , X. , Renard , M. and Rouxel , T. 1998 . Genes for race-specific resistance against blackleg disease in Brassica napus L . Plant Breed. , 117 : 373 – 378 .
- Balesdent , M.H. , Attard , A. , Ansan-Melayah , D. , Delourme , R. , Renard , M. and Rouxel , T. 2001 . Genetic control and host range of avirulence toward Brassica napus cultivars Quinta and Jet Neuf in Leptosphaeria maculans . Phytopathology , 91 : 70 – 76 .
- Balesdent , M.H. , Attard , A. , Khun , M.L. and Rouxel , T. 2002 . New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans . Phytopathology , 92 : 1122 – 1133 .
- Balesdent , M.H. , Barbetti , M.J. , Li , H. , Sivasithamparam , K. , Gout , L. and Rouxel , T. 2005 . Analysis of Leptosphaeria maculans races structure in a world-wide collection of isolates . Phytopathology , 95 : 1061 – 1071 .
- Balesdent , M.H. , Louvard , K. , Pinochet , X. and Rouxel , T. 2006 . A large-scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France . Eur. J. Plant Pathol. , 114 : 53 – 65 .
- Brun , H. , Levivier , S. , Ruer , D. , Somda , I. , Chèvre , A.M. and Renard , M. 2000 . A field method for evaluating the potential durability of new resistance sources: application to the Leptosphaeria maculans/Brassica napus pathosystem . Phytopathology , 90 : 961 – 966 .
- Brun , H. , Ruer , D. , Levivier , S. , Somda , I. , Chèvre , A.M. and Renard , M. 2001 . Presence in Leptosphaeria maculans populations of isolates virulent on resistance introgressed into Brassica napus from B. nigra B genome . Plant Pathol. , 50 : 69 – 74 .
- Chen , Y. and Fernando , W.G.D. 2006 . Prevalence of pathogenicity groups of Leptosphaeria maculans in western Canada and North Dakota, USA . Can. J. Plant Pathol. , 28 : 533 – 539 .
- Chèvre , A.M. , Barret , P. , Eber , F. , Dupuy , P. , Brun , H. , Tanguy , X. and Renard , M. 1997 . Selection of stable Brassica napus–B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 1: Identification of molecular markers, chromosomal and genomic origin of the introgression . Theor. Appl. Genet. , 95 : 1104 – 1111 .
- Chèvre , A.M. , Eber , F. , This , P. , Tanguy , X. , Brun , H. , Delseny , M. and Renard , M. 1996 . Characterization of Brassica nigra chromosomes and blackleg resistance in B. Napus–B. nigra addition lines . Plant Breed. , 115 : 113 – 118 .
- Delourme , R. , Brun , H. , Ermel , M. , Lucas , M.O. , Vallee , P. , Domin , C. , Walton , G. , Li , H. , Sivasithamparam , K. and Barbetti , M.J. 2008 . Expression of resistance to Leptosphaeria maculans in Brassica napus double haploid lines in France and Australia is influenced by location . Ann. Appl. Biol. , 153 : 259 – 269 .
- Delourme , R. , Chèvre , A.M. , Brun , H. , Rouxel , T. , Balesdent , M.H. , Dias , J.S. , Salisbury , P. , Renard , M. and Rimmer , S.R. 2006 . Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus) . Eur. J. Plant Pathol. , 114 : 41 – 52 .
- Delourme , R. , Pilet-Nayel , M.L. , Archipiano , M. , Horvais , R. , Tanguay , X.R , Rouxel , T. , Brun , H. , Renard , M. and Balesdent , M.H. 2004 . A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus . Phytopathology , 94 : 578 – 583 .
- Howlett , B.J. 2004 . Current knowledge of the interaction between Brassica napus and Leptosphaeria maculans . Can. J. Plant Pathol. , 26 : 245 – 252 .
- Kutcher , H.R. , Keri , M. , McLaren , D.L. and Rimmer , S.R. 2007 . Pathogenicity of Leptosphaeria maculans in western Canada . Can. J. Plant Pathol. , 29 : 388 – 393 .
- Kutcher , H.R. , Rimmer , S.R. , Balesdent , M.H. , Rouxel , T. and Brun , H. 2008 . Detection of specific resistance genes against Leptosphaeria maculans in Brassica napus . [Abstract]. Can. J. Plant Pathol. , 30 : 386
- Kutcher , H.R. , van den Berg , C.G.J. and Rimmer , S.R. 1993 . Variation in pathogenicity of Leptosphaeria maculans on Brassica spp. based on cotyledon and stem reactions . Can. J. Plant Pathol. , 15 : 253 – 258 .
- Leflon , M. , Brun , H. , Eber , F. , Delourme , R. , Lucas , M.O. , Vallée , P. , Ermel , M. , Balesdent , M.H. and Chèvre , A.M. 2007 . Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus . Theor. Appl. Genet. , 115 : 897 – 906 .
- Li , H. , Barbetti , M.J. and Sivasithamparam , K. 2005 . Hazard from reliance on cruciferous hosts as sources of major gene-based resistance for managing blackleg (Leptosphaeria maculans) disease . Field Crops Res. , 91 : 185 – 198 .
- Li , H. , Sivasithamparam , K. and Barbetti , M.J. 2003 . Breakdown of a Brassica rapa subsp. sylvestris pathogenicity grouping of isolates of Leptosphaeria maculans on Brassica napus cultivars and their disease reaction profiles on rapid-cycling Brassicas . Plant Dis. , 75 : 1279 – 1282 .
- Mengistu , A. , Rimmer , S.R. , Koch , E. and Williams , P.H. 1991 . Pathogenicity grouping of isolates of Leptosphaeria maculans on Brassica napus cultivars and their disease reaction profiles on rapid-cycling Brassicas . Plant Dis. , 75 : 1279 – 1282 .
- Rimmer , S.R. 2006 . Resistance genes to Leptosphaeria maculans in Brassica napus . Can. J. Plant Pathol. , 28 : S288 – S297 .
- Rimmer , S.R. 2007 . “ Chasing genes for resistance to blackleg and sclerotinia in Brassica napus ” . In Proceedings of the 12th International Rapeseed Congress Wuhan, China
- Rouxel , T. , Penaud , A. , Pinochet , X. , Brun , H. , Gout , L. , Delourme , R. , Schmit , J. and Balesdent , M.H. 2003a . A 10-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene of oilseed rape . Eur. J. Plant Pathol. , 109 : 871 – 881 .
- Rouxel , T. , Willner , E. , Coudard , L. and Balesdent , M.H. 2003b . Screening and identification of resistance to Leptosphaeria maculans (stem canker) in Brassica napus accessions . Euphytica , 133 : 219 – 231 .
- Sprague , S.J. , Balesdent , M.H. , Brun , H. , Hayden , H.L. , Marcroft , S.J. , Pinochet , X. , Rouxel , T. and Howlett , B.J. 2006 . Major gene resistance in Brassica napus (oilseed rape) is overcome by changes in virulence of populations of Leptosphaeria maculans in France and Australia . Eur. J. Plant Pathol. , 114 : 33 – 40 .
- Stachowiak , A. , Olechnowicz , J. , Jedryczka , M. , Rouxel , T. , Balesdent , M.H. , Happstadius , I. , Gladders , P. , Latunde-Dada , A. and Evans , N. 2006 . Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe . Eur. J. Plant Pathol. , 114 : 67 – 75 .
- Van de Wouw , A.P. , Marcroft , S.J. , Barbetti , M.J. , Li , H. , Salisbury , P.A. , Gout , L. , Rouxel , T. , Howlett , B.J. and Balesdent , M.H. 2008 . Dual control of avirulence in Leptosphaeria maculans towards a Brassica napus cultivar with ‘sylvestris-derived’ resistance suggests involvement of two resistance genes . Plant Pathol. , 58 : 305 – 313 .
- Yu , F. , Lydiate , D.J. and Rimmer , S.R. 2008 . Identification and mapping of a third blackleg resistance locus in Brassica napus derived from B. rapa subsp . sylvestris. Genome , 51 : 64 – 72 .