ABSTRACT
Background
MicroRNA (miR)-146a might participate in the occurrence of malignant tumor. The aim of the current investigation was to evaluate the relationship of microRNA-146a (miR-146a) rs2910164 C > G locus to the development of digestive system cancer (DSC).
Methods
We retrieved publications from PubMed, China Biology Medicine and EMBASE databases up to August 29, 2019. Finally, 56 independent case-control studies with 59,098 participants were included. The strength of the relationship between rs2910164 locus and a risk of DSC was assessed. The power value was also calculated in this study.
Results
We identified a correlation of rs2910164 locus in miR-146a with DSC development in dominant model (P = .035; power value = 0.994). MiR-146a rs2910164 locus was also identified to be correlated with a risk of DSC in Asians (GG/CG vs. CC: P = .033; power value = 0.989). Sensitivity analysis revealed that any individual study could not alter the final decision. In our study, no significant bias was found among these included studies (P > .1). The results of heterogeneity analysis suggested that small sample size (<1000 subjects), colorectal carcinoma, Asians, gastric carcinoma, esophageal squamous cell carcinoma, hepatocellular cancer, hospital-based study and high-quality score (≥7.0) subgroups contributed the heterogeneity to our findings. Galbraith radial plot determined that eleven outliers contributed to the main heterogeneity.
Conclusion
In summary, this meta-analysis highlights that rs2910164 locus might be implicated in the risk of DSC. More studies are, therefore, needed to confirm our results.
Introduction
Nowadays, malignant neoplasm of digestive system is a common burden on society which has seriously influenced individual’s survival and fitness worldwide. Digestive system cancer (DSC) included colorectal carcinoma (CRC), oral carcinoma (OC), hepatocellular cancer (HCC), esophageal carcinoma (EC), gastric carcinoma (GC), gallbladder cancer (GBC), pancreatic cancer (PC), etc. The incidence of most subtypes of DSC was occurred frequently, such as EC, GC, HCC and CRC (Bray et al. Citation2018). Although the development of carcinoma has not been fully understood, accumulating evidences indicate that cancer is a result of complex interaction between multiple environmental factors and individual’s genes.
In eukaryotes, microRNAs (miRs) are about 22 single-strand nucleotide acid which regulates related gene expression. Many investigations have suggested that miRs are important for controlling various functions of body. An abnormal expression of miR might be implicated in various human diseases. Accumulating evidences have indicated that miRs are implicated in growth and migration, inflammatory response, infection, immune response and cellular metabolism (Chen et al. Citation2019; Yang et al. Citation2019; Zhang et al. Citation2019a). MiR-146a is one of the common miRs which are of great importance for the roles of posttranscriptional regulatory. Investigations have suggested that miR-146a is important for process of innate immune response and inflammation, in which it acts as a vital negative regulator. In human infectious disease, pathogens must be recognized firstly, which is the necessary condition for activating the immune response (Saba et al. Citation2014). It is reported that several target loci of 3′-untranslated regions in toll-like receptors (TLRs) mRNAs are found (O’Neill et al. Citation2011). Interestingly, by using bioinformatics, a recent study has identified a potential interaction of miR-146a with TLR4 (Li and Shi Citation2013; Yang et al. Citation2011). MiR-146a is also implicated in cancer. It was reported that miR-146a facilitated oncogenesis of colorectal cancer and affected the microenvironment in tumor tissue (Cheng et al. Citation2019).
Single nucleotide polymorphisms (SNPs) in miRs could affect the stability and biological function of miR, and then influence the regulation of target gene. Rs2910164 C > G locus in miR-146a could influence the survival of CRC by regulating the cell apoptosis and the expression of cyclooxygenase-2 (Zhang et al. Citation2019b). A growing number of investigations have shown that miR-146a rs2910164 may confer the susceptibility to malignancy. Recently, many publications have explored the relationship of rs2910164 variants with DSC risk. Rs2910164 in miR-146a and its importance to the initiate of DSC have been widely explored. Some meta-analyses showed that G allele in rs2910164 polymorphism might not influence the initiate of DSC (Chen et al. Citation2014; Wang et al. Citation2012; Wu et al. Citation2013). Other published meta-analyses identified that rs2910164 variants could be implicated in the risk of DSC in Asian population (Li et al. Citation2014; Xie et al. Citation2015; Xu et al. Citation2014). As well, a meta-analysis reported that rs2910164 polymorphism conferred a risk of DSC in both Asians and Caucasians (Xie and Wang Citation2017). However, a more recent meta-analysis failed to confirm any relationship between rs2910164 and DSC risk (Xiong et al. Citation2017). Thus, the correlation of this locus with the development of DSC is more controversial. Nowadays, more studies have investigated the relationship of rs2910164 locus with DSC risk. By using a meta-analysis, pooling all eligible data might reduce the random error and increase the power of study. Finally, we could get a precise evaluation for the potential inherited correlation of rs2910164 polymorphism with DSC risk.
Materials and methods
Study researching
Using PubMed, China Biology Medicine and EMBASE databases, we searched the related studies (up to August 29, 2019). The following researching strategy was used: (microRNA-146a2 OR miR-146a2 OR rs2910164) AND (cancer OR carcinoma) AND (SNP OR polymorphism). To retrieve more related publications, the references in reviews and the original studies were also searched. In this study, there was no language restriction. According to the Table S1 PRISMA Checklist, this study was reported.
Data extraction
Two authors (L. Lv and Z. Chen) conducted data extraction independently. The eligible publication met the major included criteria: (a) assessing an association of rs2910164 with DSC risk; (b) designed as a case-control study; and (c) data could be obtained. Otherwise, the publications were excluded. The following exclusion criteria were used: (a) not case-control study; (b) only considering the prognosis of DSC; (c) review or meta-analysis; and (d) comments. If the extracted data were conflicting, another author (W. Tang) was invited to discuss until a consensus opinion was reached. The following information was extracted: source of controls, year of publication, first author, cancer type, Hardy–Weinberg equilibrium (HWE), country, ethnicity, the number of participants and genotypes.
Quality assessment
By using the Newcastle–Ottawa Quality Assessment Scale, we evaluated the quality score of the included studies. A high-quality study was defined as scores ≥ 7 stars (Wang et al. Citation2015).
Statistical methods
The correlation between this SNP and DSC risk was assessed by using odds ratios (ORs) and the corresponding 95% confidence intervals (CIs). The results were summarized in the corresponding models: homozygote comparison (GG vs. CC), recessive model (GG vs. CC/CG), dominant model (GG/CG vs. CC) and allelic model (G vs. C). I2 test and Q test were used to assess the heterogeneity. And P < .1 and/or I2 ≥ 50% were considered as the level of significance. When significant heterogeneity was observed, DerSimonian and Laird method (a random-effects model) was conducted to evaluate the association of rs2910164 with DSC (DerSimonian and Laird Citation1986; Higgins et al. Citation2003). Otherwise, a fixed-effects model (Mantel–Haenszel) was used to get a evaluation of rs2910164 variants with DSC risk (Mantel and Haenszel Citation1959). We also conducted subgroup analyses according to ethnicity, type of cancer, source of control, sample size (≥1000/<1000) and quality scores (≥7.0/<7). Galbraith radial plot was harnessed to further determine the source the heterogeneity. We carried out a sensitivity analysis to determine whether a single study could influence the final decision. Bgger’s funnel plots and the Egger’s test were done to assess the bias of publication. A P < .1 was considered as statistically significant for bias. All the P-values are two-sided. Stata12.0 software was used to conduct statistical analysis. In this study, the power value was also calculated by a Power-SampleSize software (α = 0.05) (Tang et al. Citation2013).
Results
Study characteristics
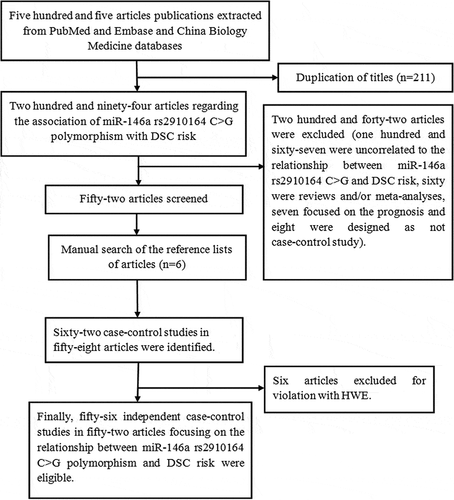
First, we retrieved 505 publications from PubMed, China Biology Medicine and EMBASE databases. After a primary filtrate, 210 duplicated articles were excluded. shows the process of the meta-analysis. Finally, 52 papers (56 independent case-control studies) involving 24,161 DSC patients and 34,937 cancer-free controls were included. Of these investigations, year of publication ranged between 2008 and 2018 and the number of participants in the eligible studies ranged from 128 to 3,585. In summary, there were 17 GC studies (Ahn et al. Citation2013; Chen et al. Citation2018; Dikeakos et al. Citation2014; Hishida et al. Citation2011; Jiang et al. Citation2016; Lin et al. Citation2019; Kupcinskas et al. Citation2014b; Ma Citation2012; Okubo et al. Citation2010; Parlayan et al. Citation2014; Pu et al. Citation2014; Rogoveanu et al. Citation2017; Soleimani et al. Citation2016; Xia et al. Citation2016; Yadegari et al. Citation2016; Zeng et al. Citation2010; Zhou et al. Citation2012a), 16 CRC studies (Chae et al. Citation2013; Chayeb et al. Citation2018; Dikaiakos et al. Citation2015; Gao et al. Citation2018; Hai-feng et al. Citation2016; Hezova et al. Citation2012; Kupcinskas et al. Citation2014a; Lv et al. Citation2013; Ma et al. Citation2013; Mao et al. Citation2014; Min et al. Citation2012; Ying et al. Citation2016), 15 HCC studies (Akkiz et al. Citation2011; Chu et al. Citation2014; Cong et al. Citation2014; Duan Lei Citation2017; Huang et al. Citation2013; Li et al. Citation2015; Xiang et al. Citation2012; Xu et al. Citation2008; Yan et al. Citation2015; Zhang et al. Citation2013; Citation2016; Zhou et al. Citation2012b), five esophageal squamous cell carcinoma (ESCC) studies (Guo et al. Citation2010; Qu et al. Citation2014; Shen et al. Citation2016; Umar et al. Citation2013; Wei et al. Citation2013), three oral squamous cell carcinoma (OSCC) studies (Chu et al. Citation2012; Palmieri et al. Citation2014; Zhang et al. Citation2017) and other carcinoma studies (one cholangiocarcinoma study (Mihalache et al. Citation2012), one GBC study (Srivastava et al. Citation2010) and one PC study (Pavlakis et al. Citation2013)). In addition, there were 42 case-control studies on Asians (Ahn et al. Citation2013; Chae et al. Citation2013; Chen et al. Citation2018; Chu et al. Citation2014, Citation2012; Cong et al. Citation2014; Duan Lei Citation2017; Gao et al. Citation2018; Guo et al. Citation2010; Hishida et al. Citation2011; Huang et al. Citation2013; Jiang et al. Citation2016; Lin et al. Citation2019; Hai-feng et al. Citation2016; Li et al. Citation2015; Lv et al. Citation2013; Ma Citation2012; Ma et al. Citation2013; Mao et al. Citation2014; Min et al. Citation2012; Okubo et al. Citation2010; Parlayan et al. Citation2014; Pu et al. Citation2014; Qu et al. Citation2014; Shen et al. Citation2016; Srivastava et al. Citation2010; Umar et al. Citation2013; Wei et al. Citation2013; Xia et al. Citation2016; Xiang et al. Citation2012; Xu et al. Citation2008; Yan et al. Citation2015; Ying et al. Citation2016; Zeng et al. Citation2010; Zhang et al. Citation2017; Citation2013; Citation2016; Zhou et al. Citation2012a, Citation2012b) and 14 case-control studies on Caucasians (Akkiz et al. Citation2011; Chayeb et al. Citation2018; Dikaiakos et al. Citation2015; Dikeakos et al. Citation2014; Hezova et al. Citation2012; Kupcinskas et al. Citation2014a, Citation2014b; Mihalache et al. Citation2012; Palmieri et al. Citation2014; Pavlakis et al. Citation2013; Rogoveanu et al. Citation2017; Soleimani et al. Citation2016; Yadegari et al. Citation2016). Other characteristics are presented in . The distributions of genotype and allele in miR-146a rs2910164 are listed in . shows the quality assessment of this meta-analysis.
Table 1. Characteristics of the studies in meta-analysis
Table 2. Distribution of miR-146a rs2910164 C > G genotypes and alleles
Table 3. Quality assessment of the meta-analysis
Findings
lists the main findings. The results of heterogeneity tests are also summarized in . Pooling the eligible studies, we found an association of rs2910164 with DSC risk in dominant model (P = .035, ).
Table 4. Results of the meta-analysis from different comparative genetic models
Figure 2. Meta-analysis of the relationship between miR-146a rs2910164 C > G polymorphism and DSC risk (GG/CG vs. CC, random–effects model).
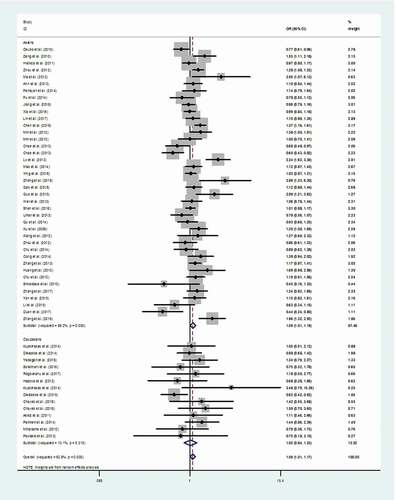
Rs2910164 locus, in Asians subgroup, was correlated with a susceptibility to DSC (GG/CG vs. CC: P = .033). In Caucasians subgroup, this SNP was also associated with a susceptibility to DSC (GG vs. CC/CG: P = .020). Additionally, rs2910164 was suggested to be associated with the occurrence of OSCC (G vs. C: P = .034 and GG/CG vs. CC: P = .022)
Sensitivity analysis
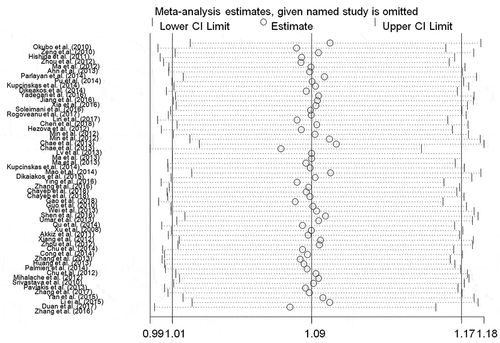
Sensitivity analysis was carried out to determine the influence of each study to the overall ORs and CIs. Our findings revealed that any individual study could not alter the ORs and CIs significantly (). These observations further suggested the correlation between rs2910164 locus and DSC risk.
Publication bias
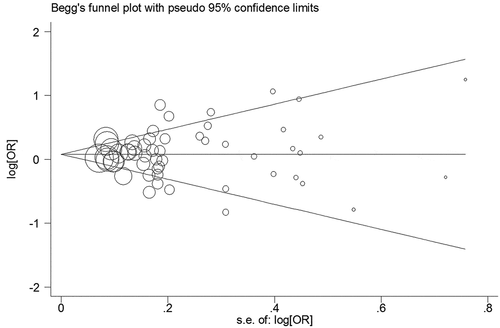
In our study, the bias of publication was assessed by using Bgger’s funnel plots and the Egger’s test. After these evaluations, no significant bias was found among these included studies ().
Heterogeneity
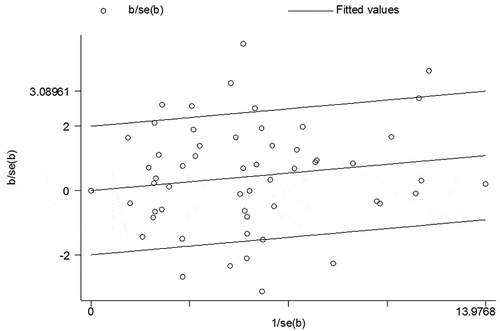
Significant heterogeneity was identified in our study. To identify the major source of heterogeneity, we conducted a heterogeneity analysis by stratified analyses. We suggested a correlation of Asians, GC, CRC, HCC, ESCC, hospital-based study, small sample size (<1000 subjects) and high-quality score (≥7.0) subgroups with significant heterogeneity. In addition, we used Galbraith radial plot to determine the heterogeneity (). Among the eligible studies, eleven outliers (Chae et al. Citation2013; Dikaiakos et al. Citation2015; Duan Lei Citation2017; Guo et al. Citation2010; Kupcinskas et al. Citation2014a; Li et al. Citation2015; Lv et al. Citation2013; Ma Citation2012; Srivastava et al. Citation2010; Zhang et al. Citation2016) were found, which contributed to the main heterogeneity.
The power of the present study (α = 0.05)
For overall comparison, the power value was 0.994 in the dominant model. It was 0.989 in dominant model for Asians. The power value of other subgroups was less than 0.8 (data were not shown).
Discussion
The risk of malignancy may be diverse among different ethnicity. A number of investigations have clarified that miRs may influence the development of DSC. Recently, rs2910164 polymorphism and its importance to the initiate of DSC has been widely explored. In the past years, several pooled-analyses have explored the relationship of rs2910164 locus with the development of DSC. However, the conflicting results have been observed. Thus, an updated meta-analysis should be carried out.
A vital characteristic of this pooled-analysis was that our study included the largest sample sizes to determine a potential relationship between rs2910164 polymorphism and DSC risk comprehensively. Here, this meta-analysis identified that rs2910164 SNP conferred an increased risk to overall DSC and Asian populations. The previous pooled-analyses have been conducted to determine a relationship of rs2910164 variants to the development of DSC. The forepassed meta-analyses showed that G allele in rs2910164 polymorphism might not influence the initiate of DSC (B. Chen et al. Citation2014; Wang et al. Citation2012; Wu et al. Citation2013). Of late, some previous published meta-analyses identified that miR-146a rs2910164 could be implicated in the risk of DSC in Asian populations (Li et al. Citation2014; Xie et al. Citation2015; Xu et al. Citation2014). In addition, another meta-analysis indicated that rs2910164 G allele increased the risk of DSC in both Asians and Caucasians (Xie and Wang Citation2017). However, a more recent meta-analysis reported that the potential relationship disappeared in neither Asians nor Caucasians (Xiong et al. Citation2017). To our knowledge, the associations were more conflicting. Thus, in this meta-analysis, we included more publications with 24,161 DSC patients and 34,937 cancer-free controls to detect the correlation between rs2910164 G allele and its importance to the initiate of DSC. And we found that rs2910164 G allele might confer risk to DSC.
A previous study identified that the expression of miR-146a promoted in acute myeloid leukemia and acute lymphoblastic leukemia cases (Wang et al. Citation2019). As well, Khorrami et al. suggested that the higher level of miR-146a was importance for the milieu of immune suppression and drug-resistant CRC cells (Khorrami et al. Citation2017). Additionally, MiR-146a was found to be correlated with the invasion and migration in CRC patients (Lu et al. Citation2017). Compared with C allele carriers, the level of miR-146a was higher in G allele carriers (Jeon et al. Citation2014; Jia et al. Citation2014; Mohamed et al. Citation2019). Here, we might speculate that the miR-146a C→G mutation could increase the expression of miR-146a and lead to immune suppression. Finally, this SNP could increase the risk of DSC.
In this meta-analysis, we observed significant heterogeneities among the eligible studies. When the source of heterogeneity was analyzed, we found that high-quality score (≥7.0), small sample size (<1000 subjects), CRC, HCC, GC, ESCC, Asians and hospital-based study subgroups increased it greatly. Additionally, in dominant model (), Galbraith radial plot and the forest plot identified eleven outliers (Chae et al. Citation2013; Dikaiakos et al. Citation2015; Duan Lei Citation2017; Guo et al. Citation2010; Kupcinskas et al. Citation2014a; Li et al. Citation2015; Lv et al. Citation2013; Ma Citation2012; Srivastava et al. Citation2010; Zhang et al. Citation2016).
In current meta-analysis, some merits should be considered. Firstly, this is a large sample size study exploring the relationship of rs2910164 locus with the susceptibility of DSC. Secondly, we only included the case-control studies which were consistent with HWE. Our findings were less bias. Thirdly, we evaluated the quality scores of the included studies. Fourthly, no significant bias of publication was found in our analysis. Finally, in this study, the power value was also calculated (α = 0.05).
The potential limitations also should be addressed. Firstly, all of the publications were performed in Caucasians and Asians, and none was conducted in other populations. Thus, our findings might be only adapted to these ethnicities. Secondly, for lack of original information of participants (e.g. family history, smoking, drinking, nutrient intake, gender, age and lifestyle), the influence of these environmental factors was not considered. Thirdly, gene–environment interactions were not performed. Finally, in this study, we only included one miR-SNP. And another SNPs in miR should not been ignored.
In summary, this study identifies that rs2910164 participates in the development of DSC. In stratified analyses, we find that this SNP also significantly increased cancer susceptibility in Asians. More studies with detailed gene-environment factors are, therefore, needed to confirm our results.
Author contribution
All authors contributed significantly to this study.
Conceived and designed the experiments: SZ, ZL
Performed the experiments: LL, HG, ZC
Analyzed the data: WT, SZ
Contributed reagents/materials/analysis tools: ZL
Wrote the manuscript: LL, HG, ZC
Other (please specify): None
Acknowledgments
We wish to thank Dr. Hao Ding (Affiliated People’s Hospital of Jiangsu University, China) for technical support.
Additional information
Funding
References
- Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT, Kwack K, Hong SP, Hwang SG, Kim NK. 2013. Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 52(Suppl 1):E39–51.
- Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O, Sandikci M. 2011. No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene. 486:104–09.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 68:394–424.
- Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ, Park JY, Jeon HS, Park JS, Choi GS. 2013. A miR-146a polymorphism (rs2910164) predicts risk of and survival from colorectal cancer. Anticancer Res. 33:3233–39.
- Chayeb V, Mahjoub S, Zitouni H, Jrah-Harzallah H, Zouari K, Letaief R, Mahjoub T. 2018. Contribution of microRNA-149, microRNA-146a, and microRNA-196a2 SNPs in colorectal cancer risk and clinicopathological features in Tunisia. Gene. 666:100–07.
- Chen BB, Cao XG, Chen XB, Ma YJ, Deng WY, Li N, Huang JX, Luo SX, Zhao EJ. 2014. MicroRNA-146a rs2910164 G/C polymorphism and gastrointestinal cancer susceptibility: a meta-analysis based on East Asian population. J Cancer Res Ther. 10(Suppl):252–55.
- Chen HS, Lu AQ, Yang PY, Liang J, Wei Y, Shang YW, Li Q. 2019. MicroRNA-28-5p regulates glioma cell proliferation, invasion and migration by targeting SphK1. Eur Rev Med Pharmacol Sci. 23:6621–28.
- Chen Y, Tang W, Liu C, Lin J, Wang Y, Zhang S, Chen G, Zheng X. 2018. miRNA-146a rs2910164 C>G polymorphism increased the risk of esophagogastric junction adenocarcinoma: a case-control study involving 2,740 participants. Cancer Manag Res. 10:1657–64.
- Cheng WC, Liao TT, Lin CC, Yuan LE, Lan HY, Lin HH, Teng HW, Chang HC, Lin CH, Yang CY, et al. 2019. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer. 145:2209–24.
- Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, Yang SF, Kuo WH. 2014. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PloS One. 9:e89930.
- Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK, Yang SF. 2012. Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PloS One. 7:e39777.
- Cong N, Chen H, Bu WZ, Li JP, Liu N, Song JL. 2014. miR-146a G>C polymorphisms and risk of hepatocellular carcinoma in a Chinese population. Tumour Biol. 35:5669–73.
- DerSimonian R, Laird N. 1986. Meta-analysis in clinical trials. Control Clin Trials. 7:177–88.
- Dikaiakos P, Gazouli M, Rizos S, Zografos G, Theodoropoulos GE. 2015. Evaluation of genetic variants in miRNAs in patients with colorectal cancer. Cancer Biomarkers. 15:157–62.
- Dikeakos P, Theodoropoulos G, Rizos S, Tzanakis N, Zografos G, Gazouli M. 2014. Association of the miR-146aC>G, miR-149T>C, and miR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Mol Biol Rep. 41:1075–80.
- Duan Lei WW-X. 2017. The association between rs2910164 in miR-146a and hepatocellular carcinoma risk: a case-control study. Biomed Res. 28:5038–40.
- Gao X, Zhu Z, Zhang S. 2018. miR-146a rs2910164 polymorphism and the risk of colorectal cancer in Chinese population. J Cancer Res Ther. 14:S97–S99.
- Guo H, Wang K, Xiong G, Hu H, Wang D, Xu X, Guan X, Yang K, Bai Y. 2010. A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. Fam Cancer. 9:599–603.
- Hai-feng ZH, Shi-yun CU, Yong-qing LI, Ying YA, Wei LA, Nian-sang LU, Jing-feng WA. 2016. Correlation between polymorphism of single nucleotide gene rs2910164 in miR-146a and incidence and metastasis of colorectal cancer. J Shanghai Jiao Tong Univ. 36:809–13.
- Hezova R, Kovarikova A, Bienertova-Vasku J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I, Vyzula R, et al. 2012. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol. 18:2827–31.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. 2003. Measuring inconsistency in meta-analyses. Bmj. 327:557–60.
- Hishida A, Matsuo K, Goto Y, Naito M, Wakai K, Tajima K, Hamajima N. 2011. Combined effect of miR-146a rs2910164 G/C polymorphism and Toll-like receptor 4 +3725 G/C polymorphism on the risk of severe gastric atrophy in Japanese. Dig Dis Sci. 56:1131–37.
- Huang QHT, LI J, Wang C, Wei Z, Li R, Zhang C. 2013. Correlation between nficroRNA-146a polymorphism and primary liver carcinoma in the Guangxi Zhuang population. Chin J of Oncol Prev and Treat. 5:100–04.
- Jeon HS, Lee YH, Lee SY, Jang JA, Choi YY, Yoo SS, Lee WK, Choi JE, Son JW, Kang YM, et al. 2014. A common polymorphism in pre-microRNA-146a is associated with lung cancer risk in a Korean population. Gene. 534:66–71.
- Jia Y, Zang A, Shang Y, Yang H, Song Z, Wang Z, Ren L, Wei Y, Hu L, Shi H, et al. 2014. MicroRNA-146a rs2910164 polymorphism is associated with susceptibility to non-small cell lung cancer in the Chinese population. Med Oncol. 31:194.
- Jiang J, Jia ZF, Cao DH, Wu YH, Sun ZW, Cao XY. 2016. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncol. 12:2215–26.
- Khorrami S, Zavaran Hosseini A, Mowla SJ, Soleimani M, Rakhshani N, Malekzadeh R. 2017. MicroRNA-146a induces immune suppression and drug-resistant colorectal cancer cells. Tumour Biol. 39:1010428317698365.
- Kupcinskas J, Bruzaite I, Juzenas S, Gyvyte U, Jonaitis L, Kiudelis G, Skieceviciene J, Leja M, Pauzas H, Tamelis A, et al. 2014a. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep. 4:5993.
- Kupcinskas J, Wex T, Link A, Leja M, Bruzaite I, Steponaitiene R, Juzenas S, Gyvyte U, Ivanauskas A, Ancans G, et al. 2014b. Gene polymorphisms of micrornas in Helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PloS One. 9:e87467.
- Li X, Li K, Wu Z. 2015. Association of four common SNPs in microRNA polymorphisms with the risk of hepatocellular carcinoma. Int J Clin Exp Pathol. 8:9560–66.
- Li Y, Shi X. 2013. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell Mol Immunol. 10:65–71.
- Li YJ, Zhang ZY, Mao YY, Jin MJ, Jing FY, Ye ZH, Chen K. 2014. A genetic variant in MiR-146a modifies digestive system cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 15:145–50.
- Lin J, Lin W, Lan B, Chen S, Wu W, Ruan J, Liu C, Tang W, Chen Y, Guo Z. 2019. Lack of association between MicroRNA-146a rs2910164 C > G polymorphism and risk of gastric carcinoma: a case-control study and a meta-analysis. Int J Clin Exp Med. 12:3810–20.
- Lu D, Yao Q, Zhan C, Le-Meng Z, Liu H, Cai Y, Tu C, Li X, Zou Y, Zhang S. 2017. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget. 8:22674–84.
- Lv M, Dong W, Li L, Zhang L, Su X, Wang L, Gao L, Zhang L. 2013. Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol. 139:1405–10.
- Ma BZJ. 2012. Investigation of the association between microRNA polymorphism and gastric cancer. Chin J Gerontol. 32:3150–52.
- Ma L, Zhu L, Gu D, Chu H, Tong N, Chen J, Zhang Z, Wang M. 2013. A genetic variant in miR-146a modifies colorectal cancer susceptibility in a Chinese population. Arch Toxicol. 87:825–33.
- Mantel N, Haenszel W. 1959. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 22:719–48.
- Mao Y, Li Y, Jing F, Cai S, Zhang Z, Li Q, Ma X, Wang J, Jin M, Chen K. 2014. Association of a genetic variant in microRNA-146a with risk of colorectal cancer: a population-based case-control study. Tumour Biol. 35:6961–67.
- Mihalache F, Hoblinger A, Acalovschi M, Sauerbruch T, Lammert F, Zimmer V. 2012. A common variant in the precursor miR-146a sequence does not predispose to cholangiocarcinoma in a large European cohort. Hepatobiliary & Pancreatic Diseases International. 11:412–17.
- Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, Oh D, Kim NK. 2012. Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 51(Suppl 1):E65–73.
- Mohamed RH, Pasha HF, Gad DM, Toam MM. 2019. miR-146a and miR-196a-2 genes polymorphisms and its circulating levels in lung cancer patients. J Biochem. 166:323–29.
- O’Neill LA, Sheedy FJ, McCoy CE. 2011. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 11:163–75.
- Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Ishizuka T, Arisawa T, Hirata I. 2010. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 15:524–31.
- Palmieri A, Carinci F, Martinelli M, Pezzetti F, Girardi A, Cura F, Rubini C, Scapoli L. 2014. Role of the MIR146A polymorphism in the origin and progression of oral squamous cell carcinoma. Eur J Oral Sci. 122:198–201.
- Parlayan C, Ikeda S, Sato N, Sawabe M, Muramatsu M, Arai T. 2014. Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pac J Cancer Prev. 15:2101–07.
- Pavlakis E, Papaconstantinou I, Gazouli M, Theodosopoulos T, Karamanolis G, Genatas K, Ladas SD. 2013. MicroRNA gene polymorphisms in pancreatic cancer. Pancreatology. 13:273–78.
- Pu JY, Dong W, Zhang L, Liang WB, Yang Y, Lv ML. 2014. No association between single nucleotide polymorphisms in pre-mirnas and the risk of gastric cancer in Chinese population. Iran J Basic Med Sci. 17:128–33.
- Qu Y, Qu H, Luo M, Wang P, Song C, Wang K, Zhang J, Dai L. 2014. MicroRNAs related polymorphisms and genetic susceptibility to esophageal squamous cell carcinoma. Mol Genet Genom. 289:1123–30.
- Rogoveanu I, Burada F, Cucu MG, Vere CC, Ioana M, Cimpeanu RA. 2017. Association of microRNA polymorphisms with the risk of gastric cancer in a romanian population. J Gastrointestinal Liver Dis. 26:231–38.
- Saba R, Sorensen DL, Booth SA. 2014. MicroRNA-146a: a dominant, negative regulator of the innate immune response. Front Immunol. 5:578.
- Shen F, Chen J, Guo S, Zhou Y, Zheng Y, Yang Y, Zhang J, Wang X, Wang C, Zhao D, et al. 2016. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol. 37:4777–84.
- Soleimani A, Ghanadi K, Noormohammadi Z, Irani S. 2016. The correlation between miR-146a C/G polymorphism and UHRF1 gene expression level in gastric tumor. J Dig Dis. 17:169–74.
- Srivastava K, Srivastava A, Mittal B. 2010. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 55:495–99.
- Tang W, Qiu H, Ding H, Sun B, Wang L, Yin J, Gu H. 2013. Association between the STK15 F31I polymorphism and cancer susceptibility: a meta-analysis involving 43,626 subjects. PloS One. 8:e82790.
- Umar M, Upadhyay R, Prakash G, Kumar S, Ghoshal UC, Mittal B. 2013. Evaluation of common genetic variants in pre-microRNA in susceptibility and prognosis of esophageal cancer. Mol Carcinog. 52(Suppl 1):E10–18.
- Wang F, Sun G, Zou Y, Fan L, Song B. 2012. Lack of association of miR-146a rs2910164 polymorphism with gastrointestinal cancers: evidence from 10206 subjects. PloS One. 7:e39623.
- Wang L, Zhang H, Lei D. 2019. microRNA-146a promotes growth of acute leukemia cells by downregulating ciliary neurotrophic factor receptor and activating JAK2/STAT3 signaling. Yonsei Med J. 60:924–34.
- Wang W, Shao Y, Tang S, Cheng X, Lian H, Qin C. 2015. Peroxisome proliferator-activated receptor-gamma (PPARgamma) Pro12Ala polymorphism and colorectal cancer (CRC) risk. Int J Clin Exp Med. 8:4066–72.
- Wei J, Zheng L, Liu S, Yin J, Wang L, Wang X, Shi Y, Shao A, Tang W, Ding G, et al. 2013. MiR-196a2 rs11614913 T > C polymorphism and risk of esophageal cancer in a Chinese population. Hum Immunol. 74:1199–205.
- Wu D, Wang F, Dai WQ, He L, Lu J, Xu L, Guo CY. 2013. The miR-146a rs2910164 G > C polymorphism and susceptibility to digestive cancer in Chinese. Asian Pac J Cancer Prev. 14:399–403.
- Xia ZG, Yin HF, Long Y, Cheng L, Yu LJ, Guo WJ, Zhu XD, Li J, Wang YN, Yang YJ, et al. 2016. Genetic variant of miR-146a rs2910164 C>G and gastric cancer susceptibility. Oncotarget. 7:34316–21.
- Xiang Y, Fan S, Cao J, Huang S, Zhang LP. 2012. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 39:7019–23.
- Xie M, Li Y, Wu J, Wu J. 2015. A risk of digestive tract neoplasms susceptibility in miR-146a and miR-196a2. Fam Cancer. 14:229–39.
- Xie WQ, Wang XF. 2017. MiR-146a rs2910164 polymorphism increases the risk of digestive system cancer: a meta-analysis. Clin Res Hepatol Gastroenterol. 41:93–102.
- Xiong X, Yan J, Li L, Li Y, Cao Y, Tu Y, Mei J. 2017. Relationship between miR-146a rs2910164 (G>C) polymorphism and digestive system cancer susceptibility: a meta-analysis. Ann Clin Lab Sci. 47:491–500.
- Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM. 2008. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 29:2126–31.
- Xu X, Yang X, Ru G, Wu Y, Zhang S, Xing C, Wu Y, Cao J. 2014. miR-146a gene polymorphism rs2910164 and the risk of digestive tumors: a meta-analysis of 21 case-control studies. Oncol Rep. 31:472–79.
- Yadegari ZS, Akrami H, Hosseini SV, Erfani N. 2016. miR-146a gene polymorphism and susceptibility to gastric cancer. Br J Biomed Sci. 73:201–03.
- Yan P, Xia M, Gao F, Tang G, Zeng H, Yang S, Zhou H, Ding D, Gong L. 2015. Predictive role of miR-146a rs2910164 (C>G), miR-149 rs2292832 (T>C), miR-196a2 rs11614913 (T>C) and miR-499 rs3746444 (T>C) in the development of hepatocellular carcinoma. Int J Clin Exp Pathol. 8:15177–83.
- Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu J, Sun Z, Shen WF. 2011. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 585:854–60.
- Yang Y, Dodbele S, Park T, Glass R, Bhat K, Sulman EP, Zhang Y, Abounader R. 2019. MicroRNA-29a inhibits glioblastoma stem cells and tumor growth by regulating the PDGF pathway. J Neurooncol. 145:23–34.
- Ying HQ, Peng HX, He BS, Pan YQ, Wang F, Sun HL, Liu X, Chen J, Lin K, Wang SK. 2016. MiR-608, pre-miR-124-1 and pre-miR26a-1 polymorphisms modify susceptibility and recurrence-free survival in surgically resected CRC individuals. Oncotarget. 7:75865–73.
- Zeng Y, Sun QM, Liu NN, Dong GH, Chen J, Yang L, Wang B. 2010. Correlation between pre-miR-146a C/G polymorphism and gastric cancer risk in Chinese population. World J Gastroenterol. 16:3578–83.
- Zhang E, Xu Z, Duan W, Huang S, Lu L. 2017. Association between polymorphisms in pre-miRNA genes and risk of oral squamous cell cancer in a Chinese population. PloS One. 12:e0176044.
- Zhang J, Gong WH, Li Y, Zhang HY, Zhang CX. 2019a. Hsa-miR-337 inhibits non-small cell lung cancer cell invasion and migration by targeting TCF7. Eur Rev Med Pharmacol Sci. 23:6548–53.
- Zhang J, Wang R, Ma YY, Chen LQ, Jin BH, Yu H, Wang JC, Gao CF, Liu J. 2013. Association between single nucleotide polymorphisms in miRNA196a-2 and miRNA146a and susceptibility to hepatocellular carcinoma in a Chinese population. Asian Pac J Cancer Prev. 14:6427–31.
- Zhang LH, Hao BB, Zhang CY, Dai XZ, Zhang F. 2016. Contributions of polymorphisms in miR146a, miR196a, and miR499 to the development of hepatocellular carcinoma. Gene Molres.
- Zhang W, Xiao J, Lu X, Liu T, Jin X, Xiao Y, He X. 2019b. PVT1 (rs13281615) and miR-146a (rs2910164) polymorphisms affect the prognosis of colon cancer by regulating COX2 expression and cell apoptosis. J Cell Physiol. 234:17538–48.
- Zhou F, Zhu H, Luo D, Wang M, Dong X, Hong Y, Lu B, Zhou Y, Zhou J, Zhang Z, et al. 2012a. A functional polymorphism in Pre-miR-146a is associated with susceptibility to gastric cancer in a Chinese population. DNA Cell Biol. 31:1290–95.
- Zhou J, Lv R, Song X, Li D, Hu X, Ying B, Wei Y, Wang L. 2012b. Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol. 31:524–30.