Abstract
By changing the three-dimensional structure, a protein can attain new functions, distinct from those of the native protein. Amyloid-forming proteins are one example, in which conformational change may lead to fibril formation and, in many cases, neurodegenerative disease. We have proposed that partial unfolding provides a mechanism to generate new and useful functional variants from a given polypeptide chain. Here we present HAMLET (Human Alpha-lactalbumin Made LEthal to Tumor cells) as an example where partial unfolding and the incorporation of cofactor create a complex with new, beneficial properties. Native α-lactalbumin functions as a substrate specifier in lactose synthesis, but when partially unfolded the protein binds oleic acid and forms the tumoricidal HAMLET complex. When the properties of HAMLET were first described they were surprising, as protein folding intermediates and especially amyloid-forming protein intermediates had been regarded as toxic conformations, but since then structural studies have supported functional diversity arising from a change in fold. The properties of HAMLET suggest a mechanism of structure-function variation, which might help the limited number of human protein genes to generate sufficient structural diversity to meet the diverse functional demands of complex organisms.
Introduction
According to Anfinsen Citation1, proteins fold to reach the global free energy minimum, where the native three-dimensional structure and its biological function are clearly defined. With the exception of a few metastable native state protein examples Citation2, recent work on acylphosphatase from Sulfolobus solfataricus Citation3, and HAMLET Citation4, folding intermediates that possess biological function are rare. Many folding intermediates in their monomeric form exhibit a tendency to aggregate, and some are subsequently converted into protofibrils or amyloid, which, apart from some notable exceptions Citation5, Citation6, are regarded as potentially toxic conformations. In contrast, the properties of HAMLET (Human Alpha-lactalbumin Made LEthal to Tumor cells) suggest that folding intermediates can attain functions of physiological importance but completely independent from its originating protein. HAMLET is a complex consisting of partially unfolded α-lactalbumin and oleic acid that kills tumor cells, while leaving healthy, differentiated cells unaffected. This review concerns the structural basis of the tumoricidal activity of HAMLET and of partially unfolded α-lactalbumin. Parallels with amyloid-forming peptides and with other unfolded protein species will also be discussed.
‘One gene–one protein–one function’
The paradigm ‘one gene, one protein, and one function’ was proposed by Tatum and Beadle based on their work on Neurospora (bread mold) in the early 1940s, earning them the Nobel Prize in Medicine in 1958 Citation7. Point mutations were introduced in Neurospora strains by X-ray irradiation, and the mutant strains were characterized by their inability to carry out specific biochemical processes. One mutant strain required ρ-aminobenzoic acid for growth, and Tatum and Beadle showed that a single gene discriminated the parent from the mutant and that this gene was essential for the synthesis of ρ-aminobenzoic acid Citation7. They concluded that a gene encodes a single protein and the ‘one gene–one protein–one function’ paradigm was born.
Abbrviations
More recently, this paradigm has been questioned, especially since the completion of the human genome sequence in 2003. The human genome was shown to contain 30,000–40,000 genes Citation8, Citation9 coding for 20,000–25,000 proteins Citation10. This number is inconsistent with function-based estimates, which propose that protein-dependent functions in the human body would require at least 100,000 proteins Citation11. Furthermore, there is a remarkably small difference in gene number between species of vastly different complexity. The genome of Caenorhabditis elegans contains 20,000 genes Citation12, which is only five times higher than bacteria like Pseudomonas aeruginosa Citation13. Given the reasonable assumption that biological complexity is reflected in the number of genes, the difference in the number of protein genes is not sufficient to explain the difference in functional complexity between humans, C. elegans, and bacteria. Implicit is the need for further structural modifications to distinguish the functional variants.
Key messages
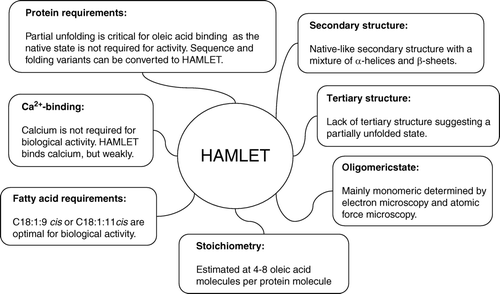
HAMLET (human α-lactalbumin made lethal to tumor cells) consists of partially unfolded α-lactalbumin and oleic acid.
The complex kills tumor cells, while leaving healthy, differentiated cells unaffected.
The properties of HAMLET suggest that partial unfolding may generate new functional variants from a given polypeptide chain.
Thus, even though alternative splicing of mRNA transcripts may generate additional diversity, it is a plausible alternative that polypeptide chains can possess more than one function in some cases. Known structural modifications that allow proteins to change their function include glycosylation, phosphorylation, and oligomerization. Such modifications may occur in response to change of cellular localization, cell type, substrate availability, or ligand binding. Based on the work with HAMLET, we propose that folding intermediates may be functionally distinct from the native, folded species and thus add to the number of functional states attained by a given polypeptide chain Citation4, Citation14.
HAMLET contains α-lactalbumin–a protein normally involved in lactose synthesis. First the protein is partially unfolded, and then the unfolded protein is bound to oleic acid, which stabilizes its three-dimensional structure. HAMLET is the first example of a protein that in its native state exhibits a well defined function but also acquires a new and beneficial activity after partial unfolding Citation4, Citation14. More recently, the term ‘moonlighting proteins’ has been introduced to describe molecules that serve one main and several additional functions, while retaining the same amino acid sequence. As discussed by Jeffery, moonlighting proteins are well folded Citation15, but more recently, intrinsically unstructured proteins were proposed to be included in the moonlighting group Citation16. These proteins lack well defined tertiary structure but can fulfill specific biological functions. HAMLET is distinct from the moonlighting proteins and from the intrinsically unfolded proteins because it undergoes a partial unfolding and remains as such to fulfill its new biological function.
Protein folding
During protein synthesis, the information contained within the mRNA is translated by the ribosome to generate a nascent polypeptide chain, which contains the complete structural information needed to fold to the native state. Many emerging nascent polypeptides are protected from misfolding or aggregation by chaperones Citation17. The majority of folding occurs in the cytosol, but, for example, secreted proteins are folded by chaperones inside the endoplasmic reticulum (ER) Citation18. Chaperones may not be essential for protein folding, as a correct protein fold can be obtained in vitro in their absence Citation1, but the protein concentrations and conditions for folding proteins in vitro are considerably different from those present within the cell. In vivo folding occurs much faster compared to refolding in vitro, especially in the case of the formation of disulfide bonds Citation19. As most of the understanding of protein folding and misfolding is based on in vitro systems with highly artificial conditions, there is a need to explore the folding conditions in vivo.
It is a great challenge to study protein folding in vitro with experimental methods due to conformational exchange in the unstable intermediates. Various biophysical methods such as hydrodynamic measurements, circular dichroism (CD), fluorescence, and nuclear magnetic resonance (NMR) spectroscopy have been applied to yield detailed structural information Citation20. Another important approach is to study changes in folding upon after substituting individual amino acid residues in a protein Citation21. In addition, computer simulation techniques have made it possible to perform simulations and to study energy landscapes of individual proteins, including α-lactalbumin Citation22.
The energy landscape
The number of possible conformations accessible by a polypeptide chain is astronomically large, and yet an unstructured polypeptide can attain to the correct fold in a short period of time Citation23. This ‘Levinthal paradox’ has led to the search of folding pathway models. The ‘classical view’ of protein folding involved linear folding events including the on-pathway, off-pathway, and sequential models Citation24. In the mid-90s, a ‘new view’ of protein folding incorporating free energy landscapes was introduced Citation25. The free energy landscape resembles a U-shaped valley, or ‘folding funnel’ Citation26, which is unique for every protein as it is defined by the amino acid sequence. The polypeptide chain is thought to adopt a continuum of folding states on the way down the folding funnel. The free energy is minimized through an increasing number of intra-molecular contacts, and the native fold of a protein is attained when reaching the most thermodynamically stable structure or the global free energy minimum.
The folding process can be more complex if the folding funnel contains various local free energy minima, or kinetic traps Citation24, and as a result folding intermediates can accumulate. The thermal energy provided within the system is usually sufficient to escape a trap to allow folding to continue down to the lowest free energy state. The properties of the HAMLET complex suggest that the binding of a cofactor may help to stabilize a kinetic trap and keep the protein from reaching the lowest free energy state. The protein thus remains in this energetically unfavorable state by incorporating the fatty acid, and in addition a new biological function is obtained.
Protein misfolding and disease
The term ‘misfolding’ refers to a change in secondary and/or tertiary structure that abolishes the functional integrity of a native, folded protein. Furthermore, the term infers that misfolding may generate harmful protein species, which damage the cells that produce them. Such variations in the folding of a polypeptide might either lead to disease by gain of toxic activity or by loss of the native biological function Citation27, potentially leading to disease. About 30% of newly synthesized polypeptides contain errors that prevent folding to the native state, and unless they are removed such misfolded species may cause a transient state of stress due to unfolded protein overload Citation28. Misfolding proteins may also be caused by mutations that permanently hinder the normal folding of the polypeptide chain. In this case, the risk of pathology increases as the unfolded protein accumulates in the tissues, causing aggregation and formation of insoluble fibrils and plaque Citation11. Native proteins contain varied secondary structural elements such as α-helices and β-sheets, but the misfolded protein aggregates invariably show a preponderance of β-sheet conformation Citation11, Citation27. The biophysical principles that rule fibril formation in vitro have been extensively studied, and significant inroads have been made into finding therapeutic measures, but much is left to be understood with regards to the in vivo conditions Citation11.
The ER employs two linked mechanisms to deal with misfolded proteins, the unfolded protein response (UPR) and ER-associated degradation (ERAD). The UPR is a stress response, which acts to remodel the ER capacity, while ERAD translocates the misfolded proteins to the cytosol for degradation by the proteasome Citation18. If these degradation systems fail, due to for example an overload of unfolded or misfolded proteins, the resulting stress response might lead to cell death. Several human diseases are caused by protein misfolding and aggregate formation Citation27, Citation29. Alzheimer's disease, transmissible spongiform encephalopathies (TSEs), and Parkinson's disease are neurodegenerative disorders included in the conformational diseases Citation30. In each case, a particular misfolded protein forms amyloid, and amyloid deposits may cause tissue damage. lists the proteins involved in these amyloid diseases so far identified (29). We give further description of amyloid-β and the prion proteins below.
Table I. Human diseases associated with formation of extracellular amyloid deposits or intracellular inclusions with amyloid-like characteristics Citation29. Reprinted, with permission, from the Annual Review of Biochemistry, Volume 75 ©2006 by Annual Reviews (www.annualreviews.org).
Amyloid-β
Alzheimer's disease is the leading cause of dementia and involves the formation of amyloid plaques in the brain. The amyloid-β (Aβ) peptide is derived from the amyloid precursor protein (APP) upon cleavage by secretases. APP is a 120-kDa membrane-associated protein, present in both neural and non-neural tissue Citation31. The plaques consist of Aβ, a 39–42-residues peptide. The hydrophobic C-terminal domain adopts a β-strand conformation and the N-terminal domain sequence permits the formation of both α-helices and β-sheets Citation32. Fibril formation is caused by a change in secondary structure to mainly β-sheet Citation33. Mutations that replace hydrophobic residues in the C-terminal for hydrophilic residues reduce the β-sheet content and decrease the ability to form fibrils Citation34. After fibrillogenesis, the fibrils eventually aggregate and form plaques, but it is believed that the fibrils themselves are cytotoxic. Geula et al. injected Aβ fibrils into brains of monkeys and discovered that the fibrils caused neuronal loss Citation35. The cytotoxic effects were only seen in old monkeys and to a much lower extent in rats, suggesting that this process is stronger in ageing brains in higher-order primates Citation35. The fibrillogenesis is inhibited in rats by short five-residue peptides, known as β-sheet breakers Citation36. A combination of the latter two studies is of great interest for the development of future therapy against Alzheimer's disease.
Prions
The prion proteins (PrPs) cause mammalian spongiform encephalopathies such as Creutzfeldt-Jakob disease in humans and bovine spongiform encephalopathy in cattle Citation37, Citation38. The prion protein (PrPc) is a monomeric, non-essential, protease-sensitive glycosylphosphatidylinositol-anchored cell surface protein Citation37. PrPc is present in neurons, but the function of the native protein is not fully understood. PrPc knock-out mice show alterations in, for example, circadian rhythm, brain copper levels, and neural stem cell differentiation. Known PrPc binding partners include copper, laminin, and laminin receptors Citation37. PrPc, which contains 40% α-helices and few β-sheets, can be converted into the cytotoxic form called PrP scrapie (PrPsc) with 30% α-helical and 45% β-sheet secondary structure Citation39. PrPc is monomeric and protease-sensitive, whereas PrPsc is multimeric and more protease-resistant Citation38. The characteristics of PrPsc as well as its ability to form amyloid fibrils associates TSEs with other conformational diseases Citation37. The elongation of PrP scrapie is accomplished by the induction of conformational change of PrPc by PrPsc to generate more PrPsc and to propagate the disease.
The principal target of prion pathology is the brain, but still most TSEs display prion replication at several extracellular locations. Orally ingested prions are absorbed in the intestines and transported to the blood and lymphoid organs where they replicate. From the spleen, muscles, appendix, or tonsils, the prions are then transported by peripheral nerves to the brain, where they cause pathology with loss of neurons. The cytotoxic process impairs brain function, including memory loss and locomotory problems Citation40.
Recently, it has been shown that prion replication can occur in vitro by cyclic amplification of protein misfolding and that inoculation of in vitro-produced prions causes scrapie-like illness Citation41.
Beneficial prions in yeast
Prion proteins have the unusual capacity to fold into two functionally distinct conformations, one of which is self-perpetuating. These self-replicating protein conformations have been proposed to constitute a kind of molecular memory that transmits genetic information Citation6 and can cause a switch of state resulting in production of heritable phenotypes Citation42.
It has also been proposed that the Sup35 protein in Saccharomyces cerevisiae can serve as an element of genetic inheritance, propagated in a similar way as hypothesized for the transmission prions in the spongiform encephalopathies. Therefore these protein factors have been called yeast prions Citation43. It has been suggested that the Sup35p protein in yeast may adopt a specific self-propagating conformation, similar to mammalian prions, giving rise to the [psi+] nonsense suppressor determinant, inherited in a non-Mendelian fashion. Sup35 molecules interact with each other through their N-terminal domains in [psi+], but not [psi-] cells. This interaction is critical for [psi+] propagation, since its disruption leads to a loss of [psi+]. Sup35p forms high molecular weight aggregates in [psi+] cells, which inhibit Sup35p activity leading to a [psi+] nonsense-suppressor phenotype Citation44. This self-perpetuating change in Sup35p conformation shows that deliberate genetic engineering can create new elements of genetic inheritance Citation43. In addition to Sup35, there are several fungal prions known. Recently, CPEB (cytoplasmic polyadenylation element-binding protein) was shown to regulate mRNA translation through an N-terminal extension that changes conformation when fused to a reporter protein in yeast Citation42.
HAMLET–a tumoricidal complex of partially unfolded α-lactalbumin and oleic acid
HAMLET is a complex of partially unfolded α-lactalbumin and oleic acid (C18:1:9cis) Citation4 that kills tumor cells and immature cells but not the healthy differentiated cells so far tested. The tumoricidal activity was discovered in casein, obtained after low pH precipitation of human milk Citation14. To identify the active component, casein was fractionated by ion exchange chromatography. The active complex showed high affinity for the matrix and was only eluted after high salt. The major component of the eluate was α-lactalbumin, and the fraction was named MAL (multimeric α-lactalbumin) due to the oligomeric nature on SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) Citation14, Citation45.
Native α-lactalbumin was shown to lack tumoricidal activity, suggesting that MAL might be structurally modified. As no posttranslational modifications were detected, we examined the conformation of α-lactalbumin in the complex, using CD spectroscopy and the fluorescent hydrophobic dye ANS (8-anilinonaphthalene-1-sulfonic acid). MAL contained partially unfolded protein, as expected from the low pH that was used to precipitate MAL from milk. In addition, the active complex contained a cofactor, which defined the activity and prevented α-lactalbumin from reverting to the native state. The cofactor was identified as oleic acid, and the conditions required for complex formation were defined by deliberate conversion of native α-lactalbumin to HAMLET in the presence of oleic acid Citation4.
Molecular properties
The identity of α-lactalbumin has been known since the late 19th century Citation48. The 14-kDa protein consists of 123 amino acids and is produced in large amounts in human breast-milk Citation49, Citation50. The first crystals were not obtained until about 65 years later Citation51, but since then many groups have determined the structure of native human α-lactalbumin Citation52–54 with similar results. The crystal structure reveals two domains, the α- and the β-domain Citation52. The large α-helical domain contains three major α-helices (amino acids 5–11, 23–34, 86–98) and two short 310-helices (amino acids 18–20, 115–118) (). The small β-domain consists of a triple-stranded antiparallel β-sheet (amino acids 40–50) and a short 310-helix (amino acids 76–82) Citation52. The protein is stabilized by four disulfide bonds Citation55 and a high-affinity calcium binding site Citation56. The disulfide bonds are spread throughout the protein (C6–120, C28–111, C61–77, C73–91) and connect the two domains.
In addition to X-ray crystallographic studies of the native state, α-lactalbumin has been important as a model of protein folding Citation57–59. The protein forms a molten globule Citation57, which is a partially unfolded state characterized by a native-like secondary structure with a loss of well defined tertiary packing Citation46, Citation47. The α-lactalbumin molten globule has been studied by several techniques including hydrogen exchange combined with NMR Citation60, Citation61, photochemically induced dynamic nuclear polarization (CIDNP) NMR Citation58, limited proteolysis Citation62, and mutational studies Citation63. The results indicate that the α-lactalbumin molten globule is characterized by a native-like, structured α-domain and a less ordered β-domain, and the α-domain has been shown to be stabilized by a hydrophobic core Citation64.
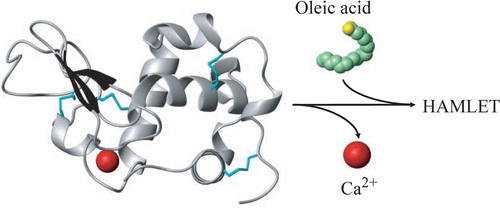
HAMLET is formed from α-lactalbumin and oleic acid in a two-step procedure (). First, the protein is partially unfolded by removal of calcium, which results in a conformational change to the apo state. Unfolding increases the affinity for oleic acid and enables the fatty acid to form a complex with the protein. By near-ultraviolet (UV) CD spectroscopy HAMLET CD spectrum resembled apo α-lactalbumin but with a decrease in signal suggesting a partially unfolded state Citation4. Increased exposure of hydrophobic domains was detected using the fluorescent hydrophobic dye ANS. The HAMLET ANS spectrum showed a blue shift compared to the native protein indicating exposure of hydrophobic surfaces enabling ANS to bind. The 1H-NMR spectrum of HAMLET showed broader peaks indicating a less-ordered protein, confirming the near-UV CD result. Oleic acid was detected in the spectrum, and the signal was broader than oleic acid alone, suggesting that oleic acid was integrated in the protein Citation4. The techniques suggested that apo α-lactalbumin is in a molten globule state Citation46, Citation47 in the HAMLET complex (discussed below).
Figure 1. Schematic of HAMLET formation from α-lactalbumin and oleic acid. Partial unfolding is achieved by removal of calcium, and the unfolded protein binds oleic acid. Structures from pdbID 1HML Citation104 and 1LFO Citation105 were modified in MOLMOL Citation106.
Sequence variation in α-lactalbumins
α-Lactalbumins are structurally conserved among species, suggesting that α-lactalbumin sequence variation among species might be used as a tool to understand the molecular basis of HAMLET formation and function Citation65. We have explored the sequence variation in naturally occurring α-lactalbumins from different species to understand the contribution of different regions of the molecule to the formation of HAMLET-like complexes with oleic acid. Human, bovine, equine, caprine, and porcine α-lactalbumin show a 71% sequence homology. The calcium-binding site is 100% conserved, but sequence differences are spread through the protein.
α-Lactalbumins were purified from human, bovine, equine, caprine, and porcine milk, and each protein was treated with EDTA (ethylenediamine tetra-acetic acid) and subjected to conversion on an oleic acid-conditioned column, according to the HAMLET conversion protocol. Each of the proteins was retained on the oleic acid-conditioned column and eluted with high salt as HAMLET-like complexes. The human protein showed a higher yield (62%) than the other species (24%–46%). The near-UV CD spectra of native human, bovine, and caprine α-lactalbumin resembled previous results Citation62, Citation66–71 with equine α-lactalbumin being the least folded protein while the porcine protein was slightly more folded. All the HAMLET-like complexes had lost tertiary structure and near-UV CD signals compared to the native protein. The tumoricidal activity of the HAMLET-like complexes was tested on lymphoma cells, and viability was determined by trypan blue exclusion: all complexes were shown to be tumoricidal, with similar activity as HAMLET, suggesting that sequence variation does not change the ability of α-lactalbumins to form active HAMLET complexes. Tumor cell viability was not affected when applying fresh milk at high concentrations, consistent with α-lactalbumin being native and oleic acid-bound in milk triglycerides Citation72.
Lipids as cofactors in HAMLET
Partially unfolded α-lactalbumin interacts specifically with longchained unsaturated fatty acids Citation70. We determined the ability of different fatty acids to form HAMLET-like complexes with apo α-lactalbumin and the tumoricidal activity of those complexes, using fatty acids differing in carbon chain length, saturation, and double bond conformation. The C18 cis fatty acids and other fatty acids with cis double bonds readily formed complexes with apo α-lactalbumin on the ion exchange matrix. The highest yield was obtained for oleic acid (C18:1:9cis) and vaccenic acid (C18:1:11cis) Citation73. In addition, complex formation with partially unfolded α-lactalbumin was achieved with palmitic acid (C16, cis) and arachidonic acid (C20, cis). In contrast, fatty acids of similar carbon chain length but with the double bond in the trans orientation were unable to form complexes, as were saturated fatty acids.
The tumoricidal activity of the different fatty acid complexes was tested on mouse leukemia cells, as the loss of cell viability and DNA fragmentation. Human α-lactalbumin in complex with C18:1:9cis (HAMLET) and C18:1:11cis killed the cells efficiently and caused DNA fragmentation, but complexes with palmitic acid (C16, cis) and arachidonic acid (C20, cis) were less tumoricidal.
Calcium-free and unfolded α-lactalbumin kills tumor cells in complex with oleic acid
α-Lactalbumin unfolds when the protein is exposed to conditions that remove the strongly bound calcium ion, such as treatment with EDTA Citation74. Unfolding with EDTA is reversible, however, as α-lactalbumin reverts to the native state if calcium is present. By mutating the calcium-binding site, both calcium binding and reversion may be prevented Citation70. The D87 mutant cannot bind calcium and thus remains partially unfolded in the presence of calcium and at natural solvent conditions. We showed that the D87 mutant forms a tumoricidal complex with oleic acid. The results suggested that the native fold is not required for the formation of the tumoricidal complex or for the tumoricidal activity. Furthermore, they excluded that calcium in α-lactalbumin is involved in the tumoricidal effect of the complex.
Mechanisms of cell death in response to HAMLET
Apoptosis and autophagy in response to HAMLET
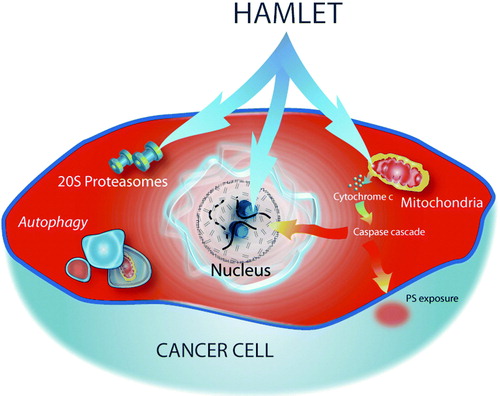
More than 40 different tumor cell lines have been exposed to HAMLET in vitro, and all were sensitive, regardless of tumor type, species, and tissue origin Citation75. The HAMLET-treated cells showed characteristics of apoptosis Citation14, and HAMLET caused mitochondrial swelling and loss of mitochondrial membrane potential Citation76, Citation77 accompanied by cytochrome C release, caspase activation Citation77, and phosphatidyl serine (PS) exposure on the cell surface. Apoptosis was not the cause of tumor cell death, however, as caspase inhibitors did not inhibit the death response Citation76–78. HAMLET was shown to kill tumor cells regardless of their Bcl-2 and p53 status Citation78, further illustrating how death in response to HAMLET differs from classical apoptosis.
In parallel with apoptosis, HAMLET triggers macroautophagy (Aits S, Gustafsson L, Hallgren O, Brest P, Gustafsson M, Trulsson M, et al., in press, Int J of Cancer, 2008). Macroautophagy occurs at basal levels in most cells but increases in response to cellular stress such as starvation Citation79, and extreme responses have been proposed as a form of programmed cell death, called autophagic/type II cell death Citation80, Citation81. After HAMLET treatment, double-membrane-enclosed vesicles, LC3 translocation and accumulation, typical of macroautophagy, were observed in tumor cells. Furthermore, inhibition of macroautophagy by Beclin-1 and Atg5 siRNAs significantly reduced HAMLET-induced cell death, consistent with the involvement of Beclin-1, LC3, and Atg5 in autophagosome formation, suggesting that autophagy is one aspect of cell death in response to HAMLET.
ER stress and proteasome response to HAMLET
HAMLET binds to the surface of tumor cells and enters the cytoplasm Citation4, Citation82–84, but native α-lactalbumin is only taken up in small amounts Citation4, Citation45, Citation83, suggesting that unfolding of α-lactalbumin and fatty acid binding are both required for uptake into tumor cells. We have hypothesized that the internalization of HAMLET may trigger the ER and proteasome responses normally activated by endogenous, unfolded proteins. Furthermore, 20S proteasomes have been proposed to degrade unfolded α-lactalbumin in vitro Citation85. Preliminary results suggested that the recognition of unfolded α-lactalbumin by the ER and by 20S proteasomes and the subsequent failure of these responses to scavenge and degrade α-lactalbumin are essential to trigger tumor cell death.
Nuclear receptors and chromatin interactions of HAMLET
HAMLET accumulates in the nuclei of tumor cells, and histones have been identified as nuclear receptors for HAMLET Citation82. By confocal microscopy, histones and HAMLET have been shown to colocalize in the nuclei of tumor cells. High-affinity interactions with histones have been documented with isolated histones, and in nuclear extracts HAMLET, histones, and DNA form virtually insoluble complexes. Healthy cells only take up small amounts of HAMLET, and there is no evidence of nuclear translocation of HAMLET in healthy cells Citation82, Citation83.
The chromatin accessibility is controlled by acetylation and deacetylation of the histone tail. Histone deacetylases (HDACs), which close the chromatin, are often overexpressed in tumor cells, and HDAC inhibitors (HDIs) are therefore used to open up the chromatin to tumoricidal agents. Recently, we have shown that HAMLET acts in synergy with HDIs, by enhancing the hyperacetylation response to the HDIs and by promoting cell death Citation86. We also showed that HAMLET binds to tailless histone proteins, in contrast to HDIs, which modify the histone tail. Future studies in tumor models should investigate if the combination of HAMLET and HDIs may be used to increase the therapeutic effect.
HAMLET and cancer therapy
Cancer therapeutics often lack tumor selectivity and cause severe side effects due to tissue toxicity. There is a need to identify new drugs that selectively kill tumor cells, and significant progress is being made in this area. Examples of new, targeted therapies are growth factor inhibitors, blockers of angiogenesis, and proapoptotic drugs. HAMLET is an interesting candidate drug, as the complex shows a high degree of selectivity for tumor cells in vitro and in vivo. The tumoricidal effect of HAMLET in vivo has been tested both in animals and in patients Citation83, Citation87, Citation88.
Human glioblastoma xenografts
A rat glioblastoma model, which reproduces the invasive growth of the human tumors, was used to test the therapeutic effect of HAMLET Citation83. Nude rats were xenotransplanted with glioblastoma cells obtained from surgical specimens of human tumors that were injected into the striatum. The tumor area was subsequently infused with HAMLET or α-lactalbumin by intracerebral convection-enhanced delivery, and the tumor size was followed by magnetic resonance imaging. HAMLET was shown to reduce the tumor size and to delay the development of pressure-related symptoms. HAMLET caused apoptosis in the tumor, as determined by TUNEL (terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling)-staining on biopsies, but there was no apoptotic response to α-lactalbumin. No toxic side effects were observed.
Placebo-controlled study of human skin papillomas
The effect of HAMLET was further studied in patients with skin papillomas Citation87. Patients with severe, therapy-resistant papillomas on hands and feet were enrolled in a placebo-controlled and double-blind study. HAMLET or saline solution was applied daily for 3 weeks, and the effect on lesion volume was recorded. This phase was followed by an open 3-week trial where all the patients received HAMLET. Finally, most of the patients were examined 2 years after the end of the study. All the HAMLET-treated patients showed a decrease in lesion volume by at least 75%, and after 2 years all lesions had resolved in 83% of the patients. We concluded that HAMLET had beneficial effects on skin papillomas without side effects.
Human bladder cancer
The papilloma study suggested that topical HAMLET administration might be useful also in cancer patients. We selected to study bladder cancer, as a variant of topical treatments are used for intravesical instillation to prevent or delay cystectomy Citation88. The response to HAMLET in bladder cancer patients was studied after intravesical application in nine patients, who received five daily HAMLET instillations prior to scheduled surgery. HAMLET caused a rapid increase in cell shedding. The shed cells showed signs of apoptosis but NaCl, phosphate buffered saline (PBS), and α-lactalbumin did not trigger cell shedding. At surgery, a reduction in tumor size was observed in eight of nine patients, and four of the patients had positive TUNEL-staining in biopsies from the remaining tumor, showing that HAMLET has a direct effect on bladder cancer tissue, in vivo Citation88.
Parallels between HAMLET, prions, and amyloid
Fibril formation
Amyloid fibrils are composed of β-strands arranged into β-sheets in which the strands are hydrogen-bonded 4.7 Å apart. In an antiparallel arrangement the repeat distance is 9.6 Å, and diffraction patterns from highly oriented amyloid fibril samples show layer lines at 9.6 Å Citation89. To examine if HAMLET might behave in a similar manner, solutions were left at various temperatures and times (days to months), and the presence of aggregates was examined by electron and atomic force microscopy. No fibril formation was detected with either of the techniques. By electron and atomic force microscopy HAMLET was shown to be mainly monomeric (unpublished data, Pettersson-Kastberg J, Malisauskas M, Morozova-Roche L, Svanborg C, 2007). Solution NMR studies also indicate that the partially folded protein–oleic acid complex is monomeric (unpublished data, Mok KH, 2008). The monomeric state in solution does not exclude the possibility that multimers might be formed upon contact with cell membranes.
Fibrillogenesis has been proposed to be a membrane-associated process Citation90–93. Fibril formation of several proteins, including lysozyme, cytochrome C, histone H1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), transthyretin, endostatin, and α-lactalbumin, was enhanced by membranes containing PS, and fluorescent lipid tracers indicated the presence of lipid in these amyloid fibrils Citation92, Citation93. The rapid fibril formation might reflect the low pH at the membrane surface Citation93 or the high proportion of hydrogen bonds in the amyloidogenic proteins that are incompletely formed Citation90. Endostatin, a tumor angiogenesis inhibitor, was shown only to form fibrils on acidic PS membranes Citation92, and since cancer cells have been reported to expose PS on the surface Citation94, they might be a target for endostatin, which increases membrane permeability Citation91, Citation92.
It is also possible that the fibrillar constituents may form complexes with membrane lipids and gain access to cells in a manner similar to HAMLET. The properties of membrane-associated HAMLET remain to be defined. Whether the effects of the fatty acids on the partially folded α-lactalbumin protein are equivalent to the membrane bilayer interactions of membrane proteins remains unknown. Our studies show that the partial unfolding of the protein must precede the contact with fatty acids, suggesting that the mode of interaction is not exclusively fatty acid-induced.
Are cofactors involved in amyloid fibril formation and disease pathogenesis?
Based on the HAMLET complex, we have proposed that cofactors may be needed to define new, bioactive folding variants. The availability of cofactors in certain tissues might also help ensure that active complexes are formed only at the site where the new function is beneficial. As unfolding may occur quite frequently Citation29, an elegant solution would be to recruit cofactors available within specific tissue environments to prevent unfolded protein molecules from becoming cytotoxic.
The need for tissue cofactors in amyloid and prion formation and toxicity has been extensively discussed. Prion aggregates contain different prion protein species varying from non-fibrillar oligomers to large amyloid plaques Citation37. These different molecular forms have been produced in vitro but have mostly not been toxic, and thus the molecular basis of toxicity is not fully understood. The aggregates are formed by the specific misfolded protein species, but other components, including other proteins or carbohydrates, may be incorporated Citation95.
While it is accepted that the change in fold is required for disease, the possible involvement of other additional proteins or nucleic acids is still being discussed Citation37, Citation38, Citation96. Three different classes of possible cofactors have been suggested Citation96. The first includes cellular receptors, which might facilitate conversion of PrPc to PrPsc and the uptake by endosomes. The known interactions with, for example, the laminin receptor and heparan sulfate support this hypothesis. A second possibility involves nucleic acids. The prion protein is known to bind to nucleic acids, and the binding results in a structural modification. The third possibility includes lipids, which could act as chaperones. This theory is supported by the fact that different lipids can stabilize α-helix- and β-sheet-enriched structures Citation96. It may be speculated that similar interactions guide the way in which oleic acid stabilizes the partially unfolded state of α-lactalbumin, which contains both α-helical and β-sheet structure, and the prion proteins.
Amyloid cytotoxicity
It is not clear how prions and amyloid fibers exert their cytotoxic effects. It is well known, for example, that aggregated Aβ fibrils kill healthy neuronal cells Citation35, but on the other hand there is increasing evidence that folding intermediates (protofibrils) can be toxic. Silveira et al. have identified aggregates with 14–28 PrP molecules as the most efficient disease inducers, but neither the small oligomers nor the larger aggregates are toxic Citation97. The lysozyme amyloids were proposed to be cytotoxic as the oligomers killed neuroblastoma cells by an apoptosis-like mechanism and the fibrils lead to a necrosis-like death Citation98. In this case, the amyloid was obtained after 12 days at pH 2.2 and 57°C. Bovine α-lactalbumin forms fibrils at pH 2.0 and 37°C Citation99. The same results were obtained with S-(carboxymethyl)-α-lactalbumin, a variant with three out of four disulfide bridges reduced. Yang et al. obtained bovine α-lactalbumin fibril formation from pH 4.5 and below. The fibrillogenesis was completely inhibited by oleic acid (640 µM) at pH 4.0–4.5, and insoluble aggregates were obtained Citation100. This did not occur at lower pH, and at neutral pH the apo protein did not form fibrils. However, GAPDH fibrils, formed under oxidizing conditions, kill cervical cancer cells ((HeLA) cells by apoptosis, but the effect on healthy cells is not known. In contrast to HAMLET, both fibrils and amyloid have been shown to kill healthy cells Citation35.
Recently, an antimicrobial peptide of bacterial origin (plantaricin A) was shown to form protein-lipid amyloid-like fibrils upon binding to negatively charged phospholipid-containing membranes. This resulted in killing of bacteria and leukemic T-cells and suggested a possible mechanistic connection between fibril formation and the cytotoxicity of plantaricin A Citation101. It is not clear if this mechanism is related to the antimicrobial activity of HAMLET Citation102, but it is important to keep in mind that HAMLET selectively kills tumor cells by apoptosis, and not by necrosis, as for example plantaricin A.
Summary
With the exception of a few metastable native state proteins, folding intermediates that possess biological function are rare. We have reviewed this question, using amyloid, prions, HAMLET, and antibacterial peptides as examples. The tumoricidal effect of HAMLET requires unfolding of α-lactalbumin and binding of oleic acid. A summary of the structural properties of HAMLET and the basis of the tumoricidal activity is shown in . In HAMLET, α-lactalbumin resembles a molten globule, as the secondary structure of α-lactalbumin is retained but the molecule lacks tertiary packing. The fatty acid interaction shows specificity, as only oleic acid-like fatty acids form active complexes with apo α-lactalbumin. The protein component can be more varied, however, as α-lactalbumin species variants form tumoricidal complexes with oleic acid Citation70.
The multi-facetted cell death response to HAMLET is illustrated in . We have used the ‘hydra’ metaphor to illustrate that HAMLET acts on different cell organelles, potentially triggering many different cell death pathways Citation103. The results suggest that the complex cellular response is advantageous, as it ensures that HAMLET may kill a broad spectrum of tumor cells, despite their resistance to programmed cell death.
Acknowledgements
The studies summarized here were performed over several years and were supported by the Sharon D. Lund foundation grant and the American Cancer Society, the Swedish Cancer Society, the Swedish Pediatric Cancer Foundation, the Medical Faculty (Lund University), the Söderberg Foundation, the Segerfalk Foundation, the French Medical Research Foundation (FRM, Paris), the Anna-Lisa and Sven-Erik Lundgren Foundation for Medical Research, the Knut Alice Wallenberg Foundation, the Lund City Jubileumsfond, the John and Augusta Persson Foundation for Medical Research, the Maggie Stephens Foundation, the Gunnar Nilsson Cancer Foundation, the Inga-Britt och Arne Lundberg foundation, the HJ Forssman Foundation for Medical Research and the Royal Physiographic Society. Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. Catharina Svanborg is a shareholder in NatImmune, which has been involved in HAMLET drug development. There has been no interaction with the company or support received for the studies described here.
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973; 181: 223–30
- Cabrita LD, Bottomley SP. How do proteins avoid becoming too stable? Biophysical studies into metastable proteins. Eur Biophys J. 2004; 33: 83–8
- Bemporad F, Gsponer J, Hopearuoho HI, Plakoutsi G, Stati G, Stefani M, et al. Biological function in a non-native partially folded state of a protein. EMBO J. 2008; 27: 1525–35
- Svensson M, Hakansson A, Mossberg AK, Linse S, Svanborg C. Conversion of α-lactalbumin to a protein inducing apoptosis. Proc Natl Acad Sci U S A. 2000; 97: 4221–6
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006; 4: e6
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005; 6: 435–50
- Beadle GW, Tatum EL. Genetic control of biochemical reactions in neurospora. Proc Natl Acad Sci U S A. 1941; 27: 499–506
- Green ED, Chakravarti A. The human genome sequence expedition: views from the ‘base camp’. Genome Res. 2001; 11: 645–51
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001; 291: 1304–51
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45.
- Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999; 24: 329–32
- Claverie JM. Gene number. What if there are only 30,000 human genes?. Science. 2001; 291: 1255–7
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000; 406: 959–64
- Hakansson A, Zhivotovsky B, Orrenius S, Sabharwal H, Svanborg C. Apoptosis induced by a human milk protein. Proc Natl Acad Sci U S A. 1995; 92: 8064–8
- Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999; 24: 8–11
- Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005; 30: 484–9
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002; 295: 1852–8
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006; 125: 443–51
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992; 355: 33–45
- Mok KH, Hore PJ. Photo-CIDNP NMR methods for studying protein folding. Methods. 2004; 34: 75–87
- Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B Biol Sci. 2001; 356: 133–45
- Vendruscolo M, Paci E, Karplus M, Dobson CM. Structures and relative free energies of partially folded states of proteins. Proc Natl Acad Sci U S A. 2003; 100: 14817–21
- Levinthal C. Are there pathways for protein folding?. J Chim Phys. 1968; 65: 44–5
- Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997; 4: 10–9
- Baldwin RL. The nature of protein folding pathways: the classical versus the new view. J Biomol NMR. 1995; 5: 103–9
- Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995; 21: 167–95
- Soto C. Protein misfolding and disease; protein refolding and therapy. FEBS Lett. 2001; 498: 204–7
- Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, et al. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002; 3: 411–21
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006; 75: 333–66
- Trzesniewska K, Brzyska M, Elbaum D. Neurodegenerative aspects of protein aggregation. Acta Neurobiol Exp (Wars) 2004; 64: 41–52
- Selkoe DJ, Podlisny MB, Joachim CL, Vickers EA, Lee G, Fritz LC, et al. Beta-amyloid precursor protein of Alzheimer disease occurs as 110- to 135-kilodalton membrane-associated proteins in neural and nonneural tissues. Proc Natl Acad Sci U S A. 1988; 85: 7341–5
- Soto C, Castano EM, Frangione B, Inestrosa NC. The α-helical to β-strand transition in the amino-terminal fragment of the amyloid ß-peptide modulates amyloid formation. J Biol Chem. 1995; 270: 3063–7
- Barrow CJ, Yasuda A, Kenny PT, Zagorski MG. Solution conformations and aggregational properties of synthetic amyloid beta-peptides of Alzheimer's disease. Analysis of circular dichroism spectra. J Mol Biol. 1992; 225: 1075–93
- Hilbich C, Kisters-Woike B, Reed J, Masters CL, Beyreuther K. Substitutions of hydrophobic amino acids reduce the amyloidogenicity of Alzheimer's disease beta A4 peptides. J Mol Biol. 1992; 228: 460–73
- Geula C, Wu CK, Saroff D, Lorenzo A, Yuan M, Yankner BA. Aging renders the brain vulnerable to amyloid beta-protein neurotoxicity. Nat Med. 1998; 4: 827–31
- Soto C, Sigurdsson EM, Morelli L, Kumar RA, Castano EM, Frangione B. β-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: implications for Alzheimer's therapy. Nat Med. 1998; 4: 822–6
- Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006; 443: 803–10
- Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998 1; 93:337–48.
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, et al. Conversion of a-helices into ß-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A. 1993; 90: 10962–6
- Aguzzi A, Heikenwalder M. Pathogenesis of prion diseases: current status and future outlook. Nat Rev Microbiol. 2006; 4: 765–75
- Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005; 121: 195–206
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003; 115: 879–91
- Lindquist S, Krobitsch S, Li L, Sondheimer N. Investigating protein conformation-based inheritance and disease in yeast. Philos Trans R Soc Lond B Biol Sci. 2001; 356: 169–76
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996; 15: 3127–34
- Svensson M, Sabharwal H, Hakansson A, Mossberg AK, Lipniunas P, Leffler H, et al. Molecular characterization of alpha-lactalbumin folding variants that induce apoptosis in tumor cells. J Biol Chem. 1999; 274: 6388–96
- Arai M, Kuwajima K. Role of the molten globule state in protein folding. Adv Protein Chem. 2000; 53: 209–82
- Ptitsyn OB. Molten globule and protein folding. Adv Protein Chem. 1995; 47: 83–229
- Halliburton WD. The proteids of milk. J Physiol. 1890; 11: 448–63
- Heine WE, Klein PD, Reeds PJ. The importance of α-lactalbumin in infant nutrition. J Nutr. 1991; 121: 277–83
- Lonnerdal B, Forsum E. Casein content of human milk. Am J Clin Nutr. 1985; 41: 113–20
- Zweig G, Block RJ. Studies on bovine whey proteins. III. The preparation of crystalline a-lactalbumin and ß-lactoglobulin ferrilactin. Arch Biochem Biophys. 1954; 51: 200–7
- Acharya KR, Ren JS, Stuart DI, Phillips DC, Fenna RE. Crystal structure of human α-lactalbumin at 1.7 A resolution. J Mol Biol. 1991; 221: 571–81
- Chandra N, Brew K, Acharya KR. Structural evidence for the presence of a secondary calcium binding site in human α-lactalbumin. Biochemistry. 1998; 37: 4767–72
- Fenna RE. Crystallization of human α-lactalbumin. J Mol Biol. 1982; 161: 211–5
- Vanaman TC, Brew K, Hill RL. The disulfide bonds of bovine a-lactalbumin. J Biol Chem. 1970; 245: 4583–90
- Hiraoka Y, Segawa T, Kuwajima K, Sugai S, Murai N. α-Lactalbumin: a calcium metalloprotein. Biochem Biophys Res Commun. 1980; 95: 1098–104
- Kuwajima K. The molten globule state of α-lactalbumin. FASEB J. 1996; 10: 102–9
- Mok KH, Nagashima T, Day IJ, Hore PJ, Dobson CM. Multiple subsets of side-chain packing in partially folded states of α-lactalbumins. Proc Natl Acad Sci U S A. 2005; 102: 8899–904
- Peng ZY, Kim PS. A protein dissection study of a molten globule. Biochemistry. 1994; 33: 2136–41
- Schulman BA, Redfield C, Peng ZY, Dobson CM, Kim PS. Different subdomains are most protected from hydrogen exchange in the molten globule and native states of human α-lactalbumin. J Mol Biol. 1995; 253: 651–7
- Chyan CL, Wormald C, Dobson CM, Evans PA, Baum J. Structure and stability of the molten globule state of guinea-pig α-lactalbumin: a hydrogen exchange study. Biochemistry. 1993; 32: 5681–91
- Polverino de Laureto P, Frare E, Gottardo R, Fontana A. Molten globule of bovine α-lactalbumin at neutral pH induced by heat, trifluoroethanol, and oleic acid: a comparative analysis by circular dichroism spectroscopy and limited proteolysis. Proteins. 2002; 49: 385–97
- Wu LC, Peng ZY, Kim PS. Bipartite structure of the α-lactalbumin molten globule. Nat Struct Biol. 1995; 2: 281–6
- Wu LC, Kim PS. A specific hydrophobic core in the α-lactalbumin molten globule. J Mol Biol. 1998; 280: 175–82
- Pettersson J, Mossberg AK, Svanborg C. α-Lactalbumin species variation, HAMLET formation, and tumor cell death. Biochem Biophys Res Commun. 2006; 345: 260–70
- Chedad A, Van Dael H. Kinetics of folding and unfolding of goat α-lactalbumin. Proteins. 2004; 57: 345–56
- Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov S, et al. α-Lactalbumin: compact state with fluctuating tertiary structure?. FEBS Lett. 1981; 136: 311–5
- Peng ZY, Wu LC, Schulman BA, Kim PS. Does the molten globule have a native-like tertiary fold?. Philos Trans R Soc Lond B Biol Sci. 1995; 348: 43–7
- Permyakov SE, Uversky VN, Veprintsev DB, Cherskaya AM, Brooks CL, Permyakov EA, et al. Mutating aspartate in the calcium-binding site of α-lactalbumin: effects on the protein stability and cation binding. Protein Eng. 2001; 14: 785–9
- Svensson M, Fast J, Mossberg AK, Duringer C, Gustafsson L, Hallgren O, et al. α-Lactalbumin unfolding is not sufficient to cause apoptosis, but is required for the conversion to HAMLET (human α-lactalbumin made lethal to tumor cells). Protein Sci. 2003; 12: 2794–804
- Wu LC, Schulman BA, Peng ZY, Kim PS. Disulfide determinants of calcium-induced packing in α-lactalbumin. Biochemistry. 1996; 35: 859–63
- Jensen RG. The lipids in human milk. Prog Lipid Res. 1996; 35: 53–92
- Svensson M, Mossberg AK, Pettersson J, Linse S, Svanborg C. Lipids as cofactors in protein folding: Stereo-specific lipid-protein interactions are required to form HAMLET (human α-lactalbumin made lethal to tumor cells). Protein Sci. 2003; 12: 2805–14
- Polverino de Laureto P, Scaramella E, Frigo M, Wondrich FG, De Filippis V, Zambonin M, et al. Limited proteolysis of bovine α-lactalbumin: isolation and characterization of protein domains. Protein Sci. 1999; 8: 2290–303
- Svanborg C, Agerstam H, Aronson A, Bjerkvig R, Duringer C, Fischer W, et al. HAMLET kills tumor cells by an apoptosis-like mechanism–cellular, molecular, and therapeutic aspects. Adv Cancer Res. 2003; 88: 1–29
- Kohler C, Gogvadze V, Hakansson A, Svanborg C, Orrenius S, Zhivotovsky B. A folding variant of human α-lactalbumin induces mitochondrial permeability transition in isolated mitochondria. Eur J Biochem. 2001; 268: 186–91
- Kohler C, Hakansson A, Svanborg C, Orrenius S, Zhivotovsky B. Protease activation in apoptosis induced by MAL. Exp Cell Res. 1999; 249: 260–8
- Hallgren O, Gustafsson L, Irjala H, Selivanova G, Orrenius S, Svanborg C. HAMLET triggers apoptosis but tumor cell death is independent of caspases, Bcl-2 and p53. Apoptosis. 2006; 11: 221–33
- Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12 Suppl 2005; 2: 1509–18
- Baehrecke EH. Autophagy: dual roles in life and death?. Nat Rev Mol Cell Biol. 2005; 6: 505–10
- Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death?. Autophagy. 2005; 1: 66–74
- Duringer C, Hamiche A, Gustafsson L, Kimura H, Svanborg C. HAMLET interacts with histones and chromatin in tumor cell nuclei. J Biol Chem. 2003; 278: 42131–5
- Fischer W, Gustafsson L, Mossberg AK, Gronli J, Mork S, Bjerkvig R, et al. Human α-lactalbumin made lethal to tumor cells (HAMLET) kills human glioblastoma cells in brain xenografts by an apoptosis-like mechanism and prolongs survival. Cancer Res. 2004; 64: 2105–12
- Hakansson A, Andreasson J, Zhivotovsky B, Karpman D, Orrenius S, Svanborg C. Multimeric α-lactalbumin from human milk induces apoptosis through a direct effect on cell nuclei. Exp Cell Res. 1999; 246: 451–60
- Wenzel T, Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat Struct Biol. 1995; 2: 199–204
- Brest P, Gustafsson M, Mossberg AK, Gustafsson L, Duringer C, Hamiche A, et al. Histone deacetylase inhibitors promote the tumoricidal effect of HAMLET. Cancer Res. 2007; 67: 11327–34
- Gustafsson L, Leijonhufvud I, Aronsson A, Mossberg AK, Svanborg C. Treatment of skin papillomas with topical α-lactalbumin-oleic acid. N Engl J Med. 2004; 350: 2663–72
- Mossberg AK, Wullt B, Gustafsson L, Mansson W, Ljunggren E, Svanborg C. Bladder cancers respond to intravesical instillation of HAMLET (human a-lactalbumin made lethal to tumor cells). Int J Cancer. 2007; 121: 1352–9
- Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci U S A. 2005; 102: 315–20
- Fernandez A, Berry RS. Proteins with H-bond packing defects are highly interactive with lipid bilayers: implications for amyloidogenesis. Proc Natl Acad Sci U S A. 2003; 100: 2391–6
- Sparr E, Engel MF, Sakharov DV, Sprong M, Jacobs J, de Kruijff B, et al. Islet amyloid polypeptide-induced membrane leakage involves uptake of lipids by forming amyloid fibers. FEBS Lett. 2004; 577: 117–20
- Zhao H, Jutila A, Nurminen T, Wickstrom SA, Keski-Oja J, Kinnunen PK. Binding of endostatin to phosphatidylserine-containing membranes and formation of amyloid-like fibers. Biochemistry. 2005; 44: 2857–63
- Zhao H, Tuominen EK, Kinnunen PK. Formation of amyloid fibers triggered by phosphatidylserine-containing membranes. Biochemistry. 2004; 43: 10302–7
- Utsugi T, Schroit AJ, Connor J, Bucana CD, Fidler IJ. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 1991; 51: 3062–6
- Dobson CM. Protein folding and misfolding. Nature. 2003; 426: 884–90
- Fasano C, Campana V, Zurzolo C. Prions: protein only or something more? Overview of potential prion cofactors. J Mol Neurosci. 2006; 29: 195–214
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature. 2005; 437: 257–61
- Gharibyan AL, Zamotin V, Yanamandra K, Moskaleva OS, Margulis BA, Kostanyan IA, et al. Lysozyme amyloid oligomers and fibrils induce cellular death via different apoptotic/necrotic pathways. J Mol Biol. 2007; 365: 1337–49
- Goers J, Permyakov SE, Permyakov EA, Uversky VN, Fink AL. Conformational prerequisites for α-lactalbumin fibrillation. Biochemistry. 2002; 41: 12546–51
- Yang F, Jr, Zhang M, Zhou BR, Chen J, Liang Y. Oleic acid inhibits amyloid formation of the intermediate of α-lactalbumin at moderately acidic pH. J Mol Biol. 2006; 362: 821–34
- Zhao H, Sood R, Jutila A, Bose S, Fimland G, Nissen-Meyer J, et al. Interaction of the antimicrobial peptide pheromone plantaricin A with model membranes: implications for a novel mechanism of action. Biochim Biophys Acta. 2006; 1758: 1461–74
- Hakansson A, Svensson M, Mossberg AK, Sabharwal H, Linse S, Lazou I, et al. A folding variant of α-lactalbumin with bactericidal activity against Streptococcus pneumoniae. Molecular microbiology. 2000; 35: 589–600
- Mok KH, Pettersson J, Orrenius S, Svanborg C. HAMLET, protein folding, and tumor cell death. Biochem Biophys Res Commun. 2007; 354: 1–7
- Ren J, Stuart DI, Acharya KR. α-lactalbumin possesses a distinct zinc binding site. J Biol Chem. 1993; 268: 19292–8
- Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates. J Biol Chem. 1997; 272: 7140–50
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5, 29–32.