Abstract
The article presents an experimental study on multiclass Support Vector Machine (SVM) methods over a cardiac arrhythmia dataset that has missing attribute values for electrocardiogram (ECG) diagnostic application. The presence of an incomplete dataset and high data dimensionality can affect the performance of classifiers. Imputation of missing data and discriminant analysis are commonly used as preprocessing techniques in such large datasets. The article proposes experiments to evaluate performance of One-Against-All (OAA) and One-Against-One (OAO) approaches in kernel multiclass SVM for a heartbeat classification problem with imputation and dimension reduction techniques. The results indicate that the OAA approach has superiority over OAO in multiclass SVM for ECG data analysis with missing values.
INTRODUCTION
The electrocardiogram (ECG) is a noninvasive diagnostic method that is an effective, simple, low-cost procedure to determine the conditions of electrical activities of the heart (Liang, Lukkarinen, and Hartimo Citation1997; Polat, Akdemir, and Güneş Citation2008) and cardiovascular system by measuring rate and regularity of heartbeats. The ECG technique can be applied in healthcare systems as well as in medical biometric modality (Agrafioti, Gao, and Hatzinakos Citation2011) for security systems. An ECG is a valuable information source of the heart rhythm functioning and can be used to diagnose precise evidence of specific heart disturbances and arrhythmias (Polat, Akdemir, and Güneş Citation2008) in healthcare systems. Developing methods for automatic ECG heartbeat classification in healthcare systems can assist to improve early detection and monitoring of heart disease disorders in the initial phase.
The clinical data analysis systems deal with significant problems that can adversely affect medical diagnoses and exact predictive treatment. Missing values and high-dimensional datasets occur frequently in applied statistical data analysis, including in healthcare systems. A number of relevant specific techniques have been investigated in order to determine missing values in incomplete data, redundancy, and irrelevant features with minimal loss of information (Hlalele, Nelwamondo, and Marwala Citation2009).
When observed data on an attribute is not present in a dataset, the dataset encounters an incomplete or missing data problem. The nine different handling methods to solve missing attributes values are described and compared by Grzymala-Busse et al. (Citation2001). Imputation methods are a common technique for handling missing data, which can be helpful for precision and controlling bias (Pelckmans et al. Citation2005). On the other hand, insignificant attributes, which reduce efficiency of learning statistical algorithms are a general problem in huge datasets. Dimension reduction is a process that reduces dimension of input data vectors to reduced dimensionality by linear combinations of original features. Dimensionality reduction is essential to facilitate efficient data compression onto proper dimension, data visualization, and classification (Van Der Maaten, Postma, and Van Den Herik Citation2008). Discriminant analysis functions have been introduced as statistical analysis methods that can be applied efficiently to learn a multiple continuous dependent attribute by predictor attributes for dimensional reduction and pattern recognition.
The presence of missing values and high-dimensional data in statistical analysis can affect computational time and accuracy of supervised and unsupervised classification methods significantly. Modeling in the presence of missing and insignificant attributes can be done efficiently by kernel machines (Vapnik, Golowich, and Smola Citation1997). Support Vector Machines (SVMs) can be employed as an optimal and robust kernel classification method based on statistical learning theory to classify observations when no input data for each variable are missing and they have significant features.
This article presents an experimental study to design an accurate medical diagnostic system for diagnosis and classification of cardiac data using multiclass SVM classification with kernel methods where a dataset has missing values and is of high data dimensionality. The heartbeats dataset from UCI repository (Bache and Lichman Citation2013) consists of ECG samples having missing attributes instances with three classes: normal, arrhythmia, and unclassified heartbeat signal recordings. The proposed methods consistof multiclass support vector machines and data imputing techniques to construct a predictive model from the ECG dataset in order to classify a subject into the determined classes.
The organization of the article is given as follows. The next section reviews Support Vector Machine (SVM) and its variant C-SVC and SVC binary classification methods. “Preprocessing Techniques” gives a brief overview of relevant techniques required to deal with missing values and high-dimensional data in the ECG dataset. In “Experiments,” the experiments for the ECG multiclass classification problem using kernel SVM classifiers in OAA and OAO approaches are described. “Conclusions” are given in the final section.
SUPPORT VECTOR MACHINES
Support vector machine is a very robust nonparametric classification algorithm based on statistical learning theory in machine learning for solving classification and regression problems (Vapnik, Golowich, and Smola Citation1997). The fundamental concepts and theories of SVM have been introduced by Vapnik (Citation1995) as a binary classification predictive technique to attain a predictive accuracy with reduced generalization error. The SVM methods for binary classification problems are based on constructing a separating hyperplane with maximum margin between two classes in data space or feature space for linear and nonlinear SVMs.
For a binary classification problem, we assume that we have training data points (,
), i = 1, …, N,
where
, and
{+1,−1} is the class label. shows a schematic of a linear SVM having two separable classes such as {+1, −1}. Let the training data consist of N data samples (
,
), …, (
,
),
, and
{+1,−1}. To separate these two classes, SVMs determine the optimal separating hyperplane having good generalization ability. The SVM design is based on a construction of an optimal separating hyperplane (maximum margin) for a dataset as a decision boundary among several possible hyperplanes in input space or feature space for the binary classification problem, as shown in (Tan, Steinbach, and Kumar Citation2006). The optimal hyperplane is designed with the maximal margin from the linear separator to the nearest positive and negative data points (Tan, Steinbach, and Kumar Citation2006; Boser, Guyon, and Vapnik Citation1992). Each decision boundary is associated with a pair of hyperplanes.
FIGURE 1 A schematic of linear SVM classification (Tan, Steinbach, and Kumar Citation2006).
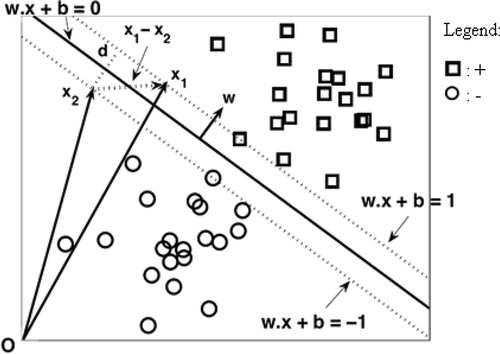
There is a trade-off between the size of the margin and the number of training errors in SVM methods for the linear decision boundary. In other words, constructing a decision boundary with maximal margin minimizes the generalization errors and performs well on the testing instances (Tan, Steinbach, and Kumar Citation2006). A linear decision boundary classifier with margin d in the two-dimensional training set is shown in and is defined as:
The decision boundary is related to two separating hyperplanes: w. x + b = +1 and w. x + b = −1 > to classify two classes (positive and negative). A weight vector w is of dimension n and b is a bias scalar term in Equation (1) as explained in .
Nonlinear classification occurs if the training samples cannot be separated by a decision function linearly in n-dimension input space with less susceptibility to overfitting. Nonlinear SVM design is done using kernel methods for training instances that have a nonlinear decision boundary in input space. The learning of the nonlinear decision boundary is done by mapping observation instances (data samples) from input space with n-dimension to an l-dimensional feature space (higher dimensional space) by kernel functions to construct optimal separating linear function for points in the feature space as follows (Tan, Steinbach, and Kumar Citation2006; Schölkopf et al. Citation2002; Abe Citation2010):
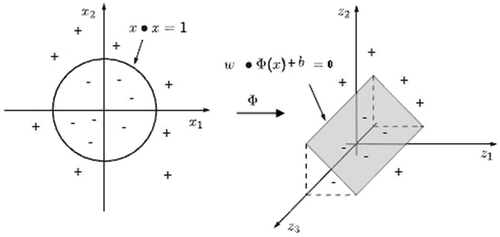
Here, denotes the inner product of two vectors. (Hamel Citation2009) represents this transformation.
FIGURE 2 A schematic of nonlinear SVM classification (Hamel Citation2009).
For the symmetric function satisfying the following Hilbert–Schmidt theorem for any input dataset
(input space) (Abe Citation2010),
There exists a transformation function, , that maps any input samples
in input space into feature space and
satisties the following condition (Abe Citation2010):
If the Equation (4) is satisfied, then
Equation (3) or (5) is known as Mercer’s condition, and the function used in Equation (3) as Mercer kernel (Abe Citation2010).
The kernel method is described as the kernel trick that applies in classification and training instead of evaluation of the transformation
.
One of the advantages SVM is to enhance generalization performance and reduction of overfitting by convenient selection of kernels, so choosing kernels for a particular application is more essential. The common kernels that are employed in SVM are as follows(Schölkopf and Smola Citation2002; Vaerenbergh Citation2009):
Linear kernel:
Polynomial kernels:
Radial basis function kernels (RBF). Boser, Guyon, and Vapnik (Citation1992) define Gaussian basis function kernels as the following formulation:
where
is a positive parameter for controlling the radius (Abe Citation2010). The RBF kernel is defined through the Gaussian kernel if
=1 as
Hyperbolic tangent kernel:
The polynomal kernel enables data to be mapped into a finite dimension in order to construct a linear classifier. The RBF kernels transform the data into infinite dimension space where the data can separate linearly. The RBF kernel has advantages over other kernels with respect to performance (Hejazi and Singh Citation2012; Hejazi and Singh Citation2013). The RBF kernel method will be employed with nonlinear multiclass SVM for ECG signal diagnosis in the experimental results section.
C-Support Vector Binary Classification
C-Support vector classification is known as the C-SVC binary classification method and is formulated as the primal constrained optimization problem. C-SVC has been proposed by Boser et al. (Citation1992) and Vapnik and Cortes (Citation1995) (Chang and Lin Citation2011). The constrained primal optimization problem for C-SVC is described as , i = 1, 2, …, N and
as
The decision function can be derived from the dual optimization problem as follows:
The threshold can be obtained using the following equation:
The goal of the classification problem is the choice of an optimal separable hyperplane, also known as the decision boundary through Equations (8) and (9).
v-Support Vector Binary Classification
A binary classification technique is -Support Vector Classification, known as
-SVC. It has been proposed by Schölkopf et al. in 2000 (Chang and Lin Citation2011). C in C-SVC is a constant parameter and there is no a priori technique to choose it, so a parameter
instead of parameter C is considered to control the number of margin errors and support vectors (SVs). Parameter
is an upper bound on the fraction of margin errors, and a lower bound on the fraction of SVs (Schölkopf and Smola Citation2002; Chang and Lin Citation2011). The primal objective function in Equation (3) is modified with term
as the margin parameter in the following way:
If , then the margin between two classes is
, but if variable
, then the margin error applies to samples with training errors with a condition (
) or resides within the margin if (
; Schölkopf and Smola Citation2002). The decision boundary can be derived as
The bias term b and margin parameter are calculated by using two sets
of identical size of s (s
), which consist of support vectors
with respect to
and
:
Only b is required for a decision boundary in Equation (11).
Multiclass Classification
Many real-world problems occur wherein the input samples are distributed to more than two classes as a set of (Tan, Steinbach, and Kumar Citation2006). Multiclass classification is a supervised learning problem that has been proposed by effective extension of binary classification (Hsu and Lin Citation2002). The multiclass problems are more complex and more computationally expensive than binary classification problems due to combinations and ties among the different classes (Hsu and Lin Citation2002). Therefore, there are different approaches for decomposing the multiclass problem into several binary problems. There are several methods to achieve decision functions for multiclass SVMs, such as One-Against-All (OAA), One-Against-One (OAO) (Tan, Steinbach, and Kumar Citation2006; Weston and Watkins Citation1998). The present work considers only OAO and OAA methods for ECG data analysis.
The multiclass SVM design is to train decision functions for N samples, typically with noise through decomposition of data points to several binary problems: , where
Rn,
and
represents the class label of the samples. Several approaches have been developed to handle multiclass SVM problems.
The OAA approach, according to Vapnik’s formulation (Vapnik Citation2000), decomposes a multiclass problem (k class labels) into k binary class problems. For any class , a binary problem is constructed in which all data points that belong to
are labeled as positive samples, whereas the remaining instances of other classes are known as negative samples (Tan, Steinbach, and Kumar Citation2006; Abe Citation2010).
The OAO is another approach that is employed to solve multiclass problems. The OAO converts multiclass problems (k class labels) into binary classifiers where every binary classifier is constructed to distinguish between a pair of classes,
(Tan, Steinbach, and Kumar Citation2006; Abe Citation2010; Chen, Lin, and Schölkopf Citation2005). Samples that do not belong to either
or
are thrown out while training the binary classifier for
(Tan, Steinbach, and Kumar Citation2006). There exist two strategies to implement the OAO approach (Chen, Lin, and Schölkopf Citation2005). The first strategy is that the parameter selection is conducted for any binary classes to have the best parameters. The parameter selection of any binary SVM method is done by cross-validation. But the second way defines the identical parameters for all
classifiers.
Test data are classified by combining predictions constructed by the binary classifiers in both OAA and OAO approaches. A voting strategy usually applies to combine the predictions, and the class that receives the maximum number of votes is dedicated to the test data (Tan, Steinbach, and Kumar Citation2006; Chen, Lin, and Schölkopf Citation2005). Another possible scheme is to transform the binary classifiers’ output into probability estimates and then assign the test samples to the class with the highest probability (Tan, Steinbach, and Kumar Citation2006).
The binary classifiers of C-SVC, -SVC with RBF kernel are employed in our experimental results for OAA and OAO approaches to solve multiclass ECG diagnostic problems. The selection of appropriate C,
and kernel parameter (
) are determined by conducting experiments to achieve higher performance.
PREPROCESSING TECHNIQUES
Datasets consisting of missing attribute values and high data dimension require preprocessing to have desirable samples and significant features before assigning input instances for training to classification methods.
Missing values attributes in real-world datasets are generally encountered in signal processing, statistics, and machine learning methods (Anderson and Gupta Citation2011) in healthcare systems (Pelckmans et al. Citation2005). A dataset has missing attribute values if no data value is stored for the attributes in any observation. Datasets with missing features’ values can result in bias and can affect the accuracy of classification methods and leads to overfitting (Zhu et al. Citation2011). There are various techniques used to handle missing values in order to have better performance of test data in a supervised classification.
A typical method to handle missing values is to ignore completely the observations that at least one of the attribute’s values is missing. The approach has less risk whenever a dataset has a sufficient large number of samples and deals with minimal loss of instances without any structural change in missing data. Otherwise, ignoring missing data leads to making more biased estimates due to loss of too much significant information that may be valuable (Stanimirova, Daszykowski, and Walczak Citation2007). The imputation method can be an effective technique to reduce and control bias for handling missing values without discarding incomplete observations. Imputation methods replace missing attribute values in a dataset with statistical calculation of all values of complete samples (Pelckmans et al. Citation2005) of a class like mean and median imputation. The mean imputation deals with some drawbacks as negative bias when the dataset is skewed, so median imputation provides robustness (Acuña and Rodriguez Citation2004). Nearest neighbor imputation is also a missing value technique that finds missing data of a sample imputed by a number of samples that have similarity to the sample of interest (Karhunen Citation2011). The similarity of two samples is specified by distance methods such as Manhattan, Euclidean, Pearson, and so on.
When a dataset has a large number of attributes for each sample, it is useful to detect a set of linear combinations of attributes containing some properties in correlation, covariance, or variance. Dimension reduction is a well-known preprocessing technique that reduces data dimensionality through a transformation of observations from the original d-dimensional space to a k-dimensional subspace (lower dimensional space) with a minimum loss of information. Dimension reduction and feature selection enhances the performance of classifiers (Kim, Street, and Menczer Citation2002) and overcomes generally overfitting the problem when observations are complete.
Principal component analysis (PCA) is a classical data analysis method for linear dimension reduction and feature selection. PCA is usually used to analyze data based on a set of principal components as eigenvectors to delete redundancy in input data by distinguishing correlations between instances (Jolliffe Citation2002; Hlalele, Nelwamondo, and Marwala Citation2009). Each principal component gives different information about the data variability. PCA in the presence of missing values leads to a difficulty in estimation of bias terms and presenting covariance matrix of input data for eigenvectors. This study considers the effectiveness of imputation along with PCA techniques in multiclass SVM classification on an ECG dataset having missing data and high data dimensionality.
EXPERIMENTS
The choice of the kernels has serious effects on the performance (Müller et al. Citation2001) of SVM classification. Normally, kernel choice is made for adequate and required performance by conducting experiments.
In this section, we evaluate a performance comparison of multiclass SVM classification on both OAO and OAA approaches with the RBF kernel method using the Library of Support Vector Machines known as LIBSVM (Chang and Lin Citation2011). We present the experimental results on a cardiac arrhythmia dataset that includes missing attribute values, from the UCI repository (Bache and Lichman Citation2013). The preprocessing techniques are carried out on the cardiac arrhythmia dataset in order to achieve a dataset without any missing values and errors. Mean and median imputation and PCA functions are performed for preprocessing the given dataset before consideration of multiclass SVM classification. It can also compare the effect of different imputation and discriminant analysis functions in performance of multiclass SVMs in both OAA and OAO approaches.
The cardiac arrhythmia dataset consists of three classes with 279 attributes and containing 384 missing attributes samples and 68 complete observations. The number of samples of the normal class in the given dataset is 452, and arrhythmia cardiac samples are 185; the rest are unclassified. Five features have missing values in the dataset and they are P, heart rate, QRST, T, and J. Each heartbeat makes a different deflection on the ECG pulse as series of positive and negative waves consist of points P, QRS complex and T. The J point is the junction between the termination of the QRS complex and the beginning of the ST segment.
Mean imputation (MI) and median imputation (MDI) are named as new datasets, which have been imputed with samples by mean and median with no data dimensionality reduction. Performing PCA on the MI and MDI datasets leads to making new datasets MIPCA and MDIPCA with subspace of attributes. The PCA algorithm on the input data of datasets has variance percentages of 95% in principal components. Each new dataset has 452 samples, which are split; 317 are training instances and others are test samples. The number of training and testing samples and features for all datasets (MI, MIPCA, MDI, MDIPCA) are shown as #Tr, #Te and #Fe in the tables.
The RBF kernel parameter, and the complexity C (regularization parameter) and
are set for performance evaluation of multiclass C-SVC and
-SVC classifiers in both OAA and OAO approaches. The performance depends on the range of value of the parameters that is chosen by conducting experimental study. The value of
of the RBF kernel method is set to 0.1, and the suitable values of C and quantile parameter
are 10 and 0.01.
Performance of Multiclass Classifiers using OAA Approach
This section considers the performance comparison of multiclass C-SVC, -SVC classification for OAA strategy on each of the datasets MI, MIPCA, MDI, and MDIPCA.
It can be seen from that median imputation is able to impute better than mean technique because of increasing accuracy of the training data and less generalization error in test samples as well as control bias of the datasets.
TABLE 1 A Performance Comparison of Multiclass Classifiers with OAA Approach
The results obtained indicate that a combination of PCA with mean and median imputation techniques does not yield an optimal model to do preprocessing of missing values and errors in ECG data analysis. The multiclass SVM classifiers in the model have poor classification performance in test data and more overfitting. Selection of relevant and significant attributes and determining how to encode features for the classifier can have a large influence on the classifier’s performance to extract a good model.
The simulation results also demonstrate that multiclass C-SVC classification is more accurate than multiclass -SVC classifier in the OAA approach. Although performance on training data for
-SVC is better than for C-SVC, the C-SVC classifier results in fewer misclassification and generalization errors in the test data.
Performance of Multiclass Classifiers Using OAO Approach
The section presents the performance comparison of multiclass classification C-SVC and -SVC in the OAO approach for the ECG classification problem on datasets MI, MIPCA, MDI, and MDIPCA.
The experimental results given in and show the effect of missing values imputation techniques and their combination with PCA on the accuracy of multiclass SVM classifiers. The classification performance of the MDI dataset is better than that of the other datasets in both OAA and OAO approaches, despite having more features. The performance of the MDI dataset is higher because of reduced training errors and cost-sensitive misclassification in training samples of the dataset. However, test accuracy of the MDI dataset increases according to reduction of the false negative (FN) and false positive (FP) instances (misclassification data) according to the predictive accuracy formula of . Therefore, overfitting is less due to reducing both training and testing errors in samples of the MDI dataset.
TABLE 2 A Performance Comparison of Multiclass Classifiers with OAO Approach
It can be seen from and that dimension reduction with PCA cannot be effective in the classification performance of multiclass SVM classifiers when missing data are imputed with mean and median. A sufficient number of significant attributes and missing attribute values, and deciding how to estimate PCA covariance matrix for multiclass SVM classifiers, can affect bias of the term and reducing performance of SVM classifiers in MIPCA and MIDPCA datasets. So, a combination of PCA with mean and median imputation techniques is not an optimal preprocessing model for a multiclass ECG problem performed with kernel SVM classifiers.
The simulation results also show that the best result is for the multiclass C-SVC classifier because of a low number of generalization errors in test samples as well as lower error in training data of all datasets. In other words, the multiclass -SVC classifier cannot be convenient for ECG data analysis for both OAA and OAO approaches because of higher ovefitting than the C-SVC classification method in all datasets.
We can conclude from the results shown in and that the OAA approach is better than the OAO strategy in multiclass SVM classification for the ECG classification problem. According to experimental results, reducing the generalization error of test data leads to less overfitting in all datasets. This is the reasons for the superiority of the OAA approach in ECG diagnosis application. The experimental results obtained from multiclass SVM classifiers indicate that OAO has a more complex algorithm ( than the OAA approach, but computing time for the training data in multiclass SVM classifiers in the OAO approach is less than it is in the OAA approach.
CONCLUSIONS
This study has investigated a multiclass problem with support vector machine methods based on an RBF kernel method for both One-Against-All and One-Against-One approaches on a cardiac arrhythmia dataset containing missing attributes values. The work has considered the effect of preprocessing techniques of mean and median imputation and PCA for handling missing data and dimensionality reduction of imputed data in classification performance. The simulation results demonstrate superiority of OAA for ECG classification over the OAO approach. Experimental results illustrate the suitability of SVM methods for ECG data analysis for diagnostic applications because of their generalization capability. This capability generally provides them with higher classification accuracies and a lower sensitivity to noisy datasets.
REFERENCES
- Abe, S. 2010. Support vector machines for pattern classification ( 2nd ed.). London, UK: Springer-Verlag London Limited.
- Acuña, E., and C. Rodriguez 2004. The treatment of missing values and its effect in the classifier accuracy. In Classification, clustering and data mining applications, eds. D. Banks et al., 639–48. Berlin, Heidelberg: Springer-Verlag.
- Agrafioti, F., J. Gao, and D. Hatzinakos 2011. Heart biometrics: Theory, methods and applications. In Biometrics, ed. J. Yang 199–216. InTech. doi: 10.5772/18113
- Anderson, H. S., and M. R. Gupta. 2011. Expected kernel for missing features in support vector machines. In Statistical signal processing workshop (SSP), 2011 IEEE, 285–88. IEEE Conference Publications.
- Bache, K., and K. Lichman 2013. UCI machine learning repository: Data sets. Irvine CA: University of California, School of Information and Computer Science. http://archive.ics.uci.edu/ml (accessed June 1, 2013).
- Boser, B. E., I. M. Guyon, and V. N. Vapnik 1992. A training algorithm for optimal margin classifiers. In Proceedings of the 5th conference on computational learning theory (CoLT 1992), 144–52. New York, NY: ACM. doi:10.1145/130385.130401.
- Chang, C., and C. Lin 2011. LIBSVM: A library for support vector machines. Taipie, Taiwan: National Taiwan University. Initial version 2001 Last updated: May 20.
- Chen, P.-H., C.-J. Lin, and B. Schölkopf. 2005. A tutorial on?-Support vector machines. Applied Stochastic Models in Business and Industry 21: 111–36. doi:10.1002/asmb.537.
- Grzymala-Busse, J. W., and M. Hu. 2001. Comparison of several approaches to missing attribute values in data mining. Lecture Notes in Computer Science Volume 2005 378–85. doi: 10.1007/3-540-45554-X_46.
- Hamel, L. 2009. Knowledge discovery with support vector machines. Hoboken, NJ: Wiley & Sons.
- Hejazi, M., and Y. P. Singh. 2012. Credit data fraud detection using kernel methods with support vector machine. Journal of Advanced Computer Science and Technology Research 2: 35–49.
- Hejazi, M., and Y. P. Singh. 2013. One-class support vector machines approach to anomaly detection. Applied Artificial Intelligence: An International Journal 27: 351–66.
- Hlalele, N., F. Nelwamondo, and T. Marwala. 2009. Imputation of missing data using PCA, neuro-fuzzy and genetic algorithms. In Advances in neuro-information processing, LNCS 5507:485–92. Berlin, Heidelberg: Springer-Verlag.
- Hsu, C.-W., and C.-J. Lin. 2002. A comparison of methods for multi-class support vector machines. IEEE Transactions on Neural Networks 13(2): 415–25. doi: 10.1109/72.991427.
- Jolliffe, I. T. 2002. Principal component analysis (2nd ed.). New York, NY: Springer.
- Karhunen, J. 2011. Robust PCA methods for complete and missing data. Finland: University School of Science, Department of Information and Computer Science.
- Kim, Y. S., W. N. Street, and F. Menczer 2002. Feature selection in data mining. In Data mining: Opportunities and challenges, eds. J. Wang, 80–105. Hershey, PA: Idea Group.
- Liang, H., S. Lukkarinen, and I. Hartimo. 1997. Heart sound segmentation algorithm based on heart sound envelogram. In Computers in cardiology, 105–108. IEEE. doi: 10.1109/CIC.1997.647841.
- Muller, K.-R., S. Mika, G. Ratsch, K. Tsuda, and B. Scholkopf. 2001. An introduction to kernel-based learning algorithms. IEEE Transactions on Neural Networks 12(2):181–201. doi: 10.1109/72.914517.
- Pelckmans, K., J. De Brabanter, J. A. K. Suykens, and B. De Moor. 2005. Handling missing values in support vector machine classifiers. Neural Networks 18(5–6):684–92. doi: 10.1016/j.neunet.2005.06.025.
- Polat, K., B. Akdemir, and S. Güneş. 2008. Computer aided diagnosis of ECG data on the least square support vector machine. Digital Signal Processing 18:25–32. doi: 10.1016/j.dsp.2007.05.006.
- Schölkopf, B., and A. Smola 2002. Learning with kernels support vector machines, regularization, optimization, and beyond. Cambridge, MA: MIT Press.
- Stanimirova, I., M. Daszykowski, and B. Walczak. 2007. Dealing with missing values and outliers in principal component analysis. Talanta 72: 172–78. doi:10.1016/j.talanta.2006.10.011.
- Tan, P. N., M. Steinbach, and V. Kumar. 2006. Introduction to data mining. Boston, MA: Pearson Addison-Wesley.
- Vaerenbergh, S. V. 2009. Kernel methods for nonlinear identification equalization and separation of signals (PhD thesis, Cantabria University).
- Van Der Maaten, L. J. P., E. O. Postma, and H. J. Van Den Herik. 2008. Dimensionality reduction: A comparative review. Maastricht, LK: Maastricht University.
- Vapnik, V. N. 1995. The nature of statistical learning theory. New York, NY: Springer-Verlag.
- Vapnik, V. N. 2000. The nature of statistical learning theory (2nd ed.). New York, NY: Springer-Verlag.
- Vapnik, V. N., S. E. Golowich, and A. Smola 1997. Support vector method for function approximation, regression estimation, and signal processing. In Advances in neural information processing systems, vol. 9, eds. M. Mozer, M. Jordan, and T. Petsche, 281–87. Cambridge, MA: MIT Press. doi:10.1.1.41.3139.
- Weston, J., and C. Watkins 1998. Multi-class support vector machines. In Proceedings of the 6th European symposium on artificial neural networks, 259–66. Bruges, Belgium: ESANN. doi:10.1.1.50.9594.
- Zhu, X., S. Zhang, Z. Jin, Z. Zhang, and Z. Xu. 2011. Missing value estimation for mixed-attribute data sets. IEEE Transactions on Knowledge and Data Engineering 23(1):110–21. doi: 10.1109/TKDE.2010.99.
