Abstract
Aim
Low-grade glioma (LGG), which is the second most frequent adult brain malignancy, severely threatens patients’ health and has a high recurrence rate. Histone H3/H4 chaperone anti-silencing function 1 B (ASF1B) has a tight association with the initiation and development of tumours. The expression and regulation mechanism of ASF1B in LGG were discussed.
Methods
ASF1B expression in LGG patients as well as the association of ASF1B with overall survival and disease-free survival of LGG patients were predicted by GEPIA database. The independent prognostic value of ASF1B in LGG patients was investigated by TCGA database. RT-qPCR, together with western blot was applied for the assessment of ASF1B in LGG cell lines. After ASF1B expression was inhibited, CCK8 and colony formation assays judged cell proliferation. Flow cytometry analysis and TUNEL assay appraised cell cycle as well as apoptosis. Cell migratory and invasive capacities were measured by wound healing as well as Transwell assays. Western blot tested the expression of proliferation-, cycle-, apoptosis-, and metastasis-associated proteins. STRING and GeneMANIA database predicted the relationship between ASF1B and tousled-like kinase 1 (TLK1). ChIP assay testified the affinity of ASF1B with TLK1. Subsequently, TLK1 was overexpressed and ASF1B expression interfered, and the functional assays were executed.
Results
ASF1B was discovered to be increased in LGG tissues and cells and indicates an unfavourable prognosis for LGG patients. ASF1B was not an independent prognostic factor for LGG. ASF1B deficiency obstructed the proliferation, cell cycle as well as metastasis of LGG cells, and induced cell death, which might be realized through the interaction with TLK1.
Conclusion
The interaction between ASF1B and TLK1 promoted the malignant progression of LGG.
TLK1 interacts with ASF1B.
Interference with ASF1B inhibits the proliferative, invasive and migratory capabilities and induces the cycle arrest, along with the apoptosis of LGG cells.
The interaction between ASF1B and TLK1 promotes the malignant progression of LGG.
Key messages
Introduction
Gliomas can be sorted into four grades in light of their malignancy [Citation1]. Gliomas at grade II and grade III are known as lower-grade gliomas (LGG) [Citation2]. LGG, which is a kind of adult brain tumour, is more common in young adults than in older people. Early manifestations of LGG patients include headache, vomiting, vision changes and increased intracranial pressure with the rapid growth of tumours, etc [Citation3]. In addition, LGG has a high recurrence rate and poses a great threat to LGG patients’ health. Currently, LGG is treated with surgery combined with chemotherapy and radiotherapy. However, surgery can only achieve gross resection, and many glioma cells infiltrate into the normal brain tissue, making total tumour removal impossible [Citation3]. Radiotherapy side effects, chemotherapy toxicity, “multi-drug resistance” and other problems remain challenging in LGG treatment [Citation4]. Targeted gene therapy of LGG is the most exciting research area. Therefore, the search for biomarkers for LGG and the exploration of the mechanism of this disease is of tremendous significance to the diagnosis and treatment of LGG.
Histone H3/H4 chaperone anti-silencing function 1 (ASF1) participates in DNA-related processes together with transcriptional regulation [Citation5]. ASF1 is composed of two subtypes, including ASF1A and ASF1B. ASF1B is closely implicated in the initiation and development of numerous tumours through regulating cell proliferation [Citation6]. A previous study has illuminated that enhancive ASF1B expression has a relation with the poor prognosis of lung adenocarcinoma [Citation7]. ASF1B promoted the advancement of cervical cancer via stabilizing CDK9 [Citation8]. In renal clear cell carcinoma, PKMYT1 was up-regulated and PKMYT1 was positively correlated with the anti-silencing effects of ASF1B gene [Citation9]. ASF1B motivated the AKT signalling to facilitate cell proliferation and migration in renal clear cell carcinoma [Citation10]. ASF1B suppression enhanced the apoptotic ability of prostate cancer cells by inactivating PI3K/Akt pathway [Citation11]. In addition, gene expression profiles showed a significant increase in ASF1B expression following traumatic brain injury [Citation12]. However, the influences of ASF1B on LGG have not been uncovered.
STRING and GeneMANIA databases predicted the possible interaction between ASF1B and tousled-like kinase 1(TLK1). Being a serine/threonine protein kinase, TLK1 regulated DNA replication as well as DNA repair pathways. Previous literature has shown that the knockdown of TLK1 inhibited the survival of glioblastoma polymorphic cells [Citation13]. miR-16 depletion promoted tumour advancement by activating TLK1 in oral squamous cell carcinoma [Citation14]. TLK1 was overexpressed in both cholangiocarcinoma and cholangiocarcinoma cells and silencing of TLK1 enhanced cisplatin-stimulated DNA damage [Citation15].
Therefore, it is reasonable to hypothesize that the interaction between ASF1B and TLK1 in LGG may affect the malignant progression of LGG. This paper is intended to discuss the role and regulatory mechanism of ASF1B in LGG.
Materials and methods
Database
GEPIA database (http://gepia.cancer-pku.cn) predicted ASF1B expression in LGG tissues as well as its correlation with overall survival or disease-free survival of LGG patients [Citation16]. STRING (https://www.string-db.org/) [Citation17] and GeneMANIA (http://genemania.org/) databases predicted the correlation between ASF1B and TLK1 [Citation18].
Cell culture
LGG cell lines Res186, Res259, and Hs683 cells were supplied by the Shanghai Institute of Cell Biology and Chinese Academy of Sciences (Shanghai, China). Normal human astrocytes (NHA) were provided by Shanghai Hongshun Biotechnology Co., Ltd (Shanghai, China). All cells were cultivated in DMEM medium (Sigma) with 10% FBS (Sigma) as well as 1% antibiotics (Sigma) which were placed in a humidified atmosphere at 37 °C containing 5% CO2. The culture medium was replaced every 2 days.
RT-qPCR
RNA was isolated via Trizol reagent (Guangzhou Shuoheng Biotechnology Co., Ltd, Guangzhou, China) in light of the standard protocol. With the application of Brilliant II Fast SYBR green QPCR master mix (Agilent Technologies, Santa Clara, CA, USA), reverse transcriptase reactions were operated. RT-qPCR was implemented with SYBR Green Master Mix (Beijing Baiao Laibo Technology Co., Ltd.) in 7500 FAST Real-Time PCR System (Bio-Rad Co., USA). Comparative Ct method was employed for the calculation of mRNA expression [Citation19]. The following were the primer sequences: ASF1B forward: 5′-GCAGGATGCTGAGGAACCAT-3′, reverse: 5′-TTCAGGGTCCCAGTTGCTTC-3′; TLK1 forward: 5′-ACTGGAAGTACGGGCAGTTG-3′, reverse: 5′- CTGTGGGAGGTTTGCGTTTG-3′; GAPDH: forward: 5′-AATGGGCAGCCGTTAGGAAA-3′, reverse 5′-GCGCCCAATACGACCAAATC-3′.
Western blot
Proteins were isolated by RIPA lysis buffer (Shanghai Zeye Biotechnology Co., Ltd.). After the subjection to 10% SDS-PAGE, the transferring of proteins to PVDF membranes was operated. Subsequently, the membranes which were impeded with 5% skim milk was cultivated with primary antibodies overnight at 4 °C, following which was the cultivation with secondary antibody conjugated with the horseradish peroxidase for 2 h. Bands were then imaged by an enhanced chemiluminescent (ECL) kit (Bio-Rad Laboratories, Inc, CA, USA). GADPH served as a control to standardize the results. Protein bands were analysed by ImageJ software.
Cell transfection
Short hairpin RNA ASF1B (sh-ASF1B) and its empty vector (sh-NC), and TLK1-specific pcDNA overexpression vector (Ov-TLK1) and its empty vector (Ov-NC) were purchased from Sangon Biotech (Shanghai, China). After cells (1 × 104cells/well) that inoculated into 6-well plates reached 70% confluence, transduction of Ov-NC (10 µl/ml) and Ov-TLK1 (10 µl/ml) or sh-ASF1B (10 µl/ml) and sh-NC (10 µl/ml) was implemented utilizing Lipofectamine 2000 (Shenzhen Ziker Biotechnology Co., Ltd, Shenzhen, China) [Citation20]. Cells were collected following 48 h of transfection. Transduction efficacy was examined with RT-qPCR as well as western blot.
Cell counting kit-8
Cell Counting kit-8 (CCK-8, MedChem Express LLC.) was adopted for the evaluation of cell viability in light of the instruction. After the supplementation of CCK-8 solution (10 μl), cells were incubated for 4 h. A microplate reader (Promega, WI, USA) was to analyse the OD value at 450 nm. After that, cell viability was evaluated with GraphPad Prism 7.0.
Colony formation assay
Following corresponding induction, Res186 cells that were plated in 10-cm plates were stained with 0.05% crystal violet solution for imaging as well as colony counting 14 days later.
Flow cytometry
Flow cytometry was employed for the appraisement of the cell cycle in line with the manufacturer’s protocol. Following corresponding induction, 70% EtOH was to immobilize cells for 15 min before PBS washing. Cell pellets were suspended in 500 µl PI solution (BD Biosciences, CA) before 40 min of cultivation at 37 °C. For flow cytometry analysis, cells were pelleted with 500 µl PBS.
Tunel
The treated cells were subjected to 10 min of fixation with 4% paraformaldehyde. In situ cell death detection kit (Shanghai Hengfei Biotechnology Co., Ltd, Shanghai, China) was applied for the assessment of cell apoptotic rate.
Wound healing
Res186 cells that were placed into 6-well plates were cultivated overnight till 100% cell fusion was achieved. The initial culture medium was replaced with DMEM containing mitomycin for a further 12 h of cultivation. With the help of a pipette tip, wounds were formed, after which the images were obtained [Citation21].
Transwell
The capability of cells to invade was appraised by employing ECMatrix gel (Chemicon)-coated transwell inserts. The upper chamber was filled with 5 × 104 cells which were collected in serum-starved DMEM. The bottom chamber was composed of medium with 10% FBS. 24 h later, residual cells in the upper chambers were discarded. After that, 0.5% crystal violet was added for 10 min of staining and 70% ethanol was utilized for immobilization. Furthermore, six fields that were selected at random were captured and observed (100× magnification).
Chromatin immunoprecipitation
Based on the Chromatin immunoprecipitation (ChIP kit, Millipore) protocol, ChIP assay was executed. Following cross-linking with 1% formaldehyde, DNA and protein were isolated by SDS lysis buffer, and sheared via sonication. ASF1B antibody (Abcam) was used for immunoprecipitation. RT-qPCR was conducted after the purification of precipitated DNA [Citation22].
Statistical analysis
All data that manifested in the format of means ± SD got were analysed using SPSS 19. Student’s t-test, one-way ANOVA, along with Tukey’s post hoc test was adopted for the demonstration of comparison among groups. Using Cox proportional risk regression for analysis, the risk ratio (HR) and 95% confidence interval (95% CI) were calculated. Differences were considered to be significant at p value less than 0.05. Each experiment was independently repeated three times.
Results
ASF1B is highly expressed in LGG tissues and has an association with the poor prognosis of LGG patients
GEPIA database showed that ASF1B had higher levels in patients suffering from LGG (). Moreover, ASF1B elevation was prominently concerned with the low overall survival rate of LGG patients (). Increased ASF1B expression was markedly related to low disease-free survival of LGG patients (). Thus, we performed univariate and multivariate Cox regression analysis using age, sex, new tumours developed after the initial treatment, tumour status, radiation therapy and ASF1B expression as variables. From and , ASF1B expression was significantly associated with patient survival, while it could not be an independent factor for the results of univariate and multivariate Cox regression analysis. However, due to the significant differential expression of ASF1B in tissues and its relationship with patient prognosis, we decided further explore whether ASF1B can influence the function of LGG cells. Western blot and RT-qPCR results showed that ASF1B expression was remarkably ascended in LGG cell lines by contrast with NHA (). Res186 cells were chosen for follow-up experiments as ASF1B had the highest level in Res186 cells.
Figure 1. ASF1B is highly expressed in LGG tissues and is associated with poor prognosis of LGG patients. (A) GEPIA database showed that ASF1B was highly expressed in LGG patients. (B) GEPIA database showed that high ASF1B expression was significantly associated with low overall survival rate in LGG patients. (C) GEPIA database showed that high ASF1B expression was significantly associated with low disease-free survival rate of LGG patients. (D and E) RT-qPCR and Western blot were used to detect the expression of ASF1B in LGG cell lines. **p < 0.01, ***p < 0.001 vs NHA.
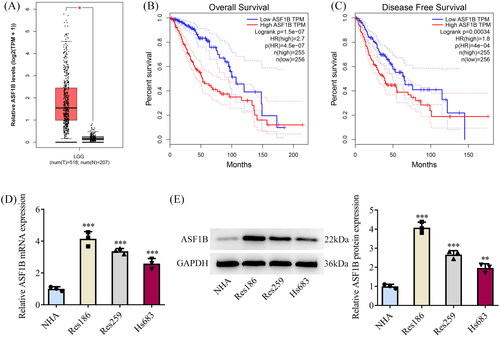
Table 1. Logistic regression analysis of ASF1B expression (univariate Cox analysis).
Table 2. Logistic regression analysis of ASF1B expression (multivariate Cox analysis).
ASF1B interference inhibits the proliferative capability of LGG cells
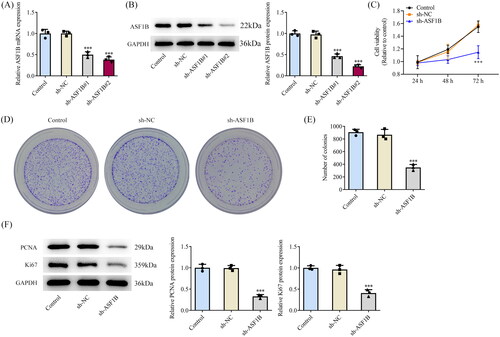
ASF1B interference plasmid was constructed and its transfection efficiency was detected by RT-qPCR as well as Western blot (). Sh-ASF1B#2 remarkably diminished ASF1B expression, so sh-ASF1B #2 was chosen for follow-up experiments. CCK8 results corroborated that relative to sh-NC group, cell viability in sh-ASF1B group was distinctly declined (). Colony formation assay results exhibited that cell proliferative ability was noticeably weakened following ASF1B interference (). It was also manifested that PCNA and Ki67 expressions were cut down notably after ASF1B interference ().
Figure 2. Interference with ASF1B inhibits proliferation of LGG cells. (A and B) RT-qPCR and Western blot were used to detect the expression of ASF1B after transfection. C.CCK-8 detected the cell viability. (D and E) Colony formation assay was used to detect the cell proliferation. (F) Western blot was used to detect the expression of PCNA and Ki67. ***p < 0.001 vs sh-NC.
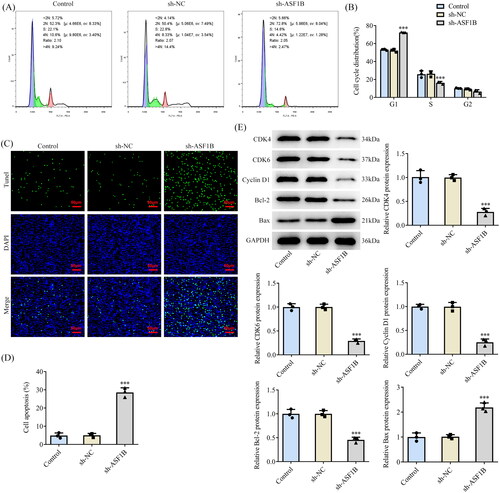
ASF1B interference induces cell cycle arrest and apoptosis in LGG
Flow cytometry showed that cell cycle arrest occurred after ASF1B interference (). Relative to the sh-NC group, the apoptosis of sh-ASF1B group was dramatically strengthened by sh-ASF1B (). As depicted, the expression of cell cycle-related proteins CDK4, CDK6, Cyclin D1 and anti-apoptotic protein Bcl-2 in sh-ASF1B group were apparently lessened, while the expression of pro-apoptotic protein Bax expression was clearly aggrandized when compared to the sh-NC group.
Figure 3. Interference with ASF1B induces cell cycle arrest and apoptosis in LGG. (A and B) Cell cycle was detected by flow cytometry. (C and D) TUNEL assay detected cell apoptosis. (E) Western blot was used to detect the expression of cell cycle and apoptosis-related proteins. ***p < 0.001 vs sh-NC.
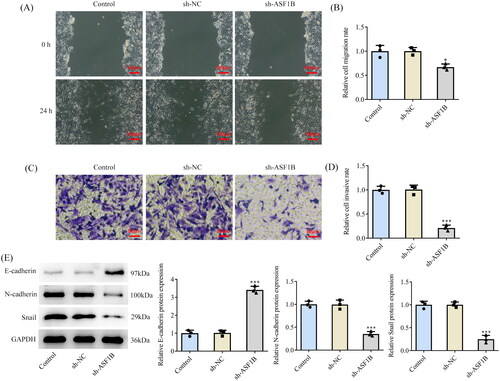
ASF1B interference inhibits the invasive and migratory capabilities of LGG cells
Results from wound healing () and Transwell () assays substantiated that cell invasive as well as migratory capacities in sh-ASF1B group were observably attenuated in contrast with the sh-NC group. It was also found that E-cadherin expression was ascended while N-cadherin and Snail expression were diminished after ASF1B interference ().
Figure 4. Interference with ASF1B inhibits invasion and migration of LGG cells. (A and B) Wound healing was used to detect the cell migration. (C and D) Transwell was used to detect the cell invasion. (E) Western blot was used to detect the expression of cell migration and invasion-related proteins. *p < 0.05, ***p < 0.001 vs sh-NC.
TLK1 interacts with ASF1B
STRING and GeneMANIA databases uncovered potential interactions between ASF1B and TLK1 (). In addition, TLK1 expression was noticeably fortified in Res186 cells (). The targeted binding of ASF1B and TLK1 was verified by ChIP assay (). In addition, inhibition of ASF1B expression significantly reduced the expression of TLK1 in Res186 cells ().
Figure 5. TLK1 interacts with ASF1B. STRING (A) and GeneMANIA (B) databases uncovered potential interactions between ASF1B and TLK1. (C and D) RT-qPCR and Western blot were used to detect the expression of TLK1 in LGG cells. ***p < 0.001 vs NHA. (E and F) The targeted binding of ASF1B and TLK1 was verified by ChIP assay. ***p < 0.001 vs IgG. (G and H) RT-qPCR and Western blot were used to detect the expression of TLK1 after depletion of ASF1B. ***p < 0.001 vs sh-NC.
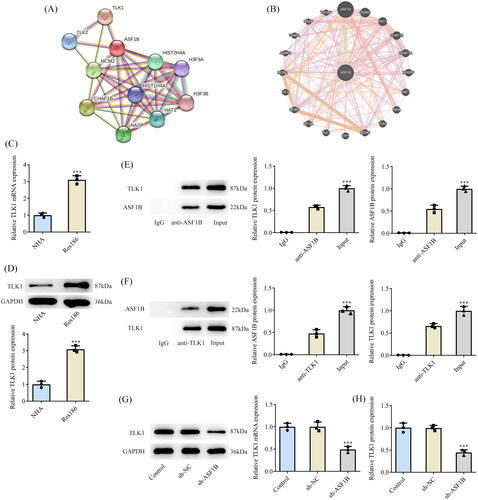
Upregulation of TLK1 partially reverses the effects of ASF1B interference on LGG cells
Next, the regulation mechanism of ASF1B in LGG was discussed and TLK1 overexpression plasmid was constructed, RT-qPCR, and Western blot was applied to examine the transduction efficacy (). CCK8 results elaborated that relative to sh-ASF1B + Ov-NC group, cell viability in sh-ASF1B + Ov-TLK1 group was evidently aggravated (). The proliferation ability of sh-ASF1B + Ov-TLK1 group was markedly increased (), accompanied by aggrandized PCNA and Ki67 expression relative to sh-ASF1B + Ov-NC group (). Relative to sh-ASF1B + Ov-NC group, cell cycle arrest was reversed and apoptosis was conspicuously cut down in sh-ASF1B + Ov-TLK1 group (). As ) demonstrated, the expression of CDK4, CDK6, Cyclin D1 and Bcl-2 were augmented in the sh-ASF1B + Ov-TLK1 group, while Bax expression was reduced by contrast with the sh-ASF1B + Ov-NC group. Wound healing and Transwell results uncovered that cell migration () and invasion () were notably facilitated in sh-ASF1B + Ov-TLK1 group relative to the sh-ASF1B + Ov-NC group. Western blot analysis of invasion and migration-related proteins exposed that E-cadherin expression in the sh-ASF1B + OV-TLK1 group was cut down, while N-cadherin and Snail expression were fortified by contrast with the sh-ASF1B + Ov-NC group ().
Figure 6. Upregulation of TLK1 partially reverses the effect of ASF1B interference on cell proliferation. (A and B) RT-qPCR and Western blot were used to detect the expression of TLK1 after transfection. ***p < 0.001 vs Ov-NC. (C) CCK-8 detected the cell viability. (D and E) Colony formation assay was used to detect the cell proliferation. F. Western blot was used to detect the expression of PCNA and Ki67. *p < 0.05, ***p < 0.001 vs Control; #p < 0.05, ##p < 0.01 vs sh-ASF1B + Ov-NC.
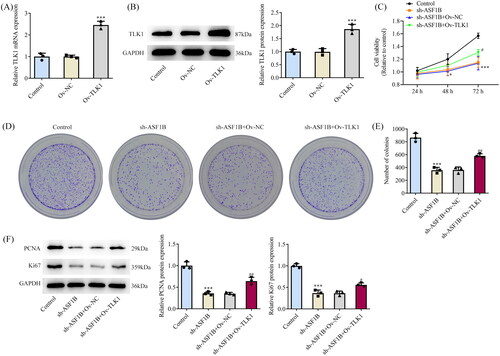
Figure 7. Upregulation of TLK1 partially reverses the effects of ASF1B interference on cell apoptosis. (A and B) Cell cycle was detected by flow cytometry. (C and D) TUNEL assay detected cell apoptosis. E and F. Western blot was used to detect the expression of cell cycle and apoptosis-related proteins. ***p < 0.001 vs Control; #p < 0.05, ###p < 0.001 vs sh-ASF1B + Ov-NC.
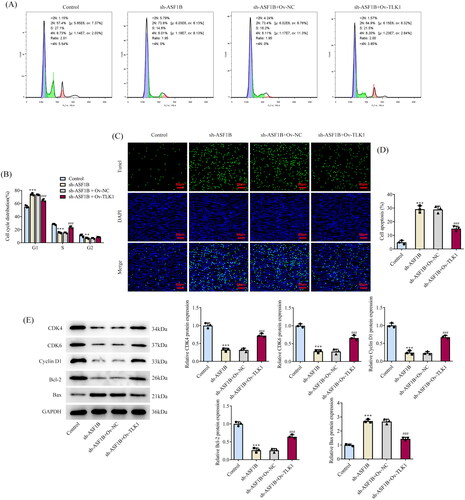
Figure 8. Upregulation of TLK1 partially reverses the effects of ASF1B interference on cell migration and invasion. (A and B) Wound healing was used to detect the cell migration. (C and D) Transwell was used to detect the cell invasion. (E) Western blot was used to detect the expression of migration and invasion-related proteins. ***p < 0.001 vs Control; ##p < 0.01, ###p < 0.001 vs sh-ASF1B + Ov-NC.
Discussion
Glioma is the most prevalent brain malignancy. LGG belongs to grade II-III glioma and is not as aggressive as high-grade glioma [Citation23]. However, LGG is more aggressive and will inevitably develop into high-grade gliomas with the passage of time [Citation24]. Recent studies on LGG have focused on molecular genetics, but the mechanisms of the migration, invasion, together with apoptosis in LGG remain elusive. Therefore, it is very important to further study the relevant mechanisms covering migration, invasion, and apoptosis in LGG.
Zhu et al. [Citation25] found through The Cancer Genome Atlas database (TCGA), genotypic tissue expression (GTEx), and the Chinese Gliomas Genome Atlas database (CGGA) that the expression of ASF1B in glioma tissue was significantly higher than that in normal tissue, and the increased expression of ASF1B was associated with poor prognosis of glioma patients. In this study, through GEPIA database, ASF1B was noticed to possess elevated expression in LGG tissues, and enhancive ASF1B expression had noticeable relation with unfavorable overall survival and disease-free survival rate of LGG patients. Clearly, our results are consistent with those of Zhu et al. Subsequently, ASF1B expression was also prominently increased in LGG cells. A previous study showed that ASF1B is overexpressed in multiple cancers and is closely associated with cancer survival [Citation26]. Next, it was found that interference with ASF1B in Res186 cells could remarkably halt cell proliferation, metastasis and stimulate cell cycle arrest and apoptosis in LGG in our study. ASF1B could enhance the migratory as well as invasive capabilities of lung cancer cells via the modulation of p53-mediated epithelial-mesenchymal transformation signalling pathway [Citation27]. MiR-214 inhibited the proliferative ability and promoted the apoptotic rate of myeloma cells by down-regulating ASF1B [Citation28]. ASF1B possessed fortified expression, and ASF1B absence could accelerate apoptosis, together with cell cycle arrest in hepatocellular carcinoma [Citation29]. In addition, ASF1B was also a prognostic marker for breast cancer as well as lung adenocarcinoma, and occupied an important position in the initiation and progression of tumours [Citation7,Citation30]. However, the impacts of ASF1B and its regulatory mechanism in LGG have not been reported. Moreover, gene expression profiles showed a significant increase in ASF1B expression following cerebral trauma injury [Citation12]. Therefore, our results, together with previously published studies, suggest that ASF1B acts as a cancer-promoting factor in the development of multiple cancers, providing a basis for the use of ASF1B as a therapeutic target for LGG or other cancers.
Next, the regulatory mechanism of ASF1B in LGG cells was deeply probed into. Potential interactions between ASF1B and TLK1 were revealed through relevant databases. Subsequently, the transcriptional regulation of TLK1 by ASF1B was confirmed through experiments in this study. It was confirmed that TLK1 mainly functioned dependent on mediating the H3/H4 histone chaperone ASF1, and ASF1B is phosphorylated by TLKs during DNA replication on its C-terminal tail residues S169 and S198 [Citation31]. TLK phosphorylation of ASF1 regulated its stability, while in vertebrates, TLK1-mediated phosphorylation of several sites on the C-terminal tail of ASF1 promoted its binding affinity for the histone H3/H4 heterodimer [Citation32]. So in LGG, does ASF1B play a regulatory role in tumour by regulating TLK1? Our results showed that ASF1B and TLK1 observably had a positive correlation in LGG patients and TLK1 was discovered to be distinctly overexpressed in LGG cells. It was shown that TLK1 expression was elevated in glioma tissues, and had correlation with large tumour volume and higher grade of glioma [Citation33]. Our findings are consistent with those of others. Finally, we found that TLK1 elevation partially reversed the impacts of ASF1B interference on cell viability, metastasis, cell cycle as well as apoptosis in LGG. The results of Ibrahim et al. [Citation13] also showed that knockdown of TLK1 inhibited the survival of glioblastoma polymorphic cells. Overexpression of TLK1 can significantly promote the growth, migration, and invasion of glioma cells, and inhibit cell apoptosis [Citation33]. Over all, this study helped us better understand the molecular mechanism by which ASF1B regulated LGG occurrence and development.
There are some disadvantages in this paper as well. First of all, only Res186 cell line was selected in our experiments and other LGG cell lines need to be chosen for validation in future experiments. Second, only in vitro cell experiments were carried out and animal experiments in vivo will be performed in the future to verify this finding.
Conclusion
In conclusion, this paper confirms that the interaction between ASF1B and TLK1 promotes the malignant progression of LGG, which lays a theoretical foundation for the discussion of targeted therapy for LGG.
Ethical statement
Our article does not experiment on animals and humans, so no ethical approval is required.
Consent for publication
All the authors agreed to be published.
Author contributions
Zifa Zhang and Shuming Liu wrote the manuscript and analyzed the data. Zifa Zhang performed the experiments and supervised the study. Shuming Liu searched the literature and revised the manuscript for important intellectual content. Zifa Zhang and Shuming Liu confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The analysed data sets generated during the present study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913.
- Yang Y, Yan LF, Zhang X, et al. Glioma grading on conventional MR images: a deep learning study with transfer learning. Front Neurosci. 2018;12(804):804.
- Youssef G, Miller JJ. Lower grade gliomas. Curr Neurol Neurosci Rep. 2020;20(7):21.
- McDuff SGR, Dietrich J, Atkins KM, et al. Radiation and chemotherapy for high-risk lower grade gliomas: choosing between temozolomide and PCV. Cancer Med. 2020;9(1):3–11.
- Paul PK, Rabaglia ME, Wang CY, et al. Histone chaperone ASF1B promotes human beta-cell proliferation via recruitment of histone H3.3. Cell Cycle. 2016;15(23):3191–3202.
- Abascal F, Corpet A, Gurard-Levin ZA, et al. Subfunctionalization via adaptive evolution influenced by genomic context: the case of histone chaperones ASF1a and ASF1b. Mol Biol Evol. 2013;30(8):1853–1866.
- Feng Z, Zhang J, Zheng Y, et al. Elevated expression of ASF1B correlates with poor prognosis in human lung adenocarcinoma. Per Med. 2021;18(2):115–127.
- Liu X, Song J, Zhang Y, et al. ASF1B promotes cervical cancer progression through stabilization of CDK9. Cell Death Dis. 2020;11(8):705.
- Chen P, Zhang Z, Chen X. Overexpression of PKMYT1 facilitates tumour development and is correlated with poor prognosis in clear cell renal cell carcinoma. Med Sci Monit. 2020;26:e926755.
- Jiangqiao Z, Tao Q, Zhongbao C, et al. Anti-silencing function 1B histone chaperone promotes cell proliferation and migration via activation of the AKT pathway in clear cell renal cell carcinoma. Biochem Biophys Res Commun. 2019;511(1):165–172.
- Han G, Zhang X, Liu P, et al. Knockdown of anti-silencing function 1B histone chaperone induces cell apoptosis via repressing PI3K/Akt pathway in prostate cancer. Int J Oncol. 2018;53(5):2056–2066.
- Zhao ZJ, Wei DP, Zheng RZ, et al. The gene coexpression analysis identifies functional modules dynamically changed after traumatic brain injury. Comput Math Methods Med. 2021;2021:5511598.
- Ibrahim K, Abdul Murad NA, Harun R, et al. Knockdown of tousledlike kinase 1 inhibits survival of glioblastoma multiforme cells. Int J Mol Med. 2020;46(2):685–699.
- Hu S, Wang H, Yan D, et al. Loss of miR-16 contributes to tumour progression by activation of tousled-like kinase 1 in oral squamous cell carcinoma. Cell Cycle. 2018;17(18):2284–2295.
- Takayama Y, Kokuryo T, Yokoyama Y, et al. Silencing of tousled-like kinase 1 sensitizes cholangiocarcinoma cells to cisplatin-induced apoptosis. Cancer Lett. 2010;296(1):27–34.
- Li C, Tang Z, Zhang W, et al. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–W246.
- Szklarczyk D, Franceschini A, Wyder S, et al. String v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43 issue):D447–452.
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Ferrandon S, DeVecchio J, Duraes L, et al. CoA Synthase (COASY) mediates radiation resistance via PI3K signaling in rectal cancer. Cancer Res. 2020;80(2):334–346.
- Yang IH, Jung JY, Kim SH, et al. ABT-263 exhibits apoptosis-inducing potential in oral cancer cells by targeting C/EBP-homologous protein. Cell Oncol. 2019;42(3):357–368.
- Zhangyuan G, Wang F, Zhang H, et al. VersicanV1 promotes proliferation and metastasis of hepatocellular carcinoma through the activation of EGFR-PI3K-AKT pathway. Oncogene. 2020;39(6):1213–1230.
- Morshed RA, Young JS, Hervey-Jumper SL, et al. The management of low-grade gliomas in adults. J Neurosurg Sci. 2019;63(4):450–457.
- Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58.
- Zhu H, Ouyang H, Pan X, et al. Increased ASF1B expression correlates with poor prognosis in patients with gliomas. Front Oncol. 2022;12:912101.
- Zhang W, Gao Z, Guan M, et al. ASF1B Promotes oncogenesis in lung adenocarcinoma and other cancer types. Front Oncol. 2021;11(731547):731547.
- Wang W, Xiao L, Pan D, et al. ASF1B enhances migration and invasion of lung cancers cell via regulating the P53-mediated epithelial-mesenchymal transformation (EMT) signalling pathway. Neoplasma. 2022;69(2):361–369.
- Misiewicz-Krzeminska I, Sarasquete ME, Quwaider D, et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica. 2013;98(4):640–648.
- Ouyang X, Lv L, Zhao Y, et al. ASF1B serves as a potential therapeutic target by influencing cell cycle and proliferation in hepatocellular carcinoma. Front Oncol. 2021;11(801506):801506.
- Montes de Oca R, Gurard-Levin ZA, Berger F, et al. The histone chaperone HJURP is a new independent prognostic marker for luminal a breast carcinoma. Mol Oncol. 2015;9(3):657–674.
- Segura-Bayona S, Stracker TH. The tousled-like kinases regulate genome and epigenome stability: implications in development and disease. Cell Mol Life Sci. 2019;76(19):3827–3841.
- Klimovskaia IM, Young C, Stromme CB, et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat Commun. 2014;5:3394.
- Wang Z, Chen X, Liang Q, et al. Inhibiting of circ-TLK1 inhibits the progression of glioma through down-regulating PANX1 via targeting miR-17-5p. J Mol Histol. 2021;52(5):1007–1020.
