ABSTRACT
This paper pursues the history of biology and technology in tandem. It focuses on DNA’s materiality regardless of informational properties. My emphasis on ‘making’ integrates attention to cultures of work in material histories of biology with analyses of the development of technical apparatuses and machines. When it comes to the history of DNA synthesis our materials are as much chemical as they are biological, which means that there is really a third history present, one that also needs to be drawn in, but on its own terms. I demonstrate the ways in which different chemistries have been combined with different technologies, all together affording different arrangements of personnel and biological science. It is a history of how synthesised DNA first came to be, became desired, and became a commodity, available for inclusion in a wide variety of experiments and experimental systems. This method could be replicated for other ‘experimental commodities’.
1. Introduction
How has DNA been made, how was it made into an experimental commodity, and why does this all matter? We can begin, perhaps a little conservatively, by appreciating that how DNA is made in organisms became a central research question in modern biology particularly once the significance of these molecules for heredity and development was recognised.Footnote1 It is also true that today virtually everything that goes on in a molecular biology laboratory (broadly defined) depends on the ability to acquire precisely defined lengths of DNA, and that annual sales of synthesised DNA run into the hundreds of millions of dollars. For more than half a century synthesised deoxyribonucleic acid has been both a target of investigation and a laboratory tool.Footnote2 Only some aspects of this history, which spans the length of the twentieth century into the twenty-first and implicates all corners of the globe, can be covered here. It is to be hoped that this paper will help inspire a deeper and more global investigation of DNA synthesis.
In truth ‘DNA synthesis’ is an inadequate term, because this is something that can occur in a number of different ways, the differences or similarities between which open up ground for competition as to ‘true’ synthesis, or erase differences that matter fundamentally by extending the word synthesis beyond its useful bounds. I cannot say ‘non-biological synthesis’ because the question of what does or does not constitute biology recurs throughout. Nor can I say ‘chemical synthesis’, not only because that would beg the same question as biology, but also because the differences between the kinds of early apparatus used for these purposes and the eventual technologies that commercialised them for the consumer cannot be reduced to chemistry alone. And indeed the chemistry by which DNA is made can itself be different: for that part of the story this is precisely the point. At a workshop dedicated to the history of DNA synthesis I made use of the term, ‘mechano-chemical’ to capture the various combinations of instruments and chemical approaches that can make DNA, but this term received as many furrowed brows as it did wrinkled up noses.Footnote3 For now I shall resort to saying DNA synthesis and allow you to gather the nuances and differences that this masks as we go.
In carving a space for DNA synthesis I am also creating its broader context, that of the history of ‘making’ DNA. My interest and focus on making came as a direct result of my collaboration with social scientists on the Engineering Life project.Footnote4 On this side of the problem, we might worry that referring to all kinds of DNA production as ‘making’ is inappropriate, perhaps because making demands a maker. Consider the research of persons such as Arthur Kornberg, whose work determining the conditions necessary to ensure the successful completion of enzymatic actions that build DNA in a test tube were international headline-grabbing news.Footnote5 Kornberg did not ‘make’ anything, we might argue, rather he facilitated the translocation of phenomena that typically occur in one place, the cell, to another, the test tube.Footnote6 But this kind of putative distinction is precisely the stuff of history.
That DNA synthesis and its ways of making DNA much more clearly implicated actors as designer-makers was part of the polemic surrounding these technologies at their origin, and contributed to inspiring new cultures of biological science and technology. This influence is today most clearly visible in the community of synthetic biologists, but was also visible in earlier parts of chemistry, molecular biology, industrial biology, and perhaps also within other subcultures of biological science. In this article I do not intend to write a history of synthetic biology or only of synthetic biology. Instead this paper provides the foundations for a new integrated historiographical approach, one working at a level removed from the particulars of synthetic biology, but which nevertheless has very direct implications for synthetic biology and its historians.Footnote7 Emphasising that from the outset the methods of making DNA discussed here were directly associated with notions of design and deliberateness, I am not saying we need to adopt these valuations and interpretations. Rather that we need to be alert to them and their history. These kinds of consideration are all the more urgent in a paper that aims to draw together histories of biology and technology, in ways that I hope are somewhat novel, but which are also inspired by and build on a range of historical discussions and models.Footnote8
In his path-breaking book The Uses of Life, Robert Bud argued that there is more to biological technology than biotechnology, the corollary being that biotechnology represents only one distinct culture of biology and technology together.Footnote9 Despite these conclusions opening up a vast field of view, he is most commonly cited as merely demonstrating that biotechnology has a long history, which is an unhappily narrow appreciation of his results, and potentially an incorrect one. I have tried to take The Uses of Life to heart, by allowing different actors to have their own understanding of how their biological materials are more or less manufactured, while also imposing my own historiographical category of ‘making’ which will be appealed to throughout. A key pay-off for doing so is that we get to watch as DNA synthesis eventually does come to be assimilated into ‘biotechnology’, in ways that pay no attention to its prehistory, precisely as we would expect in a strict future-oriented biotechnological mode, but which we would miss if only starting with or looking for biotech.Footnote10 A history of making also allows me to walk through territory often carved up according to structure and information without allowing either of these terms, or their combination, to constrain the narrative.Footnote11
In Culturing Life Hannah Landecker recognizes actors in the past and present rendering their biological materials in technological terms. Her account is of scientific actors remaking biology as technology. At the same time she emphasises the need for us, as contemporary analysts, to alienate ourselves from these interpretations, to make them strange again. Alienation facilitates history-making by resisting the assumptions of actors as facts of the matter, making it easier to see the social, epistemic and political work that these characterisations achieved and continue to achieve. I agree with the need to do this, but I intend to do so symmetrically, with all of the actors and interlocutors who claim to have things to say on behalf of biology. By contrast, one of Landecker’s strategies is to counter technological renderings of biology with her own, in which biology’s plasticity and temporal features are emphasised, so that an alternative biological discourse is made possible, through life no less. I do not adopt this strategy. The alternative view of biology that Landecker is putting together would be better looked for within the cast of characters that my historical work hopes to explore and explain, rather than be included in the assumptions undergirding this paper. Setting life aside is also a way in which I can make my history more immediately available to a wider range of philosophers and sociologists of science, who may find the case useful for discussions of affordance, bio-objects, the materiality of data practices, biological engineering, integration of the philosophy of biology and chemistry, or narrative knowledge.Footnote12
Angela Creager’s integrative efforts between biology and chemistry have been hugely important for building the historiographical landscape present in this paper.Footnote13 Though she does not explicitly call for additional integrative work between historians of biology and technology, this is effectively what she does in some prominent places, as in her chapter on the development of the ultracentrifuge and its incorporation into experimental systems. In addition, Creager’s attention to the means and methods of making diverse things also resonate in my case. A potential difference between our approaches, though not a hard-and-fast one, is that I am decidedly warier of the need for informational interpretations of genes and DNA for the purposes of driving historical accounts forward. I interpret the dominance of an information framework as having helped marginalize the cases discussed here, and no doubt many other aspects of biological making that have mattered on their own (not-necessarily informational) terms.
Given all of the overlapping and interrelated biological, technological, chemical and engineered components that make up a history of DNA synthesis, it has received remarkably little historical attention. The work of Har Gobind Khorana is the most thoroughly explored thus far, but most historians have worked at a conceptual scale where the genetic ‘code’ looms large, leaving much more to be done.Footnote14 In Membranes to Molecular Machines Matthias Grote indeed pushes us further, arguing that the importance of Khorana’s research has been understood much too narrowly thanks to the existing historiographical bias towards information and code breaking. Grote also points out that Khorana’s research interests have typically excluded him from a historiography that has remained focussed on molecular genetics and metabolic chemistry.Footnote15 The informational aspect of the history of DNA has been allowed to stand in for too much of this history. At the same time, the many roles played by DNA synthesis have been underappreciated because historians have typically treated all the ways of making DNA that emerged in the second half of the twentieth century as part of an amorphous biotechnological mass. Perhaps they have ultimately been right to do so- this judgement will always depend on the question being pursued. My questions were: how can we write histories of biology that keep material-semiotics and economics close at hand?; How can we create space for engineering – as a profession, a body of knowledge, an additional or alternative epistemology, etc. and not just as something appealed to rhetorically – in the history of biology?; How can historiography of biology and technology learn from one another? In order to keep these histories within the purview of the history of biology, we need to decenter heredity from biological history, a move recently advocated for by Angela Creager, and decenter DNA from its own history, a move similar to Eden Medina’s recent call to decenter the computer from histories of computing.Footnote16
The approach of commodity histories is particularly attractive for thinking across biology and technology at once.Footnote17 The foundations for commodities histories in biological science have been well laid in accounts of the creation of research organisms,Footnote18 the making of an international marketplace for biological information,Footnote19 histories of various international businesses of breeding,Footnote20 and of course in histories of biotechnology.Footnote21 The notion of an experimental commodity should be immediately recognisable to historians of science who have already attended to the commodification of scientific life.Footnote22 It should also be recognisable to contemporary scientists surrounded by commercial flyers, promotional tote bags, purchase orders, and receipts. It refers to those resources that are more or less vital to the daily operation of an experimental system, the majority of which are sold by specialist suppliers, which in biological science have come to include specified lengths of DNA of various different sizes. But those specialist suppliers did not always exist.
Methodologically this paper is based on a number of resources: oral history interviews with key figures from throughout the period, some choosing to remain anonymous; the presentations given at a workshop on the history of DNA synthesis organised between the Engineering Life project at the University of Edinburgh and the Science History Institute in Philadelphia; a small amount of archival investigation relating in particular to Applied Biosystems, Biogen, Celltech, and Vega Biotechnologies; and reviews of literature in chemistry and biology. The interview quotations have been edited to improve clarity, and in some cases expanded to include additional information received subsequent to the interview. The latter occurred on request by the interview subjects when they felt their original account was unhelpfully limited, but was only agreed to in cases where the additional information changed little of the tone or context of discussion.
2. Origins of the DNA synthesis knowledge community
The 1950s: in which chemists learn to make DNA in new technical apparatuses
This section walks historians of biology from an area they know well into new conceptual and analytical territory. My understanding of the origins and emergence of a DNA synthesis knowledge community is heavily dependent on the historicizing work of actors themselves and oral history interviews.Footnote23 The term ‘knowledge community’ is taken from Ann Johnson’s research on the history of engineering.Footnote24 Her analysis of how a new community of technical experts, scientists, and engineers can be grown up around a particular question or problem, which she dubbs the ‘attractor’, is directly applicable to the history of DNA synthesis. At the outset some of those most implicated in the history of synthesising DNA did not think of themselves really as ‘making’ it, and instead understood their work and its significance in other terms. Over time the larger goal of synthesising DNA became more nuanced thanks to additional goals, and become more clearly about making. These additional goals include that its synthesis be accomplished: more quickly; more reliably; more cheaply; more easily; less wastefully; and so on. By their nature, these kinds of additional goals required a combination of chemistry and technology in order for them to be surmounted, and so new technical apparatuses were designed. Over time the range of actors within the nucleic acids knowledge community who were prepared to tackle these problems became more sharply defined, and thus an even smaller professional community of DNA synthesisers emerged, precisely as Johnson argues for the case of antilock braking systems. This process is begun here and then further elaborated in Sections 3 and 4, below.
The first chemical synthesis of DNA nucleotides was completed in Cambridge, UK, in the laboratory of chemist Alexander Todd. Todd presents numerous interesting and important paths into the history of twentieth century science, linking chemical research to medicine (particularly through his method for the synthesis of vitamin B1, of considerable value to Hoffman-La Roche), planning and reconstruction during and subsequent to the Second World War, and science in national and imperial projects. He gained a considerable amount of recognition in his lifetime: awarded a knighthood in 1954; a Nobel in 1957; becoming Baron Todd of Trumpington in 1962.Footnote25 Soraya de Chadarevian’s history of molecular biology demonstrates that Todd was a consistently supportive figure for those at the University of Cambridge seeking to increase the institutional presence of biophysics and eventually molecular biology. We learn that Todd was asked to give his blessing to Watson and Crick’s model for the structure of DNA molecules prior to its publication, to avoid potential embarrassment before the chemical community. And in a long footnote, de Chadarevian also explains that Todd’s interests in nucleotides helped ensure they were subject to X-ray analysis in the Cavendish meaning that ‘detailed structural data of the nucleotides were available and of great use to Watson and Crick while they were working on the structure of DNA’.Footnote26 We are now in a position to look well beyond the helix. What really mattered about the nucleic acid research in Todd’s laboratory was that these chemists were learning how to make DNA, though Todd himself did not reduce his activities to these terms. Colin Reese, who joined the Todd lab in 1953 to complete a PhD, explains Todd’s route to nucleic acid synthesis as follows:
One of Todd’s main aims, which may have developed from his interest in the chemistry of the B group of vitamins, was the synthesis of the nucleotide coenzymes, and this work was highlighted in the citation when he received the Nobel Prize for Chemistry in 1957. However, the scope of the nucleotide research in Todd’s laboratory was very broad indeed and much fundamental research on nucleoside chemistry and chemical phosphorylation was carried out in it.Footnote27
So it would be wrong to single DNA out, even amongst the nucleic acids. Rather these were only some of the molecules of interest, co-enzymes taking precedent due to their offering a more likely route to therapeutic advance. Nevertheless, the synthesis of DNA amongst these many other things is central to subsequent developments in the history of biology.
The Todd lab’s synthetic work produced its own characterisation of DNA, which in the language of chemistry relied on notions of strength and weakness, mild reactivity and violent reactivity, stability and instability, yield, reaction steps, and so on. His work can be readily placed within Lily Kay’s international historiography as another investigator of biological ‘specificity’.Footnote28 But better appreciating the distinctiveness of the work of a synthetic chemist can also trouble any primarily informational view of either the history of molecular biology or the nature of specificity. We can begin to understand how by briefly comparing and contrasting Todd’s research with two other chemists equally invested in the structure and function of DNA at this time, but by other methods and with different aims. The first, Kurt G. Stern, was at this same time working in the Department of Chemistry of the Polytechnic Institute in New York.Footnote29 There adopting the assumption that DNA was indeed the material of heredity, alongside a number of additional constraints regarding how cellular development through such a material would have to work, he began to derive theories of gene structure. While there is clearly an informational component to Stern’s perspective, which Kay draws our attention to, it also matters that he was working out a set of characteristics designed to capture a material that he had very much at hand (his obituary lists the making of instruments to purify enzymes and nucleic acids amongst his accomplishments) and, in addition, that his informational thinking was never removed from a world of material.
If the polynucleotides are formed by the condensation of tetranucleotide units, it is obvious that each such building block, upon joining up with the chain, has the choice of ‘head to tail’ and ‘tail to tail’. In this manner ‘modulated’ nucleic acid chains incorporating many different gene codes could be created by the cell.Footnote30
While an informational component is clearly present, I want to emphasise that the material discourse is equally well worth preserving, particularly as people like Todd and Stern were learning to acquire and manipulate DNA ‘building blocks’ at this time, precisely as organic chemists and biochemists had already been doing for many other substances for decades.Footnote31 As for the informational never being divorced from the material, when Stern turns to explain his views on how genes must be structured, including photographs of physical models he had made, he leans on an analogy with technologies for capturing voice recordings, including photographs of the tracks made into wax surfaces by a recording stylus.Footnote32 This, I argue, is a material imaginary of DNA as much as an informational one.
The second, Erwin Chargaff, is a figure already well known to historians of biology thanks to his ‘rule’.Footnote33 He is important first as a significant member of the early DNA synthesis knowledge community in his own right, going on to co-edit the 1955 two volume The Nucleic Acids: Chemistry and Biology, and also for the further support he offers to my decentering of heredity and emphasis on making.Footnote34 Best known for his research determining the ratios of different nucleosides contained within different samples of DNA, comparing and contrasting these between species, Chargaff too placed DNA as one group of molecules amongst many.Footnote35 Nevertheless, a key aspect of his research was concerned with the making of quantities of DNA nucleosides, achieved by improving methods for enzymatically degrading lengths of DNA sourced from organisms. Precisely this kind of research was also underway in the Todd lab at this time, though with the additional ambition of assembling the degraded nucleosides into new dinucleotides. In the same way that Todd did not necessarily think of these aims as learning to make DNA, though he and his staff clearly were doing so, likewise Chargaff was learning to make the raw materials for DNA synthesis, though his own interests lay in understanding the spatial arrangement of molecules in the cell. The laboratory life of organic chemists and biochemists brought them into contact with DNA as a substrate for which they had particular authority and control. We can cement their inter-relationship with DNA by further pursuing Chargaff and Todd’s degradation of DNA, which also brings commodities and key technologies into view.
Nobody working on the chemistry of DNA could do so without access to raw materials. To complete the work in the paper just cited, Chargaff had to get hold of calf thymus and liver, ox liver, sheep liver, etc. Todd’s lab meanwhile, in 1952, published their own method for scaling up the production of nucleosides, one based on using herring sperm as the initial raw materials from which DNA nucleosides could be extracted. ‘Herring-sperm deoxyribonucleic acid (180g.; 7.3% of P) of commercial origin (Isaac Spencer and Co. Ltd., Aberdeen) was dissolved in warm water’. An acknowledgement also went to ‘Messrs. Gea Ltd., Copenhagen, for a generous gift of deoxyribonuclease’.Footnote36 So DNA raw materials and the degrading agent were by this time already commodities that could be pumped into a laboratory’s experimental system. The first company mentioned, Isaac Spencer and Co. Ltd., had been founded in the 1880s to manufacture cod liver oils from the fishing trade, but were clearly prepared to diversify.Footnote37 The second was a Danish pharmaceutical manufacturer. One of the current paper’s largest gaps in knowledge is that I have not been able to track when and where different suppliers of nucleoside raw materials emerged, and how a market developed.Footnote38 I have however managed to learn how these materials came to be used, how they have changed over time, and the close relationship between technical methods for the extraction of nucleosides or nucleotides from organic sources and their subsequent synthesis. Even in the Todd laboratory, where processes for making DNA nucleosides and nucleotides through degradation were being refined and improved, it quickly made more sense for companies to take care of this work and then sell the nucleosides to laboratory workers. In some cases this was for the purposes of developing methods for their synthesis. We must also recognise the importance of column chromatography, which enabled not only the purification of these nucleosides, but would also go on to form essential technical apparatus for synthesis.
The configurations of biology and chemistry I have just outlined make no sense outside of a technological repertoire.Footnote39 I have been very fortunate to meet the historian Apostolos Gerontas, who has recently completed the first deep study of the emergence and eventual development of chromatographic technique.Footnote40 Chromatography is essentially a method for the separation of different chemical substances, but as we shall see, it also goes on to serve a number of other functions. Gerontas shows that though the technique was first developed by Mikhail Tsvet around the turn of the twentieth century, it was not until the 1930s that it really began to be taken up widely by organic chemists, who had otherwise remained committed to chemical methods of separation and degradation.Footnote41 Todd himself reflects on his work’s dependency on these technologies in his Nobel lecture, given in December 1957.Footnote42 Combining the chromatograph with the spectrophotometer – which measured the capacity for a substance to absorb light – provided the chemist with methods for getting hold of the material they wanted and also checking that they really did have it. This is precisely the kind of work that the Todd lab undertook in their research leading to the synthesis of DNA. In the process, as I have tried to explain in this first section, DNA nucleosides and nucleotides were given further characteristics, in this instance by 1) their rate of diffusion through filtering materials in a chromatographic column, and 2) their capacity to absorb light. The early DNA synthesis knowledge community was composed of a broad and heterogeneous group, all invested in DNA synthesis for a wide variety of reasons. In the next section, as the nucleic acids community grows, so does the ambition to synthesise become sharper and more competitive, so that Ann Johnson’s process towards professionalisation gets underway.
3. Successful syntheses: but success by what measure?
The late 1950s and 1960s: In which DNA synthesis increasingly becomes an end in itself
The previous section provided a broader comparative context for the work of the Todd lab, without actually getting into the successful synthesis of a dinucleotide. This was published in 1955 in paper number 32 of Todd’s nucleotide series, work carried out largely by one of his laboratory research staff, Adolf Michael Michelson.Footnote43 We should note that these researchers occupied a field effectively identified as belonging to Todd. One’s aim was to be incorporated as a co-author on a paper inside Todd’s nucleotide series, or the series of papers on phosphorylation, and so on. The extent to which all of the outputs of the Todd lab are habitually located in his person is testimony to his effective management of IP-broad priority claims.Footnote44 This circumstance also points to the extent to which the directions and strategies for research were largely routine, which does not in the least undermine them as difficult or draining. Systematically altering major and minor features of a reaction, searching for and trialling different chemicals to act as reagents or blocking agents etc., developing novel pathways between steps, measuring inputs and outputs, dealing with mistakes and experimental failures, intricate apparatuses, this was the daily work of learning to synthesise DNA. Which brings us to motivation. There already existed methods for the making of DNA that we could think of as more biological. ‘Hitherto the synthesis of simple nucleotides by phosphorylation of nucleosides, using other than enzymic methods, has not been very practicable owing to the limitations of the methods used’.Footnote45 The point, for Todd’s lab, was to create methods for synthesis that did not rely on enzymatics, which to the organic chemists’ mind was something like cheating. Or if not cheating, then less worthwhile, because it missed out on the key pay-off for the synthetic chemist; that the successful completion of a total synthesis effectively proved the correctness of your theoretical structure, and kept you from needing difficult-to-attain starting materials, such as enzymes. Which brings us to evaluation. Todd explains in his autobiography that he had little to no interest in actually pursuing polynucleotide synthesis because the nature of the work was too repetitive and boring.Footnote46 Michelson was largely left to his labours, and despite the Todd and Michelson paper becoming a common rallying point for those building a history of DNA synthesis, the actual method developed, dependent on phosphotriesters, was initially only of use for making dinucleotides (two base pairs).Footnote47 Instead, an altogether different kind of chemical approach gathered momentum, in part thanks to its immediate capacity to create longer oligonucleotides.
In the mid 1950s, one of Todd’s researchers, George Kenner, went to pursue postdoctoral work at the ETH in Zurich in the lab of Vladimir Prelog. There he met and became friendly with other postdocs interested in chemical synthesis of biological molecules, including Har Gobind Khorana. Having convinced Khorana to return with him to Cambridge to further his protein synthesis chemistry, Khorana ends up becoming interested there in the nucleotide work being pursued by Todd and Michelson. However, rather than continue working on phosphotriester chemistry Khorana moves on to develop a chemistry of his own, based on phosphodiesters, leaving the UK to take up a post at Vancouver in the Fisheries Research Board of Canada. Using his independent chemical method, first published in 1956, Khorana found ways to make small trimers of DNA chemically, which could then be linked up with enzymes.Footnote48 It was by this mixed approach that his lab completed the first synthesis of a whole gene.
In moving towards the later 1950s and early 1960s, in which we find a growth of interest surrounding Khorana’s alternative phosphodiester approach, our textual sources can begin to be informed with participants from oral history interviews, such as Marv Caruthers, Josef Jiricny and Curt Becker. Caruthers’ contributions to the history of DNA synthesis are amongst the most well-known, particularly through his co-founding of the company Applied Biosystems (henceforth, ABI) and development of the phosphoramidite chemistry at its centre, the latter constituting the third and final chemical strategy for the synthesis of DNA covered in this paper. Jiricny’s example is no less illuminating, providing as he does a concrete example of the kind of organic chemist who had begun to be asked to make DNA more routinely for biologists, using the phosphotriester strategies, and who by the end of the 1970s was looking for ways to professionalise this practice. Becker meanwhile was a person on the ground at the origins of ABI, playing key roles in the development of their products, particularly how to best commodify DNA using the phosphoramidite chemistry, working with Bill Efcavitch of ABI on development of their first commercial synthesier. In meeting these different transition points, with the fortunes of phosphotriester, phosphodiester and phosphoramidite methods ebbing and flowing, it is worth being explicit about the levels of competition surrounding these different chemistries.
Competition between different approaches and strategies is not some extra element of the history of chemistry but a fundamental feature. All participants and interlocutors have some kind of view or another about the proper ranking of chemical approaches and their permutations. Beyond the everyday work of chemists, this competition is also evidence of the growing specialisation of synthetic chemists within the nucleic acids research community. What had been the annual Gordon Research Conference on Proteins and Nucleic Acids, first organised in 1950 (with Kurt Stern as co-chair), became in 1962 two separate conferences, one on Proteins and the other focussed exclusively on Nucleic Acids. Around 10 years later the specialist journal Nucleic Acids Research would be launched. Of course many organic chemists, biologists, biophysicists, and biochemists would continue to work across the putative divisions, bouncing around different molecules. But a key part of the reason for tracks of specialisation to emerge like this at all, was that a very real race was on regarding who could synthesize best. While the improvement of synthesis methods was not the only kind of work underway in these areas, it was nevertheless the case that improving or suggesting paths towards better synthesis (better as meaning more reliable, more efficient, easier, faster, higher yielding), was a clear and attractive way in which to organise one’s lab work and explain the value of one’s results. The fact of competitiveness between chemical methods therefore forms part of the historiographical explanation for the eventual professionalisation of DNA synthesis. The suggested historiographical argument, that actors picked out efficiencies and dedicated themselves to improvements toward recognised shared goals, will be familiar to historians of technology and engineering. The critiques these historians have developed concerning the privileging of efficiencies, the need for engineering to recognise the politics of its epistemic and design goals, and that technologies do not work deterministically, are likewise necessarily born in mind.Footnote49
In addition, I argue that the kind of epistemological programme built up around the improvement of methods for the synthesis of DNA went on to have another life in biology. Synthetic chemistry’s epistemic goals and values, which we will soon see integrated into technologies for biology, came to be broadly embraced by synthetic biology, which most commonly understands itself to be making biology better, better as meaning more reliable, more efficient, easier, faster, higher yielding.Footnote50 In pursuing a historiographical integration of biology and technology, I have ended up bringing together elements that matter for the more specific history of synthetic biology. In this respect my argument is that synthetic biology locates and sees itself in its materials. While historians may expect much more evidence for such an argument than has been supplied here, or can be supplied in a single paper, this is the direction of argument that my integration of the historiography of biology and technology has taken me. Khorana’s synthesis of a gene is an excellent place to find further evidence of biology’s debt to chemistry, and the material-semiotic significance of synthesised DNA for biologists.
Soon after determining his own method for synthesis, Khorana began organising his lab around a larger project, to synthesise a full gene. Caruthers joined the lab roughly ten years after Khorana first published his independent synthesis method:
MC: These oligonucleotides were simple ones. Initially the yeast alanine tRNA gene was proposed to be synthesized using what’s now called PCR, but this was before PCR was invented by Kary Mullis. In other words Khorana actually developed PCR 10-12 years before Kary Mullis. The way Khorana was originally going to do this synthesis was to prepare two 20mers with 10 base pair overlaps and then carry out repair synthesis which would generate two 30mers. Then he was going to denature these 30mers, add new 20mers to each end by hybridization, and extend so as to generate 40mers. Further extensions using this strategy would generate the tRNA gene. Therefore the total synthesis was predicated upon chemically synthesizing 20mers.The only enzyme that was required was E. Coli DNA polymerase. This was the strategy as he explained it to me and was the approach I submitted as a postdoctoral fellowship grant application for research in his laboratory. As expected, this grant was funded. However by the time I joined the group as a postdoc, T4 DNA ligase and kinase had been discovered. As a consequence we were able to synthesize this gene using 12mers, which were easier to prepare. Our total strategy therefore changed. A small section of the gene was synthesized with 20mers, because that’s how we started, and the rest with 12mers, or in some cases oligomers even less than 12 in length.Footnote51
Caruthers also charts an increase in the amount of strategizing between making the first gene, yeast alanine tRNA, and the second, E.coli tyrosine suppressor tRNA.
MC: That was our strategy for the first gene: Using chemical synthesis followed by initially DNA polymerase, but later T4 kinase and T4 ligase in order to finish the project. For the next gene, the E. Coli tyrosine suppressor tRNA gene, we knew much more about how to strategize our chemical and enzymatic synthesis. For example we knew that the best ligation strategy was to join oligonucleotides having 5ʹ-pyrimidine to 3ʹ-pyrimidine. Therefore wherever possible we developed a synthesis plan where an oligonucleotide had pyrimidines at the 3ʹ and 5ʹ ends. In other words we designed our chemistry mainly to emphasize a synthesis strategy that would maximize overall yields and limit various chemical and enzymatic problems. And we put hard to synthesise nucleotides inside, so we didn’t have to do so much, so there was a fair amount of planning for the second one. And then on top of that I got involved, and so did Hans van de Sande, working with a faculty member over in Chem Engineering on formulating a computer program designed to synthesize genes based upon everything we had learned during the preparation of the yeast alanine and tyrosine suppressor genes. This was really a good exercise. We developed a computer program that would maximise our chemistry. We published that in JACS. I learned a lot about computer programming when combined with synthesis and how to integrate these two technologies.Footnote52
We see how difficult it was to get anything like a 20mer, or even the shorter 12mers they eventually landed on; the crucial role of enzymatics, the decision to put sequence lengths that were hard to synthesise ‘inside’, i.e. allow the enzymatics to fill in these sections by instead synthesising their complementary (and easier to synthesise) oligo; and the eventual increase in ‘planning’ of synthesis strategies, even resulting in collaboration with an engineer on a computer programme – DINASYN – to aid in making choices for sections of synthesis and strategies for their assembly.Footnote53 This is not a synthetic biology project, and to label it as such would be to appreciate it narrowly and anachronistically. Nevertheless, the key practical and epistemic features that these scientists focus on and emphasised are some of the very same commonly emphasised in synthetic biology today.
Here I can also take the opportunity to highlight global interest in methods for the improvement of synthetic DNA at this time.
MC: There were laboratories elsewhere all over the world that were probing how to build synthetic DNA. These included Alexander Todd, Michael Gait, and Colin Reese in England, Robert Teoule in France, Wolfgang Pfleiderer, Fritz Eckstein, Hubert Koster, Hartmut Seliger, Hans Fritz, and Friedrich Cramer in Germany, Kjeld Norris in Denmark, Maciej Wiewiórowski and Wojciech Stec in Poland, Mikhail Kolosov, Yu Ovchinnikov, and Zoe Shabarova in Russia, Wang De-Bao and Wang Yu in China, Saran Narang, Thomas Nielson, and Kelvin Ogilvie in Canada, Jacque van Boom in the Netherlands, Tsujiaki Hata, Mario Ikehara, and Eiko Ohtsuka in Japan, Jacek Smrt in Czechoslovakia, and Robert Letsinger and Keichi Itakura in the United States. There were a few labs in the US but not too many.Footnote54
MC: There was a conference that Hubert Köster hosted at a resort outside Hamburg in 1980. He literally had everybody in the world who was developing methodologies for DNA and RNA chemical synthesis at that meeting.Footnote55
Attention to this meeting, the 1980 ‘International Symposium on Chemical Synthesis of Nucleic Acids’ in Hamburg, alongside the annual international Gordon Conference meetings on nucleic acids, would be two of the best ways in which the history of DNA synthesis could be immediately made more global. I have not yet done this work, and it would require collaboration, but it would be essential to place what I have found so far in proper context.
4. Getting biology hooked on synthesis
The late 1960s, 70s and early 80s: In which chemists bring DNA to biology by diverse means, machines, and business models
Thus far I have focussed on those for whom synthetic DNA was a central research interest. But enrolling the average biochemist or biologist would prove to be a job of work.
MC: They sort of ignored it. Now that’s not a fair statement. Because people were interested but they couldn’t imagine how they were ever going to use it … Arthur Kornberg was very much interested in what we were doing and often discussed how to use synthetic DNA to solve biological problems … On occasions I have emphasized this observation by telling a story that happened to me at the 1975 Nucleic Acids Gordon Conference. This conference was five years prior to the time when we first published our break through research on DNA synthesis. I was asked to review DNA synthesis … that evening I was sitting around a table having a round of beers with maybe 8 or 10 other scientists you know, and one of them who is now a member of the US Academy of Sciences, looked at me and he said “Marv, why do you want to learn how to synthesise DNA?” He said “Khorana used synthetic DNA to solve the genetic code, and now he’s got his gene, but what else are you going to do with it?” He said “You’re a bright guy why don’t you go and do something more interesting?” … and that was sort of the general philosophy at the time, kinda “well you know people are going to dabble making synthetic DNA but what are we ever going to do with it?”
At the origins it was most common for a chemist to take on biological questions themselves, using synthetic DNA in the process, with biologists coming to be inspired or learn by example. In this section I show how synthetic DNA, already on its way to becoming a desirable input for experimenters, finally became an experimental commodity. Different chemistries, thanks to their features, invited different kinds of embodiment in machinery, different arrangements of lab personnel, and therefore different kinds of biological research. While ABI famously won the competition for market dominance, we can situate it amongst all its competitors by recognising how their 380A DNA synthesis machine, the chemistry it relied on, and the cost of its reagents, embodied particular lab personnel arrangements and biological aspirations. We can also better explain the success of their machine as arriving to supply a commodity that had already been made amenable, desirable, and desired over the previous 20 or so years, rather than actually launching this desirability itself.
One way in which to develop the historical characterisation of experimental commodities is by attention to the business models that emerged around them. Here the kind of object that a business model is taken to be is as much an epistemic enterprise as it is a commercial one.Footnote56 The analysis I borrow for these purposes has identified four (and no more than four) essential business models which, as ideal types, can be used to explore cases without conflating the actual business with the model it assimilates to.Footnote57 The four in question are: product model; solutions model; matchmaking model; and multi-sided model. The way in which DNA has been commodified has been diverse, and has changed over time. In the present paper only the first two, products and solutions, will appear, but matchmaking and multi-sided DNA synthesis businesses have most certainly emerged in the period beyond my focus of study. Adopting these categories helps focus my analysis, but also builds a further much needed path between the history of technology and biology in business history,Footnote58 and outward to histories of capitalism.Footnote59
Last, in this section we also reach the key difference between chemists who were DNA synthesisers, and those who went further, to become part of the professional DNA synthesis knowledge community. The latter, as Ann Johnson describes, elected to become embroiled in the process of designing and marketing products, while the former did not. Of course all chemical investigations still remained crucial for improving existing methods and searching for new ones, but the additional status of professional, as in Johnson’s case, makes sense provided we reserve it for that narrower group of people getting products to market, and who in the process created a new additional professional reputation that was going to be risked and tested on new terms.
4.1. Phosphotriester chemistry and the product business model
The fortunes of phosphotriester chemistry had been revived in the mid 1960s thanks to the intervention of Bob Letsinger, whose lab learnt how to affix the first nucleotide of a sequence to a solid support, vastly increasing the overall length of polynucloetide which this chemistry could achieve. Between the mid 1960s and the end of the 1970s phosphotriester chemistry came to dominate the scene. Decades of familiarity with phosphotriester chemistry, and the ready availability of the apparatuses and components needed to establish this chemistry in a single functioning unit on the lab bench, made the path between it and its embodiment in a bespoke DNA synthesis machine by the end of the 70s relatively straightforward. This area had also been subject to little commercially significant patenting activity. A search of the USPTO for ‘phosphotriester’ between 1940 and 1990 does return a few examples from the mid 1960s onwards, followed by a larger number from the 1980s onwards, but none give the appearance of offering a market monopoly on the making and synthesis of triesters.Footnote60 This is no doubt because phosphotriester chemistry always invited too many permutations for any given approach to provide a straightforward route to patentable, widely applying, and nonobvious methods, or at least we can conclude that such a patent culture was not successfully established in this chemistry. The kinds of machine that could be built to incorporate this chemistry were also small and simple. I interpret these machines as embodying the product business model, because in a very straightforward sense the consumer was being sold, in a single transaction, a standardised product designed to fit into their existing working lives. In these instances DNA was an experimental commodity under your personal and immediate control, because once you bought the machine the sky was the limit, provided your sky did not need to extend very far beyond 10-15mers. But the latter was more than enough for certain applications, particularly if you intended to ligate small oligos into larger polynucleotides. While not cheap, they were by and large priced within the reach of most labs regularly synthesising DNA, and could be used with almost any variety of chemistry and reagent.
Following the revival of interest in phosphotriester chemistry, organic chemists around the world continued attempting to improve it further.Footnote61 By the end of the 1970s, some had begun to develop these familiar bench-side rigs into something automatable, packed into a single instrument. Josef Jiricny was one such person, taking responsibility for the making of DNA for his own research and for nearby biologists.
JJ: I studied chemistry in Birmingham [1970-73] … .The reason I studied chemistry is because I emigrated when I was 18 from Prague, and of course as a refugee there was no chance of getting into a medical school. So I thought well, chemistry was close enough, but I realised that I needed some kind of biological angle to what I was doing, so when I finished my degree in Birmingham I then did a PhD in London [1973-76] … When I finished my PhD I really decided to try and change my direction from synthetic organic chemistry to something that’s more kind of medically or biologically applied. So I applied for a job with Colin Reese, who was then the Daniel Professor of Organic Chemistry at King’s College London in the Strand … I was able to work part time on this synthesis of oligonucleotides.Footnote62
Here he describes the process of putting together an automated system, photographed in .
Image 1. Josef Jiricny’s automated synthesis machine, using the phosphotriester approach.Pulled together with various parts from around the Imperial Cancer Research Fund. I am very grateful to Dr. Jiricny for permission to publish.
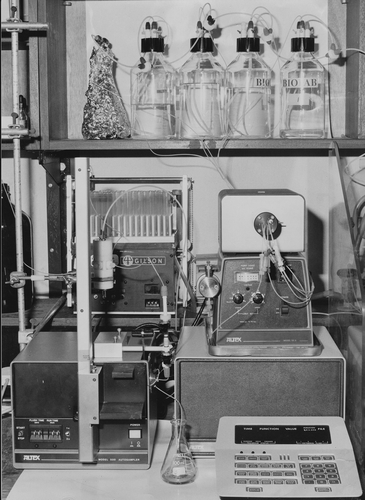
I have left the quote as intact as possible, because I know of no other description like it.
JJ: I was offered a postdoc at ICRF [Imperial Cancer Research Fund], in the Chemistry department. I was then wanting to synthesize a piece of DNA that was modified by some carcinogen, I wanted to get into cancer research and this had never been done. People treated mice with carcinogens, and treated cells with carcinogens, but nobody actually knew what were the exact structures of the adducts in the DNA … so I decided to synthesise the whole piece of DNA with this modification on it and then find out what the biological consequences of that modification were … I started developing, using Colin Reese’s chemistry, on solid phase, and it was working reasonably well. So I thought “why not automate it?”, because I was on the 4th floor in Lincoln’s Inn fields, and there … . [were]lots of machines which most of the time they weren’t using because the first thing they did with them was take them apart and try to improve them or modify them or whatever. A lot of the time the machines ended up not working after this little intermezzo. [They] had this old HPLC [High Precision Liquid Chromatography] which was sort of outdated … we were talking over coffee, and I was saying I would like to see if I can somehow automate the cycle because you essentially had to just write a very simple programme, you had the coupling reaction which needed to be recycled, joining the two, to send the compound through the column many times until it reacted. But then you needed to wash it off, to monitor how good the coupling was, to prepare the column for the next step and then inject the next nucleotide.Footnote63
Key features include: the presence of a chemist using synthetic DNA to answer biological problems; the ongoing significance of chromatography for the history of making DNA (here HPLC enabling automation); the overlap and interplay between synthesising one kind of molecule, amino acids, and another, nucleic acids; the need to begin understanding the inner workings of technologies in detail; and the growing interest in accessing synthetic DNA amongst biochemists and biologists, particularly those working in fields closely aligned to pharmaceuticals research. We can also glimpse the very widely and thoroughly established practice amongst synthetic chemists of pursuing experimental stepwise synthesis of modifications of a molecule of interest, to understand how these modifications change its properties. Such an experimental strategy is celebrated in central parts of synthetic biology.Footnote64 Though Jiricny was approached by companies interested in packaging this all into a single machine, this plan never came to fruition, for at precisely this time ABI’s DNA synthesiser, based on yet another entirely different chemistry, was released. In the face of its capabilities, Jiricny instead convinced ICRF to purchase one of these machines with the ambition of becoming manager of ICRF’s DNA synthesis core facility. Jiricny’s story may not be unique, but it is a concrete example of someone transitioning from being a chemist who can synthesise DNA, to aspiring to make it more professionally in bespoke machinery.
Others did make it all the way to packing phosphotriester chemistry into a marketable DNA synthesis machine. Leon Barstow, President of Vega Biotechnologies, emphasised how his machine could be considered superior to its competitors, including that of API, precisely because its phosphotriester chemistry increased an individual user’s control over their experimental system, including the reagents used and the kinds of chemical approach applied. In an undated essay, seemingly written during 1981, which was deposited at the Smithsonian with an example of the Vega machine (originally priced at around $50,000), Barstow laid out his stall: ‘Until automation, DNA fragments – usually 15 base units in length – were exceedingly expensive and formed the bottleneck to most bioengineering development projects. The preparation of a DNA fragment by classical solution methods could take from 3 to 6 months and cost from $25,000 to $50, 000ʹ.Footnote65 This was all now to change thanks to the availability of machines like his. ‘When I first began designing the VEGA DNA synthesizer in the spring of 1980, I visited most of the academic groups in the world that were doing research in solid phase DNA synthesis. One of the things I found was that most of the academic groups had their own unique approach and that each group was changing its approach on an almost weekly basis. Thus, flexibility was essential if the machines were to be able to accommodate changes in chemistry as they were introduced’. It is obviously the case that Barstow’s essay, as with any promotional material, is mainly serving his interests, and I have not yet found testimonials from clients that could speak to his machine’s functionality or reliability. But that’s not the point here. Rather it is to capture aspects of the ways in which DNA became a commodity. In this case, the central features were user control and customisable chemistry: ‘the phosphotriester method has been changed more than 30 times in the past two years. On the VEGA machine, all of these changes were made either through simple keyboard entry or, at worst, through a straightforward software change … The VEGA model 280 DNA synthesizer consists of a Micro-processor/Controller, a Chemistry unit, and a Printer … The technician simply fills out the form, indicating by alphanumeric codes the solvent or reagent, volume, reaction time, and disposal port. Once a protocol has been established, it can be stored on a floppy disk and reused at a future time’. These machines also aspired to give any bench worker their own device, suitable for self-sufficiency in the design of experiments requiring short polynucleotides. After all, it could not synthesise many sequences simultaneously.
Of course another way in which to commodify synthetic DNA in the phosphotriester approach was to reorganise one’s personnel, rather than buy a machine. This is what Genentech did.
MC: The phosphotriester chemistry was quite labour intensive because people made these tri-nucleotide blocks. For example for my first trip to Genentech, and I gave a seminar there, they had these walk in refrigerators filled with all 64 trinucleotide blocks, and then depending on what you were making chemically, you’d pull this block and this block to make your 6mer. And then this one and this one to make your other 6mer, and then you’d join them together. So they had technicians whose only job was to make trinucleotides. They had a group of I’d say 6 or 7 people who were strictly making oligonucleotides.Footnote66
Just as the first computers were people, so have our first DNA synthesisers been. In the next and final examples, we will see how other chemistries and technologies served alternative ambitions and required different organisations of laboratory personnel.
4.2. Phosphoramidite chemistry and the solutions business model
We have not yet properly introduced the phosphoramidite approach to DNA synthesis. Its sudden arrival here at the end of my story replicates its arrival into the DNA synthesis knowledge community. In stark contrast to the phosphotriester approach, it was known by virtually nobody outside those who developed it into a patented chemical process for the synthesising of DNA. Indeed the patent ambitions of Marv Caruthers and Serge Beaucage ensured that the international community was kept in the dark until Caruthers’ lab was entirely ready. Far from being a chemical system that everyone was familiar with, phosphoramidites were forbidding. Those who had tried working with phosphoramidites, such as Jiricny, had soon given up because the average level of moisture in the lab always activated the amidites quicker than they could be used in the intended reaction, no matter how much effort went in to keeping them sealed off (in our interview Jiricny confessed to thinking that the climates of San Francisco and Los Angeles contributed to Caruthers’ successes, being far less damp than the UK). In terms of commodification, University of Colorado Boulder was very proactive in defending their IP rights. The Caruthers and Beaucage patent, US No. 4415732, effectively covered all phosphoramidite production and use. Boulder actively pursued litigation against infringers found embedding phosphoramidite chemistry into synthesis machines until, as Caruthers explained in our interview, the costs of litigation eventually reached the same level as the profits from their licenses, at which point they sold the patents to ABI, who chose to avoid further litigation costs by freely enabling licenses with interested parties. While this strategy reduced the royalties ABI received, it also helped to ensure their phosphoramidite chemistry became the international standard. The fact that there was no pre-existing community of practitioners ready to know how to adopt the phosphoramidite approach into their working life is the first aspect of this particular chemistry and technology that pushes it into the solutions category of business model. The transaction between patent-owning company and the customer meant that relationships were much more long term, so that customers were not simply buying a product in a one-off purchase, but were buying into an ongoing consultancy-like relationship with ABI. The company aimed to keep the machine operating according to their own designs by, for instance, voiding the warranty on any machine found to have used reagents supplied by a company other than ABI. This was not about being master of your own molecules, but a precision instrument for a precise range of tasks.
The 380A also fits into a solutions business model thanks to the expense of running the machine. The unit would cost in the region of $42,000 or $55,000 depending on how many reaction columns you chose, but really it was the cost of the reagents which ensured it could only be afforded by elite institutions and pharmaceutical companies (the kinds of organisations initially prioritised by the ABI sales force). At the outset a single reaction cycle (i.e. the addition of a single nucleotide) cost in the region of $20-25 in reagent. Indeed in the original business model, which targeted organic chemists like Jiricny, this was the primary way in which ABI intended to make profit, through reagent contracts, not the machinery. Placed inside an institute or company, the 380A was intended to form part of an organisation’s key facilities unit, a form of centralised organisation of equipment that increasingly came to be seen as necessary in science subsequent to the Second World War as new more sophisticated and expensive instruments began to enter the scientific marketplace.Footnote67 The ABI machine was designed to arrive at an institution and start solving the problems of making DNA for everyone, with implications for personnel: those who had been making it before (such as the team at Genentech) could be reassigned, or others, as in Jiricny’s case, could become the provider of synthesised DNA for a whole organisation. Of course, it only afforded this functionality thanks to its chemical capacity and technological design. Marv tells the chemistry story as follows:
MC: Mark [Matteucci] joined my lab in 1976. He wanted a PhD in organic chemistry, and so I thought “you know, nobody has really figured out how to synthetically prepare DNA yet in a meaningful way”, fast, so people could use it generally … once we got the [chlorophosphite] chemistry up and running, Mark designed and built with the help of, in the department we had a person who would repair motors and things like that, and this man helped him build a little semi-automatic machine, basically on a piece of plywood that we put together in Chemistry.Footnote68
MC: That was the first major breakthrough. The other was with Serge Beaucage who joined my laboratory as a postdoc. Mark was using 2ʹ-deoxynucleoside chlorophosphites as synthons for preparing DNA. The problem we discovered with these compounds was simple but difficult to solve. They had to be prepared immediately before use, they were unstable, and had to be stored at −70 degrees centigrade. We asked the question “how are we going to tame these compounds so we can store them in a bottle, keep them for months on a laboratory shelf, eventually activate them, and BANG they’ll react very quickly?” Serge’s breakthrough was to develop synthons that would satisfy these criteria. This he accomplished by developing synthons we called phosphoramidites.Footnote69
MC: When Serge arrived we knew we had to stabilise these compounds somehow. He had read the Russian literature, and there was some chemistry there where you could activate aminophosphenes by inserting carbon dioxide … So we started trying to reproduce that chemistry and we couldn’t do it … But along the way we found we were synthesizing what I was positive was a dinucleotide, just by that reaction … .So we were forming dinucleotides … .Serge took off in that direction and before the summer was out we had phosphoramidite chemistry.Footnote70
With regards to founding ABI:
MC: Because of these two breakthroughs, which allowed us to synthesize DNA rapidly and with a very simple process, I was getting calls almost daily by 1980 from numerous companies who wanted to collaborate on projects based upon the use of synthetic DNA … the most important development started with a phone call from Winston Salser in early February 1980. He had been approached by several venture capitalists about forming a new biotech company and wanted to know if I was interested in participating. Of course I decided to pursue this opportunity mainly because I was impressed with the quality of the other scientists who were also involved. After two meetings, at Stanford and UCLA, we decided to move forward with a new biotech company and leased space in an industrial park in Thousand Oaks, California. Eventually this company became known as Amgen. Our first meeting in Thousand Oaks was entirely focused on a discussion of potential projects that could lead to therapeutic drugs. However Lee Hood and I also proposed that we form a division that would develop a protein sequencer and DNA synthesizer based upon Lee’s and my respective research. This was not acceptable to the other scientists who wanted to be a drug development company. Consequently through separate discussions at this same meeting, Lee and I decided to proceed with a separate biotech instrument company. As a result of these discussions, Lee, I, and the venture capitalists, selected an engineer from Hewlett Packard, Sam Eletr, to form this new instrumentation company which became known as Applied Biosystems, and we were off and running.Footnote71
And the origins of the 380A:
MC: When we decided to start Applied Biosystems, I suggested to Sam Eletr that we hire Bill Efcavitch and Curt Becker from my laboratory to build the first DNA synthesizer. Bill and Curt started with a piece of plywood, some tubing, solenoid valves, a tank of nitrogen, appropriate solvents, and the reagents needed for DNA synthesis. Within a short time, they were synthesizing DNA on this platform. Consequently Sam hired John Bridger at HP to fabricate an instrument that we called the 380A DNA Synthesizer. I’d go out there about once a month, and make suggestions on this and that, but I did not work on that it was Curt and Bill.Footnote72
MC: In those days, scientists who saw the Applied Biosystems protein sequencers and DNA synthesizers would comment that these machines looked just like an HP product. They had the same grey-burgundy coloration, the same sleek look and were entirely enclosed in a smooth frame. I guess this was because they were developed by two former HP engineers-Sam Eletr and John Bridger. One important comment relates to how rapidly this team developed these DNA synthesizers. We formed Applied Biosystems in March of 1981. By December of 1982, about 18 or 19 months later, they were delivering their first commercial DNA synthesizer to my laboratory.Footnote73
Before the equipment would really take off though, one final problem had to be solved: the organic chemists. Their commitment to exploring a range of different chemistries, and reluctance to commit themselves to a machine using only phosphoramidites (because many were really only making DNA for biologists as a side-interest, their primary focus being on questions in chemistry) kept ABI unsatisfied with sales. Curt Becker, who worked closely with Bill Efcavitch as he developed the 380A, explains:
CB: There was a handful of organic chemists sprinkled around the world who were synthesising DNA mostly by triester chemistry. They all had their little twists on it and they considered their chemistry their chemistry. When we came out with the DNA synthesiser we believed … the market was those 35, 40, 50 organic chemists around the world that were synthesising for biologists in their country and around their campuses … .They resisted phosphoramidite chemistry … a lot of them tried to reproduce the results that Marv had published and had been unable to, but you didn’t just make phosphoramidites on a first time basis and everything was groovy! You had to, it was a painstaking process to put all of that package together … They were “I am not going to go and buy all these expensive reagents from Applied Biosystems when I can put my own reagents on the instruments”. I was actually tasked to go to Europe in 1984 and develop processes and cycles to use triester chemistry on the DNA synthesiser. I came back from that month and a half trip after interviewing everybody and said “no we’re not going to do that, basically what I think we should do is bypass the chemists, as a business model they’re not our market segment, we need to go right directly to the molecular biologists and biologists and sell them the DNA synthesiser. To be able to do so we’re going to need to be able to cut the cost per-cycle and we’re going to need to, to compensate for the reduction in cost per-cycle we’re going to speed up the cycle times, so we’ll give the instrument a higher throughput”.Footnote74
In order to make the ABI system now work for biologists rather than chemists, some new financial and material calculations had to be done. Rather than keeping the costs of reagents high to continue profiting from small communities of dedicated organic chemists, a move that some at ABI advocated for, instead the chemistry of the machine had to be re-thought to make it now attractive to biologists. A sweet spot needed to be found where the rapidity of a set of reaction cycles could get a biologist far enough along in their research programme, that the cost of adopting the machine and its reagents were preferable to relying on nearby organic chemists who might every now and then have some time on their hands to make DNA. Finding that sweet spot became far easier for many biologists around this time, coinciding as it did with the arrival of PCR, for which one needs a plentiful supply of synthetic DNA, at the very least for the initiation of amplification. It thereby became both possible and attractive for more biologists to take on synthesis themselves (in particular thanks to automation), but also pursue different kinds of biological work.
For instance, the kinds of biological research which really showcased this machine and the sequencer instruments also sold by ABI, were large gene library searching and synthesising operations (ideal for ABI for it meant customers needed to buy both sequencing and synthesis machines). The best examples of this approach could be found in the pharmaceuticals industry, and here Caruthers led from the front, for this approach lay behind the speed which Rasmussen emphasises in the rapid research, development, and sale of erythropoietin by Amgen, the biotech company Caruthers had been approached to found by Salser in the first place.Footnote75 Located in this equipment, and increasingly serving the biologist directly, DNA was rendered a rapid, reliable, and mass produced commodity, very much in line with the characterisation Michael Fortun develops for the more well-known example of DNA sequencing.Footnote76 However, keeping both sequencing and synthesis in view at the same time serves to rebalance our historiographical outlook, leaning away from a dominant informational view, and providing potentially more appropriate terms for a historicizing of synthetic biology. My account also reminds us of the central role of engineers in the design of these machines, and how machine-made DNA could accrue some of that profession’s esteem, particularly when multiple thousands of dollars could ride on the decision to buy one machine rather than another.
Closely identifying phosphoramidites with the solution business model does not mean that other chemistries could not also commodify DNA on these terms. I have found one example of a machine using modified phosphotriester chemistry, one currently on display at the Science Museum (London), the original creators of which were keen to emphasise could ‘compete with the rate of synthesis of the phosphite approach’. Again, I am not in a position to say anything about the truth of these claims, it is simply important that decades of competition amongst chemists recorded in journals such as Nucleic Acids Chemistry were now directly impinging on their marketing strategies and also the commodification of DNA as a service to be provided.Footnote77 Unlike ABI though, Celltech, the company who built the automated machine in question, had no intention of turning their instrument into a product.Footnote78 Rather it was their expertise and capacity in oligonucleotide and gene design and synthesis that was for sale.
In addition to central research facilities Celltech also offers custom DNA synthesis. In 1983 a separate production unit was set up to provide rapid and reliable supply of oligonucleotides for research customers. To ensure that there is no conflict of interest between ‘in-house’ and customer needs the unit has separate staff, and its own synthesisers or ‘gene machines’, designed and built in-house … .Longer DNA sequences can also be supplied, including full length genes produced by automatic synthesis and enzymatic ligation of oligonucleotides. The strategy for defining the oligonucleotides for synthesis and assembly into large DNA sequences is computer designed by Celltech. The DNA synthesis service available to customers extends from oligonucleotides to computer designed gene synthesis of ligated and cloned genes.Footnote79
This kind of business model is one of the most familiar in DNA synthesis today, and potentially bleeds into the matchmaking business model due to its emphasis on bringing the right kinds of expertise and interests together at Celltech, though there is no room to consider these possibilities here.
To recap section 4: I have argued that different formats for the automation of different chemistries embodied different expectations of biological work, and different ways to commodify DNA. In the case of Vega, DNA was a small bespoke commodity for the bench, and the business strategy was to sell small machines to individuals. In the case of Celltech, DNA design and delivery was a centralised service, the commercial strategy being to position the company as a one-stop shop for scientific and industrial clients supplying their DNA needs. Last, for ABI DNA was a commodity that could be rapidly produced at scale for those able to pay their way into the cutting edge by buying top of the line equipment and proprietary reagents, capable of multiple syntheses at once. The decades of improvements to different chemistries, and the more or less stylishly designed and developed automated equipment that delivered it, leant to any synthesised DNA the different epistemic and commercial values that produced it.
5. DNA as experimental commodity
I have explained how DNA came to be made in a variety of ways, how different chemistries came to be embodied in a range of different apparatuses, each of which produced a different relationship between DNA’s producers and consumers, resulting in different kinds of commodification. If I had told this story as primarily a biological one, I would likely have needed to ignore or marginalize the significance of the chemico-physical properties of DNA outside of the cell, and I would have instead ended up exploring synthesised DNA only as yet another interventionist tool developed in the twentieth century for experimental biology. But different DNAs produced in different ways have experienced material-semiotic adventures of their own. I have not attempted to disguise the extent to which my historical research has been directly inspired by the present and an interest in how something like synthetic biology could come to be. I have attempted to build a history of synthesised DNA addressing the material-semiotics of these molecules in a way that is particularly useful to scientists, social scientists, and philosophers of science in the present, shining a light on particular meanings and epistemic perspectives which have been present in synthesised DNA for as long as people have been taking pride in its making. By focussing on making, my historical approach has not actually needed either Molecular Biology or Synthetic Biology in order to be written. Actors have come and gone, each carving up the territory according to their own ambitions and understandings, and the historian is not required to make a commitment to any of their ontologies or epistemologies. I have used molecular biology and synthetic biology as recognisable markers, signalling to the reader where I think this history should be located in the existing historiography, but it does not actually need such terms in its telling. Demarcation problems have unhelpfully preoccupied historians, philosophers, and social scientists working in these areas. One can believe that Molecular Biology as a grand unified international endeavour never happened and that Synthetic Biology as a grand unified international endeavour isn’t happening, and still adopt my materials and analysis.
The most straightforward historiographical way in which to ensure the material properties of DNA were not marginalised by the informational, was to offset the ‘biological’ significances with ‘technological’ ones. Here another potential danger lay, as if I had treated DNA synthesis and its machines as only another case in the long history of technology, I would have risked ignoring the multiple (and often biological) significances of this substrate, and perhaps inadvertently closed down critical exploration of twentieth century biological science and technology by appearing to passively adopt the perspectives of central commercial actors. Instead, use of tools and analyses from the history of technology, and business studies, have kept critical questions open and suggested novel paths for their future investigation. It is not simply then that this story benefits from dual attention as biological and technological, but that the integrated historiographical approach achieves something more than the sum of its parts: a corrective and complementary splinting.
By orienting my study around making molecules, have I done anything particularly novel, or have I simply added one more article to the long and journalistic tradition of fetishising DNA? In response, I take it that in our time DNA is about as thoroughly fetishised as it can be, and as a result the process of historical looking can do a great deal of good, by deflating the phenomenon, making it less dazzling, and by refocusing our attentions, altering the kinds of questions we are prompted to ask in the present, and reevaluating our historiographical priorities. I would also add that up until now, our histories of molecular biology have far too heavily emphasised the history of DNA as an information carrying molecule, even as those same historians have explicitly sought to problematise the reading of DNA as primarily informational. My research has done the work of grubbying the informational sacred with the material profane.
Taking this research forward, we would next need to catalogue the variety of ways that synthesised DNA – and those other molecules which have always been investigated alongside it – have been used in biological research. For DNA this would include everything from Jiricny’s carcinogens, to research into its binding, to PCR, to biological engineering, and outwards to figures such as Nadrian Seeman.Footnote80 We could then also begin incorporating the publics of DNA synthesis, particularly the ways in which science journalists and communicators have contributed to diverse understandings of DNA synthesis, its capacities and meanings (see ), and the lives and afterlives of DNA synthesisers on public display in museums.Footnote81
Image 2. Left) New Scientist, 29th January 1981, 261. This cartoon accompanies an article on the ‘seamier’ side of DNA synthesis machines, i.e. that some of the commercial players are not to be trusted and that experience with machines has not always matched expectations. This helps evidence the kind of world that professional members of the synthesis knowledge community came to occupy. Right) New Scientist, 23rd May 1985, 22. This cartoon accompanies an article on recent improvements in synthesis capacity developed in Germany which are explicitly contrasted with the ‘limitations’ of the ABI machine. The first cartoon to feature DNA synthesis in New Scientist was published 24th October 1963, p. 224 by the cartoonist Bax, whose identity could not be uncovered.

In addition, I have not attempted to gather testimony from the users of any of these machines, and while it would be dull to pursue such evidence in order to sort out which instruments were best, it would be vastly more interesting to find out what they meant to people, and how they were interacted with (or not) in any given setting. DNA synthesis was and remains an important part of the public face of biotechnology, one worth recovering to be compared and contrasted with genetic recombination at large. The extent to which DNA synthesis is gendered is also something that should not be overlooked (though I have done so here), the communities of men here relying on close relationships for facilitating the fair and proper apportioning of credit, no doubt involving no small amount of gatekeeping. Engineering and engineers have also entered the picture. Given the significance of valves and precision liquid handling for the commodification of DNA, which are precisely the same kinds of engineering and components that matter in Ann Johnson’s history of antilock braking systems, it is possible that these two histories are really one. But this interesting possibility will have to be pursued another time. I hope to have provided foundations for a more thorough and global investigation of DNA synthesis, one exemplifying what can be gained by integrating the histories of biology and technology.
Acknowledgements
The majority of research for this paper was undertaken during my time as a Research Fellow on the Engineering Life project, supported by the European Research Council through Consolidator Grant (616,510- ENLIFE). More about that project can be found at http://www.stis.ed.ac.uk/engineeringlife
This paper was written while employed as Research Fellow on the Narrative Science project, supported by the ERC under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 694,732). More about that project can be found at https://www.narrative-science.org/
I am very grateful indeed to the support I have received from members of both projects and their respective leaders, Dr Jane Calvert and Prof. Mary S. Morgan. In addition, I am very grateful to Jane Calvert, Deborah Scott, Rob Smith, Karen Rader, Tiago Saraiva and Robert Bud for providing critical feedback on earlier drafts. I am of course very grateful to the interview participants, particularly Marv Caruthers, who were generous with their time and expertise, and who have been invaluable for making something like this project possible. Various parts of this material were presented at meetings of the International Society for the History, Philosophy and Social Studies of Biology (São Paulo, 2017), 25th International Congress of History of Science and Technology (Rio, 2017), Society for the History of Technology (St. Louis, 2018), 7th Norwegian Conference on the History of Science (Oslo, 2018) and a workshop on Bridging the Philosophy of Biology and Chemistry (Paris, 2019). I list them in order to thank all the audiences who have responded to this project as it unfolded, directly shaping the directions taken. Last but not least, many thanks to Daniel Liu for organising the panel from which this special issue has emerged, and to Karen Rader for demonstrating such excellent and generous academic and intellectual leadership, not only recognising the need for such historiographical integrations but also collaborating with the editors of History and Technology, Amy Slaton and Tiago Saraiva, in order to bring it to fruition in such an exciting and innovative way. A network of scholars is now being built around integrative and interdisciplinary questions concerning biological engineering, about which more can be learnt at www.bioengcoll.org.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
Notes
1. Rasmussen, Gene Jockeys; de Chadarevian, Designs for Life; Olby, The Path; Cobb, Life’s Greatest Secret; Morange, A History; Sarkar, The Philosophy and History; Kay, Molecular Vision; Kay, Who Wrote; Judson, Eighth Day; and Abir-Am, “Discourse of Physical Power.”
2. Rheinberger, Epistemic Things.
3. Berry, “Synthesis and the Organism.”
4. Daisy Ginsberg et al., Synthetic Aesthetics; Schyfter, “How a ‘Drive to Make’ Shapes Synthetic Biology”; Schyfter et al., “Guest editorial”; Project website: http://www.stis.ed.ac.uk/engineeringlife.
5. Bud, Uses of Life, 172.
6. This response was offered by Chedd, “Toward the Synthesis Of Life?”
7. Campos, “That was the Synthetic Biology that Was”; Keller, “What Does Synthetic Biology Have to do With Biology?” and Johnson, “From Bio-organic Chemistry.”
8. Campos, Radium; Curry, Evolution; de Chadarevian, Designs for Life; Kohler, Lords of the Fly; Creager, Life of a Virus; Rader, Making Mice; Landecker, Culturing Life; Radin, Life on Ice; Charnley, “Agricultural Science”; Hahn, Making Tobacco; Saraiva, Fascist Pigs; Wolfe Scheffler, “Following cancer viruses”; Agar and Ward, eds., Histories of Technology; Reuss and Cutcliffe, eds., The Illusory Boundary; and Jørgensen, Jørgensen and Pritchard, eds., New Natures.
9. Bud, Uses of Life.
10. For more on the temporal dimensions to the material-semiotics at work here see Berry and Palladino, “Life, Time and Organism.”
11. Abir-Am, “Themes, Genres.”
12. Calcott et al., “Engineering and biology”; Harré and Llored, “Procedures”; Leonelli, Data-Centric Biology; Morgan and Norton-Wise, “Narrative Science and Narrative Knowing”; Schummer “The Philosophy of Chemistry”; and Vermeulen, Tamminen and Webster, Bio-Objects.
13. Creager, Life of a Virus.
14. Kay, Who Wrote the Book of Life? 12–13; Morange, Molecular Biology, 137; and Judson, Eighth Day, 488.
15. Grote, Membranes to Molecular Machines. My thanks to Robert Meunier for making the connection between my research and Grote’s, and to Matthias for discussing his project with me further.
16. Medina, “Forensic Identification”; and Creager, “A Chemical Reaction.”
17. My very sincere thanks to Tiago Saraiva for making this suggestion during conversations concerning my draft. The present commodities history had been lurking underneath layers of unnecessary complication.
18. Clarke and Fujimura eds., Right Tools; and Rader, Making Mice.
19. Parry, Trading the Genome.
20. Fitzgerald, Business of Breeding; Charnley; “Experiments in Empire”; Fullilove, Profit of the Earth; Berry, “Plant Breeding Industry After”; and Hahn, Making Tobacco.
21. See note 8 above. Also Huzair and Sturdy, “Biotechnology and the Transformation of Vaccine Innovation.”
22. Gaudillière and Löwy, Invisible Industrialist; Mirowski, Science-Mart; and Thackray ed., Private Science; see also recent discussion of ‘compound histories’ for the longer history of chemistry in which at least part of my case resides, Roberts and Werret, Compound Histories.
23. Reese, “The Chemical Synthesis”; Reese, “Fifty Years”; Caruthers, “Robert Letsinger”; Caruthers, “Our Gift to Science”; Caruthers, “Gene Synthesis with H.G. Khorana”; Caruthers, “Chemical Synthesis”; Todd, Time to Remember; Sinsheimer, “Early steps”; Smith, “Synthetic DNA and Biology”; and Zhdanov and Zhenodarova, “Chemical Methods.”
24. Johnson, Hitting the Brakes. The kinds of knowledge which the community embodys are multiple, everything from tacit to social to theoretical to technical. I therefore take it that Johnson’s use of knowledge in the name ‘knowledge community’ should be read as a reclamation of the word, keeping it out of the hands of those whose understanding of knowing remains narrow, often with an emphasis on formal language, or at their strangest, even reserving it for deductive-nomological statements.
25. Brown and Kornberg, “Alexander Robertus Todd”; Shampo, Kyle, and Steensma, “Alexander Todd”; Todd, Time to Remember; and Buchanan, “Lord Todd.”
26. de Chadarevian, Designs for Life, 193.
27. Reese, “Oligo- and Poly-nucleotides,” 3851.
28. Kay, Who Wrote the Book of Life? and Todd and Baddiley, “Nucleotides: Part 1.”
29. Mark, “Kurt G. Stern.”
30. Stern, “Nucleoproteins and Gene Structure”, 944.
31. Johnson, “From Bio-organic Chemistry”; and Schumer, “Knowing through making.”
32. Stern, “Nucleoproteins and Gene Structure,” 944–945.
33. Morange, Molecular Biology, 38.
34. Chargaff and Davidson, Nucleic Acids.
35. Chargaff, “Chemical Specificity.” Chargaff’s method in the introduction of this text, what we could call ‘trial by narrative,’ should be of interest to those researchers currently scrutinising the role of narrative (as an epistemic feature, tool or set of tools) in the sciences. Morgan and Norton Wise, “Narrative Science.”
36. Andersen, Decker, and Todd, “Nucleotides: Part XIV,” 2722 and 2724.
37. https://www.heraldscotland.com/news/11901393.bucking-trends-at-spencer-coatings/; accessed on 27/10/2018.
38. Further clues are found in Sanghvi, “A Roadmap”: ‘As recently as six years ago, all the nucleotides needed for DNA synthesis were made from nucleosides isolated from natural sources, such as fish milt. A flow chart of this isolation process is shown in . Several companies, including Yamasa in Japan, Reliable in the USA and ProBioSint in Italy have used this method to produce nucleosides in metric ton quantities. It is not a rapid process (it can take 1.5 years from beginning to end) and it is very labor intensive’ (p. 20).
39. Ankeny and Leonelli, “Repertoires.” See also Srivastava and Gerson’s presentation in Berry, Synthesis and the Organism, 10–14.
40. This arose as Bettina Bock von Wülfingen asked me to chair her panel on colour in the history of science at the 2018 European Society for the History of Science Conference, in which Gerontas presented some of his work on chromatography and plant pigment. My thanks to Bettina and Apostolos!
41. Gerontas, “Reforming Separation”; and Gerontas, “From Glass to Gas.”
42. Todd, “Nobel Lecture: Synthesis in the Study,” 524.
43. 1955 paper. It was only thanks to a joint patent application made by Todd and Michelson over their method for the synthesis of ATP that I was able to learn Michelson’s full name. US PTO 2,645,637.
44. Radick and MacLeod, “Claiming Ownership.”
45. Todd, “Synthesis in the Study of Nucleotides,” 1946.
46. Todd, Time to Remember, 88.
47. Reese, “The Chemical Synthesis,” 3157.
48. Khorana et al., “A New Approach.”
49. Edgerton, “From innovation to Use”; and Staudenmaier, Technology’s Storytellers.
50. Davies et al., “Bigger, Faster, Better?” and Frow, “Making Big promises.”
51. Interview with Marv Caruthers conducted 28/11/2017 lasting 1hr 50 minutes.
52. Ibid.
53. Powers et al., “Optimal Strategies.”
54. Interview with Marv Caruthers conducted 5/9/2017 lasting 1hr 30 minutes.
55. Ibid.
56. Baden-Fuller and Morgan, “Business Models as Models.”
57. Massa, Tucci and Afuah, “A Critical Assessment”; http://businessmodelzoo.com/ accessed 30/10/2018.
58. Clarke, Science at the End; Edgerton, “Science and Technology”; and Weatherburn, “Scientific Management.”
59. Rieppel, Deringer and Lean, Science and Capitalism.
60. See for example US3440190A (1965), US4321365A (1977), US4394443A (1980), US4725677A (1983).
61. Gait ed., Oligonucleotide Synthesis.
62. Interview with Josef Jiricny via Skype 2/11/2017 lasting 1hr 4 minutes.
63. Ibid.
64. Schyfter, “Propellers and promoters.”
65. National Museum of American History, Accession File 1984.0719. Barstow, “Gene Machines.” My very sincere thanks to Luis Campos who retrieved a copy of this essay for me.
66. See note 51 above.
67. See the presentation from Scott Cole in Berry, Synthesis and the Organism, 20–25.
68. See note 54 above.
69. Ibid.
70. See note 51 above.
71. See note 54 above.
72. Ibid.
73. See note 51 above.
74. Interview with Curt Becker via Skype conducted 30/10/2018 lasting 1hr 10 minutes.
75. Rasmussen, Gene Jockeys, 131–159.
76. Fortun, “The Human Genome Project.” On sequencing as a lense on the period, García-Sancho, Biology, Computing.
77. Patel et al., “Improvements.”
78. Some of the first products that Celltech commercialised were for the monoclonal antibodies market. As Jon Agar describes, by this time the Thatcher government, and Thatcher in particular, had taken the loss of a lead on the monoclonal antibody market as a national failing. In this respect then Celltech and particularly its DNA synthesis work can be understood as rewriting and righting history. Agar, Science Policy.
79. Celltech Annual Report 1983. Archives of Science Museum, London. DNA synthesiser designed and built by Celltech, 1985–2038.
80. My thanks to Patrick McCray for suggesting Nadrian Seeman’s work as case to pursue, and also for the encouragement, criticisms, and suggestions he gave following the presentation of this work at HSS.
81. The notion that museum objects have ‘lives and afterlives’ may well originate somewhere specific, but was brought to my attention through intense discussion with Laura Volkmer, Sam Alberti, Tacye Phillipson and Niki Vermeulen. My thanks to them for being excellent intellectual co-conspirators.
Bibliography
- Abir-Am, P. “Themes, Genres and Orders of Legitimation in the Consolidation of New Scientific Disciplines: Deconstructing the Historiography of Molecular Biology.” History of Science 23 (1985): 73–117. doi:10.1177/007327538502300103.
- Abir-Am, P. “The Discourse of Physical Power and Biological Knowledge in the 1930s: A Reappraisal of the Rockefeller Foundation’s ‘policy’ in Molecular Biology.” Social Studies of Science 12 (1982): 341–382. doi:10.1177/030631282012003001.
- Agar, J. Science Policy Under Thatcher. London: UCL Press, 2019.
- Agar, J., and J. Ward, eds. Histories of Technology, the Environment, and Modern Britain. London: UCLPress, 2018.
- Andersen, W., C. A. Decker, and A. R. Todd. “Nucleotides. Part XIV. A Method Suitable for the Large-Scale Preparation of Deoxyribonucleosides from Deoxyribonucleic Acids.” Journal of the Chemical Society (1952): 2721–2724. doi:10.1039/JR9520002721.
- Ankeny, R. A., and S. Leonelli. “Repertoires: A post-Kuhnian Perspective on Scientific Change and Collaborative Research.” Studies in History and Philosophy of Science 60 (2016): 18–28. doi:doi.10.1016/j.shpsa.2016.08.003.
- Baddiley, J., and A. R. Todd. “Nucleotides. Part I. Muscle Adenylic Acid and Adenosine Diphosphate.” Journal of the Chemical Society (1947): 648–651. doi:10.1039/JR9470000648.
- Baden-Fuller, C., and M. S. Morgan. “Business Models as Models.” Long Range Planning 43 (2010): 156–171. doi:10.1016/j.lrp.2010.02.005.
- Berry, D. J. “The Plant Breeding Industry after Pure Line Theory: Lessons from the National Institute of Agricultural Botany.” Studies in History and Philosophy of Biological and Biomedical Sciences 46 (2014): 25–37. doi:doi.10.1016/j.shpsc.2014.02.006.
- Berry, D. J. “Synthesis and the Organism: Biology, Chemistry, and Engineering.” London School of Economics and Political Science Library. Accessed October 28, 2018. http://eprints.lse.ac.uk/90505/
- Berry, D. J., and P. Palladino. “Life, Time, and the Organism: Temporal Registers in the Construction of Life Forms.”Journal of the History of Biology 52 (2019). doi:10.1007/s10739-018-9513-3.
- Brown, D. M., and H. Kornberg. “Alexander Robertus Todd, O.M., Baron Todd of Trumpington. 2 October 1907-10 January 1997.” Biographical Memoirs of Fellows of the Royal Society 46 (2000): 516–532. doi:10.1098/rsbm.1999.0099.
- Buchanan, J. G. “Lord Todd.” Advances in Carbohydrate Chemistry and Biochemistry 55 (2000): 1–13.
- Bud, R. The Uses of Life: A History of Biotechnology. Cambridge: Cambridge University Press, 1993.
- Calcott, B., A. Levy, M. L. Siegal, O. S. Soyer, and A. Wagner. “Engineering and Biology: Counsel for a Continued Relationship.” Biological Theory 10 (2015): 50–59. doi:10.1007/s13752-014-0198-3.
- Campos, L. A. “That Was the Synthetic Biology that Was.” In Synthetic Biology: The Technoscience and Its Societal Consequences, edited by M. Schmidt, A. Kelle, A. Ganguli-Mitra, and H. de Vriend, 5–21. New York: Springer, 2010.
- Campos, L. A. Radium and the Secret of Life. Chicago: University of Chicago Press, 2015.
- Caruthers, M. H. “Chemical Synthesis of DNA.” Concepts in Biochemistry 66 (1989): 577–580.
- Caruthers, M. H. “Gene Synthesis with H. G. Khorana.” Resonance 17, (2012 December): 1143–1156. doi:10.1007/s12045-012-0131-7.
- Caruthers, M. H. “The Chemical Synthesis of DNA/RNA: Our Gift to Science.” The Journal of Biological Chemistry 288 (2013): 1420–1427. doi:10.1074/jbc.X112.442855.
- Caruthers, M. H. “Robert Letsinger: The Father of Synthetic DNA Chemistry.” Proceedings of the National Academy of Sciences 51 (2014 December 23): 18098–18099. doi:doi.10.1073/pnas.1420277111.
- Chargaff, E. “Chemical Specificity of Nucleic Acids and Mechanism of Their Enzymatic Degradation.” Experientia 6 (1950): 201–240. doi:10.1007/BF02173653.
- Chargaff, E., and J. N. Davidson, eds. The Nucleic Acids. 1 vol. New York: Academic Press Inc, 1955.
- Charnley, B. “Agricultural Science, Plant Breeding and the Emergence of a Mendelian System in Britain, 1880–1930.” PhD diss., University of Leeds, 2011.
- Charnley, B. “Experiments in Empire-building: Mendelian Genetics as a National, Imperial, and Global Agricultural Enterprise.” Studies in History and Philosophy of Science 44 (2013): 292–300. doi:doi.10.1016/j.shpsa.2012.11.003.
- Chedd, G. 1967. “Toward the Synthesis of Life?” New Scientist, 728, December 21.
- Clarke, A. E., and J. H. Fujimura, eds. The Right Tools for the Job: At Work in Twentieth-Century Life Sciences. Princeton: Princeton University Press, 1992.
- Clarke, S. Science at the End of Empire: Experts and the Development of the British Caribbean, 1940–1962. Manchester: Manchester University Press, 2018.
- Cobb, M. Life’s Greatest Secret: The Race to Crack the Genetic Code. London: Profile Books Ltd., 2015.
- Creager, A. N. H. The Life of a Virus: Tobacco Mosaic Virus as an Experimental Model, 1930–1965. Chicago: Chicago University Press, 2001.
- Creager, A. N. H. “A Chemical Reaction to the Historiography of Biology.” Ambix 64 (2017): 343–359. doi:10.1080/00026980.2017.1412136.
- Curry, H., and Anne. Evolution Made to Order: Plant Breeding and Technological Innovation in Twentieth-Century America. Chicago: University of Chicago Press, 2016.
- Davies, G., E. Frow, and S. Leonelli. “Bigger, Faster, Better? Rhetorics and Practices of Large-scale Research in Contemporary bioscience.” BioSocieties 8 (2013): 386–396. doi:10.1057/biosoc.2013.26.
- de Chadarevian, S. Designs for Life: Molecular Biology after World War II. Cambridge: Cambridge University Press, 2002.
- Edgerton, D. E. H. “Science and Technology in British Business History.” Business History 29 (1987): 84–103. doi:doi.10.1080/00076798700000082.
- Edgerton, D. E. H. “From Innovation to Use: Ten Eclectic Theses on the Historiography of Technology.” History and Technology 16 (1999): 111–136. doi:10.1080/07341519908581961.
- Fitzgerald, D. The Business of Breeding: Hybrid Corn in Illinois, 1890–1940. Ithaca, NY: Cornell University Press, 1990.
- Fortun, M. “The Human Genome Project and the Acceleration of Biotechnology.” In Private Science: The Biotechnology Industry and the Rise of Contemporary Molecular Biology, edited by A. Thackray, 182–201. Philadelphia: University of Pennsylvania Press, 1998.
- Frow, E. “Making Big Promises Come True? Articulating and Realizing Value in Synthetic Biology’.” BioSocieties 8 (2013): 432–448. doi:10.1057/biosoc.2013.28.
- Fullilove, C. The Profit of the Earth: The Global Seeds of American Agriculture. Chicago: University of Chicago Press, 2017.
- Gait, M. J., ed. Oligonucleotide Synthesis: A Practical Approach. Oxford: IRL Press Ltd., 1984.
- García-Sancho, M. Biology, Computing, and the History of Molecular Sequencing: From Proteins to DNA, 1945–2000. New York: Palgrave Macmillan, 2012.
- Gaudillière, J.-P., and L. Ilana. Invisible Industrialist: Manufacture and the Construction of Scientific Knowledge. Basingstoke: Palgrave Macmillan, 1998.
- Gerontas, A. “Reforming Separation: Chromatography from Liquid to Gas to High Performance Liquid.” PhD diss., Norwegian University of Science and Technology, 2013.
- Gerontas, A. “From Glass to Gas to High Performance Liquid: Chromatography from Its Inception to automation.” Unpublished manuscript in author's possession.
- Ginsberg, D., J. C. Alexandra, P. Schyfter, A. Elfick, and D. Endy, eds. Synthetic Aesthetics: Investigating Synthetic Biology’s Designs on Nature. Cambridge, Mass.: MIT Press, 2014.
- Grote, M. Membranes to Molecular Machines. Active Matter and the Remaking of Life. Chicago: University of Chicago Press, 2019.
- Hahn, B. M. Making Tobacco Bright: Creating an American Commodity, 1617-1937. Baltimore: The Johns Hopkins University Press, 2011.
- Harré, R., and J.-P. Llored. “Procedures, Products and Pictures.” Philosophy 93 (2018): 167–186. doi:10.1017/S0031819117000535.
- Huzair, F., and S. Sturdy. “Biotechnology and the Transformation of Vaccine Innovation: The Case of the Hepatitis B Vaccines.” Studies in History and Philosophy of Biological and Biomedical Sciences 64 (2017): 11–21. doi:10.1016/j.shpsc.2017.05.004.
- Johnson, A. Hitting the Breaks: Engineering Design and the Production of Knowledge. Durham and London: Duke University Press, 2009.
- Johnson, J. A. “From Bio-organic Chemistry to Molecular and Synthetic Biology: Fulfilling Emil Fischer’s Dream.” In Keynote lecture at International Workshop on the History of Chemistry, Tokyo, 2015. Accessed July 1, 2019. http://kagakushi.org/iwhc2015/papers/01.JohnsonJeffrey.pdf
- Jørgensen, D., F. A. Jørgensen, and S. B. Pritchard, eds. New Natures: Joining Environmental History with Science and Technology Studies. Pittsburgh: University of Pittsburgh Press, 2013.
- Judson, H. F. Eighth Day of Creation: Makers of the Revolution in Biology. New York: Cold Spring Harbor Laboratory Press, 1996.
- Kay, L. E. The Molecular Vision of Life: Caltech, the Rockefeller Foundation, and the Rise of the New Biology. Oxford: Oxford University Press, 1993.
- Kay, L. E. Who Wrote the Book of Life?: A History of the Genetic Code. Stanford: Stanford University Press, 2000.
- Keller, E. F. “What Does Synthetic Biology Have to Do with Biology?” BioSocieties 4 (2009): ^ 291–302. doi:10.1017/S1745855209990123.
- Khorana, H. G., G. M. Tener, J. G. Moffatt, and E. H. Pol. “A New Approach to the Synthesis of Polynucleotides.” Chemistry and Industry (1956 December 29): 1523.
- Kohler, R. E. Lords of the Fly: Drosophila Genetics and the Experimental Life. Chicago: Chicago University Press, 1994.
- Landecker, H. Culturing Life: How Cells Became Technologies. Cambridge, MA: Harvard University Press, 2007.
- Leonelli, S. Data-Centric Biology: A Philosophical Study. Chicago: University of Chicago Press, 2016.
- MacLeod, C., and G. Radick. “Claiming Ownership in the Technosciences: Patents, Priority and Productivity.” Studies in History and Philosophy of Science 44 (2013): 188–201. doi:doi.10.1016/j.shpsa.2012.11.010.
- Mark, H. “Prof. Kurt G. Stern.” Nature 177, (1956 March 24): 556. doi:10.1038/177556a0.
- Massa, L., L. T. Christopher, and A. Afuah. “A Critical Assessment of Business Model research.” Academy of Management Annals 11 (2017): 73–104. doi:10.5465/annals.2014.0072.
- Medina, E. “Forensic Identification in the Aftermath of Human Rights Crimes in Chile: A Decentered Computer History.” Technology and Culture 59 (2018): S100–S133. doi:10.1353/tech.2018.0151.
- Mirowski, P. Science-Mart: Privatizing American Science. Cambridge, MA: Harvard University Press, 2011.
- Morange, M. A History of Molecular Biology. Cambridge, MA: Harvard University Press, 2000.
- Morgan, M. S., and M. N. Wise. “Narrative Science and Narrative Knowing. Introduction to Special Issue on Narrative Science.” Studies in History and Philosophy of Science 62 (2017): 1–5. doi:doi.10.1016/j.shpsa.2017.03.005.
- Olby, R. The Path to the Double Helix: The Discovery of DNA. Seattle: University of Washington Press, 1974.
- Parry, B. Trading the Genome: Investigating the Commodification of Bio-information. New York: Columbia University Press, 2004.
- Patel, T. P., T. A. Millican, C. C. Bose, R. C. Titmas, G. A. Mock, and M. A. W. Eaton. “Improvements to Solid Phosphotriester Synthesis of Deoxyoligonucleotides.” Nucleic Acids Research 10 (1982): 5605–5620. doi:10.1093/nar/10.18.5605.
- Powers, G. J., R. L. Jones, G. A. Randall, M. H. Caruthers, J. H. van de Sander, and H. G. Khorana. “Optimal Strategies for the Chemical and Enzymatic Synthesis of Bihelical Deoxyribonucleic Acids.” Journal of the American Chemical Society 97 (1975): 875–884. doi:10.1021/ja00837a030.
- Rader, K. Making Mice: Standardizing Animals for American Biomedical Research, 1900–1955. Princeton: Princeton University Press, 2004.
- Radin, J. Life on Ice: A History of New Uses for Cold Blood. Chicago: University of Chicago Press, 2017.
- Rasmussen, N. Gene Jockeys: Life Science and the Rise of the Biotech Enterprise. Baltimore: Johns Hopkins University Pres, 2014.
- Reese, C. B. “The Chemical Synthesis of Oligo- and Poly-Nucleotides by the Phosphotriester Approach.” Tetrahedron 34 (1978): 3143–3179. doi:10.1016/0040-4020(78)87013-6.
- Reese, C. B. “Oligo- and Poly-Nucleotides: 50 Years of Chemical Synthesis.” Organic and Biomolecular Chemistry 21 (2005): 3851–3868. doi:10.1039/b510458k.
- Reuss, M., and S. H. Cutcliffe, eds. The Illusory Boundary: Environment and Technology in History. Charlottesville: University of Virginia Press, 2010.
- Rheinberger, H.-J. Towards a History of Epistemic Things: Synthesizing Proteins in the Test Tube. Stanford: Stanford University Press, 1997.
- Rieppel, L., W. Deringer, and E. Lean, eds. Osiris, Volume 33: Science and Capitalism: Entangles Histories. Chicago: University of Chicago Press, 2018.
- Roberts, L., and S. Werret. Compound Histories: Materials, Governance and Production, 1760–1840. Brill: Leiden, 2017.
- Robin, W. S. “Following Cancer Viruses through the Laboratory, Clinic, and Society.” Studies in History and Philosophy of Biological and Biomedical Sciences 48 (2014): 185–188. doi:doi.10.1016/j.shpsc.2014.09.004.
- Sanghvi, Y. S. 2007. “A Roadmap to the Assembly of Synthetic DNA from Raw Materials.” Working Papers for Synthetic Genomes: Risks and Benefits for Science and Society. https://dspace.mit.edu/handle/1721.1/39657
- Saraiva, T. Fascist Pigs: Technoscientific Organisms and the History of Fascism. Cambridge, MA: MIT Press, 2016.
- Sarkar, S., ed. The Philosophy and History of Molecular Biology: New Perspectives. Dordrecht, Boston, London: Kluwer Academic Publishers, 1996.
- Schummer, J. “The Philosophy of Chemistry.” Endeavour 27 (2003): 37–41. doi:10.1016/S0160-9327(03)00004-8.
- Schummer, J. “Knowing through Making: A Comparison between Synthetic Chemistry and Synthetic Biology.” Paper presented at ‘Bridging the Philosophies of Biology and Chemistry. University Paris Diderot, Paris, June 25–27, 2019.
- Schyfter, P. “Propellers and Promoters: Emerging Engineering Knowledge in Aeronautics and Synthetic Biology.” Engineering Studies 5 (2013): 6–25. doi:10.1080/19378629.2012.762651.
- Schyfter, P. “How a ‘drive to Make’ Shapes Synthetic Biology.” Studies in History and Philosophy of Biological and Biomedical Sciences 44 (2013): 632–640. doi:10.1016/j.shpsc.2013.05.010.
- Schyfter, P., E. Frow, and J. Calvert. “Guest Editorial- Synthetic Biology: Making Biology into an Engineering discipline.” Engineering Studies 5 (2013): 1–5. doi:10.1080/19378629.2013.763647.
- Shampo, M. A., R. A. Kyle, and D. P. Steensma. “Alexander Todd- British Nobel Laureate.” Mayo Clinic Proceedings 87 (2012): 19. doi:10.1016/j.mayocp.2011.12.009.
- Sinsheimer, R. “Early Steps on the DNA Ladder- A Recollection.” The Journal of Biological Chemistry 279 (2004): 40247–40251. doi:10.1074/jbc.X400004200.
- Smith, M. “Synthetic DNA and Biology.” Bioscience Reports 14 (1994): 51–66. doi:10.1007/BF01210301.
- Staudenmaier, J. M. Technology’s Storytellers: Reweaving the Human Fabric. Cambridge, MA: MIT Press, 1985.
- Stern, K. G. “Nucleoproteins and Gene Structure.” Yale Journal of Biology and Medicine 19, no. 6 (1947): 937–949.
- Thackray, A., ed. Private Science: Biotechnology and the Rise of the Molecular Sciences. Philadelphia: University of Pennsylvania Press, 1998.
- Todd, A. “Synthesis in the Study of Nucleotides.” Journal of the Chemical Society (1946 July): 647–653. doi:10.1039/jr9460000647.
- Todd, A. “Synthesis in the Study of Nucleotides.” In Nobel Lectures, Nobel Foundation, Chemisrty 1942–1962. Amsterdam: Elsevier Publishing Company, 1964.
- Todd, A. A Time to Remember: The Autobiography of A Chemist. Cambridge: Cambridge University Press, 1983.
- Vermeulen, N., S. Tamminen, and A. Webster. Bio-Objects: Life in the 21st Century. Farnham: Ashgate, 2012.
- Weatherburn, M. “Scientific Management at Work: The Bedaux System, Management Consulting, and Worker Efficiency in British Industry, 1914–1948.” PhD diss., Imperial College London, 2014.
- Zhdanov, R. I., and S. M. Zhenodarova. “Chemical Methods of Oligonucleotide Synthesis.” Synthesis 4 (1975): 222–245. doi:10.1055/s-1975-23714.
