ABSTRACT
A new architecture of deep neural networks, directed acyclic graph convolutional neural networks (DAG-CNNs), is used to classify heartbeats from electrocardiogram (ECG) signals into different subject-based classes. DAG-CNNs not only fuse the feature extraction and classification stages of the ECG classification into a single automated learning procedure, but also utilized multi-scale features and perform score-level fusion of multiple classifiers automatically. Therefore, DAG-CNN negates the necessity to extract hand-crafted features. In most of the current approaches, only the high level features which extracted by the last layer of CNN are used. Instead of performing feature level fusion manually and feeding the results into a classifier, the proposed multi-scale system can automatically learn different level of features, combine them and predict the output label. The results over the MIT-BIH arrhythmia benchmarks database demonstrate that the proposed system achieves a superior classification performance compared to most of the state-of-the-art methods.
Introduction
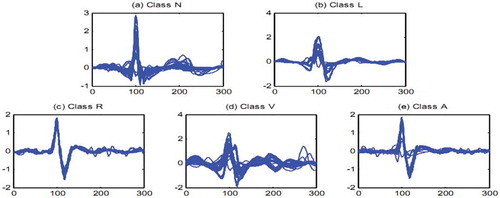
Heart’s electrical activities are captured by using some electrodes that are connected to specific points of patient’s chest. Electrocardiogram (ECG) classification is one of the most challenging tasks in heartbeat analysis. Medical centers are using ECGs in order to detect various cardiovascular diseases. By monitoring a patient’s ECG tape; expert cardiologists are able to recognize any kinds of arrhythmias which can be the cause of several serious heart diseases. In the last decade, researchers have proposed different pattern recognition systems in order to detect such arrhythmias automatically, which have been very helpful for cardiologists and hospitals. Although collecting the ECG data is easy, a lot of challenges still exist in order to extract the most useful information from the ECG signals. In , we illustrated 50 randomly selected sample signals from 5 different AAMI (AAMI Citation1987) classes, namely, non-ectopic (N), supraventricular ectopic (S), ventricular ectopic (V), fusion (F), and unknown (Q), in order to show the inter-class variation and intra-class similarity between them. To address the drawbacks of visual and manual interpretations of ECGs, researchers pursue the development of computer-aided diagnosis (CAD) systems to automatically analyse and interpret these signals.
Figure 1. Fifty randomly selected heartbeat signals for (a) Class N, (b) Class L, (c) Class R, (d) Class V, (e) Class A.
In recent years, several conventional machine learning techniques were developed for heartbeat classification from ECG signals (Alajlan et al. Citation2014; Alcaraz et al. Citation2011; Alonso-Atienza et al. Citation2014; Alvarado, Lakshminarayan, and Principe Citation2012; Homaeinezhad et al. Citation2012; Javadi et al. Citation2013; Melgani and Bazi Citation2008a). These methods consist of three main stages, namely preprocessing, feature extraction and classification. In preprocessing stage, the quality of ECG signals is enhanced by removing noise and baseline wander (Langkvist, Karlsson, and Loutfi Citation2012; Pereira et al. Citation2016; Sameni et al. Citation2007; Tracey and Miller Citation2012; Wang et al. Citation2015). Then, the ECG waveforms are segmented into their main sub-waves, namely, P wave, QRS complex and T wave (Bono et al. Citation2014; Ning and Selesnick Citation2013; Phukpattaranont Citation2015). After segmentation, different kinds of extracted features can be categorized into morphological features (De Chazal, O’Dwyer, and Reilly Citation2004; Dima et al. Citation2013), temporal-based features(De Chazal and Reilly Citation2006; Kutlu and Kuntalp Citation2012), wavelet transform derived features (Hatipoglu and Bilgin Citation2017; Yu and Chou Citation2008), high-order statistics (HOS) (Kutlu and Kuntalp Citation2012), Hermite basis function features (De Lannoy et al. Citation2012), and hidden Markov modeling (HMM) features (Chang et al. Citation2012). After feature extraction stage, popular techniques such as principal component analysis (PCA), independent component analysis (ICA), and linear discriminant analysis (LDA) are usually used for the purpose of reducing the dimensionality of the extracted features into the most discriminative ones (Martis, Acharya, and Min Citation2013; Ye, Kumar, and Coimbra Citation2012; Yu and Chou Citation2008). In the last stage of ECG classification, the features are fed into classifiers such as neural networks (NNs) (Jiang and Kong Citation2007), probabilistic NNs (Wang et al. Citation2013), recurrent NNs (Singh and Tiwari Citation2006), support vector machines (SVMs) (Alajlan et al. Citation2014; Homaeinezhad et al. Citation2012), least square SVMs (Dutta, Chatterjee, and Munshi Citation2010), path forests (Da Luz et al. Citation2013), and Gaussian processes (GPs) (Alajlan et al. Citation2014; Melgani and Bazi Citation2008b).
In recent years, CNNs have been used in many image and signal processing applications and they are shown to outperform state-of-the-art methods. CNNs are successfully utilized in biomedical image and signal processing such as histopathological images (Hatipoglu and Bilgin Citation2017), magnetic resonance (MR) images (Pereira et al. Citation2016), and X-ray images (Kallenberg et al. Citation2016). Recently, the use of CNNs is also proposed for ECG signal processing in a significant number of publications. In this respect, in (Acharya et al. Citation2017a), a 9-layers deep convolutional neural network is developed for automatic identification of 5 AAMI categories of heartbeats in ECG signals. They used original and noise removed set of signals from MIT-BIH database in their experiments. Also, the authors generated synthetic data samples to overcome the imbalance in the number of different heartbeat samples in each class. In (Acharya et al. Citation2017b), the authors used a 11-layer deep CNN for detecting normal atrial fibrillation (Afib), atrial flutter (Afl) and ventricular fibrillation (Vfib) type of ECG diseases. Two different CNNs are constructed for training such that one is used for 2 seconds and the other is used for 5 seconds durations without QRS detection. In (Luo et al. Citation2017), ECG signals were first converted into time-frequency images by using modified frequency slice wavelet transform (MFSWT) and then a deep learning method was applied for heartbeat classification. Zubair et al. used a simple 1-D CNN consisting of 3 convolution layers, 3 pooling layers and one MLP layer with output on MIT_BIH dataset in order to classify AAMI recommended heartbeat types (Zubair, Kim, and Yoon Citation2016). Pourbabea et al. proposed a CNN method to automatically learn and extract features for the identification of patients with paroxysmal atrial fibrillation(PAF) from his/her ECG signals (Pourbabaee, Roshtkhari, and Khorasani Citation2016). Following feature extraction step, a KNN classifier is employed to improve the performance of their system. In the work of Kiranyaz et al. a real time patient-specific ECG classification is proposed through using a 1-D CNN which is trained using a small number of common and patient-specific training data (Kiranyaz, Ince, and Gabbouj Citation2016).
Based on the demonstrated success of CNNs for biomedical signal and image processing, the research work presented in this article proposes a directed acyclic graph CNN for the automated diagnosis of heartbeat signal and experimentally exhibits its success on various AAMI heartbeat signals taken from the MIT-BIH database.
The ECG signals database
The MIT-BIH (Mark and Moody Citation2001) arrhythmia database is providing 48 annotated records which are collected from 47 male and female subjects; labelled non-sequenced integers from 100 to 234. Each record is about 30 minutes in length. The signals were sampled at a frequency of 360 hertz. Annotation file of each record contains useful information such as the occurrence times of R-peak locations or class of the corresponding heartbeat signals. In MIT-BIH database, there are 15 different heartbeat categories as summarized in .
Table 1. Heartbeat classes given by the MIT-BIH database along with the regrouping defined by the AAMI standard.
Assessment strategy
There are mainly two different approaches to assess the performance of a machine learning algorithm in ECG domain: class-based and subject-based. Class-based methods are applied based on the selection of various heartbeats from MIT-BIH dataset and classifying them into the associated disease categories. The subject based methods, that are more widely studied in literature, are based on the use of AAMI standard (AAMI Citation1987) which breaks down the 15 heartbeat classes into 5 sub-classes, namely, non-ectopic (N), supraventricular ectopic (S), ventricular ectopic (V), fusion (F), and unknown (Q) (De Chazal and Reilly Citation2006; Ince, Kiranyaz, and Gabbouj Citation2009; Jiang and Kong Citation2007; Kiranyaz, Ince, and Gabbouj Citation2016; Luo et al. Citation2017; Zhang et al. Citation2014). illustrates the AAMI standard based division of 15 heartbeat classes into the five above mentioned sub-classes together with the association between MIT-BIH arrhythmia dataset and AAMI standard.
Patient-specific classifier training
Considering two different patients having the same cardiac problem, their ECG signals bear significant morphological differences and this is in fact one of the fundamental difficulties of designing a common classifier that accurately detects the categories of heartbeat signals. One approach proposed to deal with this problem is the use of patient-specific classification techniques (Ince, Kiranyaz, and Gabbouj Citation2009; Kiranyaz, Ince, and Gabbouj Citation2016; Zhang et al. Citation2014). In these approaches, the system is trained by using a limited number of heartbeat samples, for example 5 minutes or 300 heartbeats, from the beginning of each individual ECG record to enhance the classification performance.
Convolutional neural networks
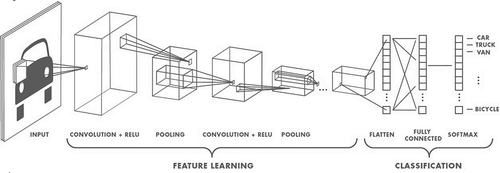
Based on the recently available literature, CNNs outperform state-of-the-art classifier systems in image processing, machine vision, speech recognition and other visual and signal processing tasks. Basically, CNNs are hierarchical neural networks constructed by a stacked combination of some common components, namely; convolution layer, rectified linear units and max pooling unit. The output of each layer is used as an input of the next layer. The output layer of a CNN architecture is a fully connected multilayer perceptron (MLP) that has the same number of output neurons as the class labels. A graphical description of a CNN architecture is given in . Brief functional descriptions of each CNN component are as follows:
Convolution layer: This layer of a CNN extracts and learns features from the input data. Convolution layer has many adaptive filters (or kernels) which may have different sizes in different layers.
Rectified linear unit layer (ReLU): Rectified linear units, or the ReLU layer, are nonlinear activation functions which perform a threshold operation to each element of the input such that any value less than zero is set to zero.
Pooling layer: In order to reduce CNN computations, this layer reduces the size of extracted features by down-sampling. This operation can be done by taking the average or the maximum number that the filter convolves around in each kernel.
Fully connected layer: The last layer of a CNN is a multi-layer perceptron (MLP) with k output neurons, where k is the number of class labels.
Figure 2. Example of CNN architecture (Mathworks online resource, Citation2017).
Directed acyclic graph-convolutional neural networks
Deep Learning is a subfield of machine learning concerned with algorithms inspired by the structure and function of the brain called artificial neural networks. Recently, deep artificial neural networks (including recurrent ones) have outperformed numerous state-of-the-art methods in solving pattern recognition and machine learning problems. A detailed review of deep learning neural networks can be found in (Schmidhuber Citation2015). The directed acyclic graph (DAG) networks can represent more complex network architectures compared to ones consisting of a linear chain of layers. DAG architecture for neural networks (NNs) have emerged from the idea of recurrent NNs that have some feedback connections from forward layers to backward ones, which give them the ability of capturing dynamic states. The main advantage of DAG-structured networks is that their forward layers can have multiple input parameters from several backward layers. This way, they can achieve different levels of signal representations. CNNs form a subset of deep learning neural networks for which some of the recently published work can be found in (Raiko, Valpola, and LeCun Citation2012; Sermanet et al. Citation2013; Szegedy et al. Citation2015). A fundamental feature of the deep learning neural networks is the use of connections between their layers, called “skip connection,” that is similar to DAG-CNNs main idea, and it is shown that these skip connections can improve the accuracy of the classification tasks significantly.
DAG-CNN was proposed by Yang and Ramanan (Yang and Ramanan Citation2015) to learn a set of multi-scale image features that are successfully used for classification of three standard scene benchmarks. They showed that the multi-scale model can be implemented as a DAG-structured feed forward CNN. By this approach, it is possible to use an end–to-end gradient-based learning for automatically extracting multi-scale features using generalized back propagation algorithm over the layers that have more than one input. In fact, all the required equations for training the network are standard CNN equations except for the Add and ReLU layers since they have multiple inputs or outputs. Let us consider the ith ReLU layer in , Let be its input,
be the output for its jth output branch (its jth child in the DAG), and assume that z is the final output of the softmax layer. The gradient of z with respect to the input of the ith ReLU layer can be computed as Equation (1):
Figure 3. Visualization of the parameter setup at i-th ReLU and k-th ADD (Yang and Ramanan Citation2015).
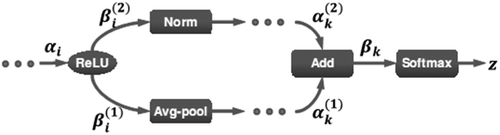
where C is the number of output edges of the ith ReLU.
For the Add layer, let βk = (
, · · ·,
represents the output of an Add layer with multiple inputs. We can compute the gradient along the layer by applying the chain rule as Equation (2):
In the convolutional layers (layer L1,L4,L7, and L10 in ), the convolution operation is computed by the Equation (3):
where y and f are the ECG signal and the applied filter, respectively, and N is the number of elements in the ECG signal y. The convolution layer’s output is represented by vector . For all layers of the DAG-CNN architecture, except ReLU and ADD layers, the Equation (4–5) are used to update biases and weights as follows:
where , λ,
and
denote the weight, bias, layer number, regularization parameter, learning rate, total number of training samples, momentum, updating step, and cost function respectively.
In DAG-CNNs, since lower layers are directly connected to the output layer through multi-scale connections, it is guaranteed that these layers’ neurons receive a strong gradient signal during learning and do not suffer from the problem of vanishing gradients.
In this study, DAG-CNNs are used for automatically extracting and combining discriminative features and classifying the ECG data into different heartbeat classes.
The proposed DAG-CNN methodology
Pre-processing
Before we fed the raw ECG signals into our proposed method, the following simple 3-step pre-processing of ECG signals is carried out: removing the base line, denoising the signals and signal segmentation. To carry out the first two pre-processing steps, Daubechies wavelet-6 filters are used (Singh and Tiwari Citation2006), whereas for segmentation of raw ECG signals into heartbeats, first the R-peaks are found by using Pan-Tompkins (Pan and Tompkins Citation1985) procedure. Consequently, 140 samples are selected from the left sides and 139 samples are selected from the right sides of the detected R-peaks. Hence, the length of sampled points is 280 for each segmented heartbeat. At the beginning of each ECG recording, if the number of sample points is less than 140 on left side of the first R-peak, that heart beat was ignored. Similarly, the last heartbeat is also ignored if the number of sample points on the right side of its R-peak is less than 139. Finally, Z-score normalization is applied on all sampled values in order to convert them to a common scale with an average of zero and standard deviation of one by using the Equation 6.
where ,
and
are signal, signal’s mean and signal’s standard deviation respectively.
Proposed DAG-CNN model
In this research work, a DAG-CNN architecture is proposed to improve the discrimination capability of a deep neural network by allowing its layers to share their learned features and work collaboratively for classification. The proposed multi-scale CNN topology applies learned features with different level of complexity in order to predict the output label with high precision.
CNNs can be used as automatic feature extractors and the learned features can be fed to classifiers like SVMs or NNs to predict the output labels. Mid-level features at intermediate layers of a CNN can be discriminative for classifying different patterns with varying complexities. However, in CNN architectures used in literature so far, these cross-layer heterogeneity features are ignored. It is clear to see that these mid-level features are already computed when the system is trained to extract high-level features, and hence, their use do not bring any extra computational burden within our proposed model. Instead of performing feature level fusion and feeding the results to a classifier, we proposed a multi-scale system by using a CNN with directed acyclic graph topology. Our proposed model can automatically learn different level of features, combine them and predict the output label.
One of the common problems with feature level fusion is the size of the feature vectors. In CNNs, the size of the learned features in intermediate layers can be very large and combining these features may cause the curse of dimensionality problem. To overcome this problem, we compute marginal activations by performing average pooling on the learned features of some layers which are used for feature level fusion.
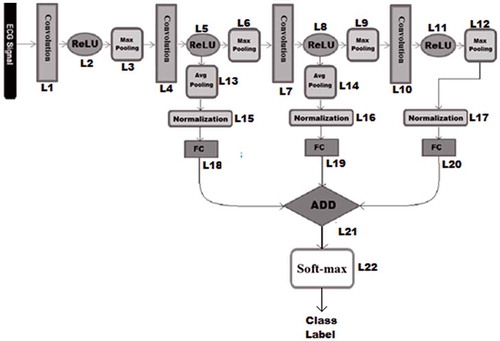
We assumed that the output of each component (convolution, ReLU, pooling, normalization, fully connected and ADD) of the proposed model is treated as a separate layer. Therefore our model has 22 layers. Our proposed model consists of a typical CNN as its base structure and some links from the intermediate and last ReLU layers. These links are connected to an average pooling layer to reduce their dimensionality. Then the extracted features are normalized and given to separate fully connected MLP layers. Each of these fully connected layers have the same number of neurons in their last layer and their number is equal to number of class labels (five different AAMI classes) and generate a score vector for each input sample. These score-vectors are added with each other, element by element and fed into the final decision layer with softmax activation function to predict the class label.
In this study, we have shown that combining different level features can improve the classification accuracy significantly. Particularly, the classification accuracy is improved when we add features learned by intermediate layers, with the exception of the low-level features of early layers that cause a decrease in classification accuracy. For the purpose of testing different combinations of feature layers and finding the best one experimentally, features of the last layer are considered as of necessary and intermediate layer features are added layer-by-layer, one at a time, in a backward fashion until no improvement observed in classification accuracy. This greedy approach ignores the features of layers closer to the input layer. Experimental evaluations as illustrated within the next section exhibited that the proposed system’s capability of fusing multi-scale features improves the accuracy of classification tasks.
Experimental results
In our experiments, following the AAMI standard, four records labelled with (102,104,107 and 217) containing paced beats with low signal quality, were removed. Also to follow AAMI recommendations the remaining 44 records are divided into two disjoint sets DS1 and DS2. Trainset (DS1) contains records labelled with 101, 106, 108, 109, 112, 114, 115, 116, 118, 119, 122, 124, 201, 203, 205, 207, 208, 209, 215, 220, 223 and 230; and the testset (DS2) contains records labelled with 100, 103, 105, 111, 113, 117, 121, 123, 200, 202, 210, 212, 213, 214, 219, 221, 222, 228, 231, 232, 233 and 234 from MIT-BIH dataset ().
Table 2. Summary of the training and testing heartbeat samples.
In , we summarize the details of the DAG-CNN model. S-shaped rectified linear unit (SReLU) (Xiaojie et al. Citation2016) has been used in our networks. Compared to other activation functions, SReLU is able to learn both convex and non-convex functions. For all the max-pooling and average-pooling layers kernel size and stride are set to 2. All of the 3 fully connected layers (FC) in are multi-layer perceptron (MLP) with 3 layers consisting of 25, 15, and 5 neurons respectively. In the last layer, the softmax function is used to generate the final decision of the system which can be one of the output classes namely N, S, V, F, and Q.
Table 3. The details of back-bone CNN architecture of DAG-CNN model.
Our proposed DAG-CNN architecture was trained through the standard backpropagation technique with a batch size of 8. In order to obtain optimum performance, the other learning parameters are set as follows: in order to prevent overfitting of training data, the regularization parameter (λ) is set to 0.2, momentum parameters which adjust the speed of learning during training is set to 0.8, and learning rate that controls the convergence of the training data is set to 0.0002 and is linearly changed according to the mean-squared error values in each five iteration. The training was performed over 40 epoch rounds. The goal of our DAG-CNN model is to classify heartbeats into five AAMI classes. summarizes beat-by-beat classification results of ECG heartbeat patterns for all test records.
Table 4. Different heartbeat types classification result.
For each of the four classes N, S, V, and F, we compared the classification performance of our system with the state-of-the-art approaches in . For this comparison we computed the four standard metrics: classification accuracy (Acc), sensitivity (Sen), specificity (Spe), and positive predictive ratio (Ppr). The equations of these common metrics are as follow: by considering TP as true positive,TN as true negative, FP as false positive, and FN as false negatives:
Table 5. Four heartbeat types classification metrics compared to the state-of-the-art (percentage, %).
Accuracy is the ratio of the number of correctly classified patterns to the total number of patterns classified:
Sensitivity is the rate of correctly classified events among all events:
Specificity is the rate of correctly classified non-events among all non-events:
and Positive predictive ratio is the rate of correctly classified events in all detected events:
For clinical applications, the sensitivity, specificity and positive predictively measurement are more relevant performance criteria because there is a large difference in the number of beats from different classes in the training/testing data.
Clinically, supraventricular ectopic beats (SVEB) and ventricular ectopic beats (VEB) are two critically abnormal and serious heartbeats. So for performance evaluations, we also present the results in terms of VEB [V class versus (N, S and F)] and SVEB [S class versus (N, V and F)]. The VEB and SVEB classification results of the proposed technique over all DS2 records are summarized in . It is observed that, the overall performance of the proposed method in VEB and SVEB detection is significantly better than most of the state-of-the-art methods and is equally well with the best method (Luo et al. Citation2017).
Table 6. SVEB and VEB Classification metrics compared to the state-of-the (percentage, %).
Conclusions
This manuscript presented a novel method based on deep learning for automatic classification of heartbeat signals. The proposed method extracts multiple-scale features automatically from different layers of convolutional neural networks (CNN) by using a directed acyclic graph CNN architecture. Experimental evaluations based on the popular MIT-BIH ECG dataset showed that fusion of multiple-scale features improves the classification performance of the overall systems and makes the proposed method competitive to its state-of-the-art alternatives. Based on the obtained experimental results, it is evident that the proposed system has significant potential to be used in real clinical environments.
References
- AAMI. 1987. Recommended practice for testing and reporting performance results of ventricular arrhythmia detection algorithms. Arlington, VA, USA: Association for the Advancement of Medical Instrumentation.
- Acharya, U.R., H. Fujita, S. L. Oh, Y. Hagiwara, J. H. Tan, and M. Adam. 2017a. Automated detection of arrhythmias using different intervals of tachycardia ECG segments with convolutional neural network. Information Sciences 405:81–90. doi:10.1016/j.ins.2017.04.012..
- Acharya, U.R., S. L. Oh, Y. Hagiwara, J. H. Tan, M. Adam, A. Gertych, and R. S. Tan. 2017b. A deep convolutional neural network model to classify heartbeats. Computers in Biology and Medicine 89:389–96. doi:10.1016/j.compbiomed.2017.08.022.
- Alajlan, N., Y. Bazi, F. Melgani, S. Malek, and M. A. Bencherif. 2014. Detection of premature ventricular contraction arrhythmias in electrocardiogram signals with kernel methods. Signalling Image Video Processing 8:931–42. doi:10.1007/s11760-012-0339-8.
- Alcaraz, R., F. Sandberg, L. L. Sörnmo, and J. J. Rieta. 2011. Classification of paroxysmal and persistent atrial fibrillation in ambulatory ECG recordings. IEEE Transactions Biomedical Engineering 58:1441–49. doi:10.1109/TBME.2011.2112658.
- Alonso-Atienza, F., E. Morgado, L. Fernandez-Martinez, A. Garcia-Alberola, and J. L. Rojo-Alvarez. 2014. Detection of life-threatening arrhythmias using feature selection and support vector machines. IEEE Transactions Biomedical Engineering 61:832–40. doi:10.1109/TBME.2013.2290800.
- Alvarado, A. S., C. Lakshminarayan, and J. C. Principe. 2012. Time-based compression and classification of heartbeats. IEEE Transactions Biomedical Engineering 59:1641–48. doi:10.1109/TBME.2012.2191407.
- Bono, V., E. B. Mazomenos, T. Chen, J. A. Rosengarten, A. Acharyya, K. Maharatna, J. M. Morgan, and N. Curzen. 2014. Development of an automated updated Selvester QRS scoring system using SWT-based QRS fractionation detection and classification. IEEE Journal Biomedical Health Information 18:193–204. doi:10.1109/JBHI.2013.2263311.
- Chang, P.-C., -J.-J. Lin, J.-C. Hsieh, and J. Weng. 2012. Myocardial infarction classification with multi-lead ECG using hidden Markov models and Gaussian mixture models. Applications Soft Computation 12:3165–75. doi:10.1016/j.asoc.2012.06.004.
- da Luz, E. J. S., T. M. Nunes, V. H. C. de Albuquerque, J. P. Papa, and D. Menotti. 2013. ECG arrhythmia classification based on optimum-path forest. Expert Systems Applications 40:3561–73. doi:10.1016/j.eswa.2012.12.063.
- de Chazal, P., M. O’Dwyer, and R. B. Reilly. 2004. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Transactions Biomedical Engineering 51:1196–206. doi:10.1109/TBME.2004.827359.
- de Chazal, P., and R. B. Reilly. 2006. A patient-adapting heartbeat classifier using ECG morphology and heartbeat interval features. IEEE Transactions Biomedical Engineering 53:2535–43. doi:10.1109/TBME.2006.883802.
- de Lannoy, G., D. Francois, J. Delbeke, and M. Verleysen. 2012. Weighted conditional random fields for supervised interpatient heartbeat classification. IEEE Transactions Biomedical Engineering 59:241–47. doi:10.1109/TBME.2011.2171037.
- Dima, S.-M., C. Panagiotou, E. B. Mazomenos, J. A. Rosengarten, K. Maharatna, and J. V. Gialelis. 2013. On the detection of myocadial scar based on ECG/VCG analysis. IEEE Transactions Biomedical Engineering 60:3399–409. doi:10.1109/TBME.2013.2279998.
- Dutta, S., A. Chatterjee, and S. Munshi. 2010. Correlation technique and least square support vector machine combine for frequency domain based ECG beat classification. Medica Engineering Physical 32:1161–69. doi:10.1016/j.medengphy.2010.08.007.
- Hatipoglu, N., and G. Bilgin. 2017. Cell segmentation in histopathological images with deep learning algorithms by utilizing spatial relationships. Medical and Biological Engineering and Computing 1–20. doi:10.1007/s11517-017-1630-1.
- Homaeinezhad, M. R., S. A. Atyabi, E. Tavakkoli, H. N. Toosi, A. Ghaffari, and R. Ebrahimpour. 2012. ECG arrhythmia recognition via a neuro-SVM–KNN hybrid classifier with virtual QRS image-based geometrical features. Expert Systems Applications 39:2047–58. doi:10.1016/j.eswa.2011.08.025.
- Ince, T., S. Kiranyaz, and M. Gabbouj. 2009. A generic and robust system for automated patient-specific classification of electrocardiogram signals. IEEE Transactions Biomedical Engineering 56:1415–26. doi:10.1109/TBME.2009.2014740.
- Javadi, M., S. A. A. A. Arani, A. Sajedin, and R. Ebrahimpour. 2013. Classification of ECG arrhythmia by a modular neural network based on mixture of experts and negatively correlated learning. Biomedical Signalling Processing Control 8:289–96. doi:10.1016/j.bspc.2012.10.005.
- Jiang, W., and S. G. Kong. 2007. Block-based neural networks for personalized ECG signal classification. IEEE Transaction Neural Networks : the Official Journal of the International Neural Network Society 18:1750–61. doi:10.1109/TNN.2007.900239.
- Kallenberg, M., K. Petersen., M. Nielsen., Y. A. Ng, P. F. Diao, C. Igel., C. M. Vachon, K. Holland., R. R. Winkel, N. Karssemeijer., and M. Lillholm. 2016. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Transactions on Medical Imaging 35 (5):1322–31. doi:10.1109/TMI.2016.2532122.
- Kiranyaz, S., T. Ince, and M. Gabbouj. 2016. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Transactions on Biomedical Engineering 63 (3):664–75. doi:10.1109/TBME.2015.2468589.
- Kutlu, Y., and D. Kuntalp. 2012. Feature extraction for ECG heartbeats using higher order statistics of WPD coefficients. Computer Methods Program Biomedical 105:257–67. doi:10.1016/j.cmpb.2011.10.002.
- Langkvist, M., L. Karlsson, and A. Loutfi. 2012. Sleep stage classification using unsupervised feature learning. Advancement Artificial Neural Systems 2012:1–9. doi:10.1155/2012/107046.
- Luo, K., J. Li, Z. Wang, and A. Cuschieri. 2017. Patient-specific deep architectural model for ECG classification. Journal of Healthcare Engineering 2017:13. Article ID 4108720. doi:10.1155/2017/4108720.
- Mark, R., and G. Moody. 2001. MIT-BIH arrhythmia database directory. [Online]. http://ecg.mit.edu/dbinfo.html
- Martis, R. J., U. R. Acharya, and L. C. Min. 2013. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomedical Signalling Processing Control 8:437–48. doi:10.1016/j.bspc.2013.01.005.
- Mathworks online resource. 2017. Convolutional Neural Networks article, Natick, Massachusetts, United States. Accessed December 12, 2017. https://www.mathworks.com/content/mathworks/www/en/discovery/convolutional-neural-network/jcr:content.html.
- Melgani, F., and Y. Bazi. 2008a. Classification of electrocardiogram signals with support vector machines and particle swarm optimization. IEEE Transactions Information Technological Biomedical 12:667–77. doi:10.1109/TITB.2008.923147.
- Melgani, F., and Y. Bazi. 2008b. Detecting premature ventricular contractions in ECG signals with Gaussian processes. Computers in Cardiology, Bologna, 237–240. doi:10.1109/CIC.2008.4749021.
- Ning, X., and I. W. Selesnick. 2013. ECG enhancement and QRS detection based on sparse derivatives. Biomedical Signalling Processing Control 8:713–23. doi:10.1016/j.bspc.2013.06.005.
- Pan, J., and W. J. Tompkins. 1985. A real-time QRS detection algorithm. IEEE Transactions on Biomedical Engineering 32 (3):230–36. doi:10.1109/TBME.1985.325532.
- Pereira, S., A. Pinto, V. Alves, C. Silva, and A. Brain. 2016. Tumor segmentation using convolutional neural networks in MRI images. IEEE Transactions on Medical Imaging 35 (5):1240–51. doi:10.1109/TMI.2016.2538465.
- Phukpattaranont, P. 2015. QRS detection algorithm based on the quadratic filter. Expert Systems Applications 42:4867–77. doi:10.1016/j.eswa.2015.02.012.
- Pourbabaee, B., M. J. Roshtkhari, and K. Khorasani. 2016. Feature leaning with deep convolutional neural networks for screening patients with paroxysmal atrial fibrillation. International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, 5057–64
- Raiko, T., H. Valpola, and Y. LeCun. 2012. Deep learning made easier by linear transformations in perceptrons. AISTATS 22:924–32.
- Sameni, R., M. B. Shamsollahi, C. Jutten, and G. D. Clifford. 2007. A nonlinear Bayesian filtering framework for ECG denoising. IEEE Transaction Biomedical Engineering 54:2172–85.
- Schmidhuber, J. 2015. Deep learning in neural networks: An overview. Neural Networks 61:85–117. doi:10.1016/j.neunet.2014.09.003.
- Sermanet, P., K. Kavukcuoglu, S. Chintala, and Y. LeCun. 2013. Pedestrian detection with unsupervised multi-stage feature learning. CVPR IEEE 3626–33. arXiv:1212.0142
- Singh, V., and A. Tiwari. 2006. Optimal selection of wavelet basis function applied to ECG signal denoising. Digital Signal Processing 16 (3):275–87. doi:10.1016/j.dsp.2005.12.003.
- Szegedy, C., W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, and A. Rabinovich. 2015. Going deeper with convolutions. CVPR 1–9. doi:10.1109/CVPR.2015.7298594.
- Tracey, B. H., and E. L. Miller. 2012. Non local means denoising of ECG signals. IEEE Transactions Biomedical Engineering 59:2383–86. doi:10.1109/TBME.2012.2208964.
- Wang, J., Y. Ye, X. Pan, and X. Gao. 2015. Parallel-type fractional zero-phase filtering for ECG signal denoising. Biomedical Signalling Processing Control 18:36–41. doi:10.1016/j.bspc.2014.10.012.
- Wang, J.-S., W.-C. Chiang, Y.-L. Hsu, and Y.-T. C. Yang. 2013. ECG arrhythmia classification using a probabilistic neural network with a feature reduction method. Neurocomputing 116:38–45. doi:10.1016/j.neucom.2011.10.045.
- Xiaojie, J., X. Chunyan, F. Jiashi, W. Yunchao, X. Junjun, and Y. Shuicheng. 2016. Deep learning with S-shaped rectified linear activation units. Proceedings of the AAAI Conference on Artificial Intelligence (AAAI), 1737–43, Phoenix, Arizona, USA. doi:10.3389/fpsyg.2016.01737
- Yang, S., and D. Ramanan. 2015. Multi-scale recognition with DAG-CNNs. Proceedings of the IEEE International Conference on Computer Vision, 1215–23
- Ye, C., B. V. K. V. Kumar, and M. T. Coimbra. 2012. Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Transactions Biomedical Engineering 59:2930–41. doi:10.1109/TBME.2012.2213253.
- Yu, S. N., and K. T. Chou. 2008. Integration of independent component analysis and neural networks for ECG beat classification. Expert Systems Applications 34:2841–46. doi:10.1016/j.eswa.2007.05.006.
- Zhang, Z., J. Dong, X. Luo, K. Choi, and X. Wu. 2014. Heartbeat classification using disease-specific feature selection. Computers in Biology and Medicine 46:79–89. doi:10.1016/j.compbiomed.2013.11.019.
- Zubair, M., J. Kim, and C. Yoon. 2016. An automated ECG beat classification system using convolutional neural networks. 6th International Conference on IT Convergence and Security (ICITCS), 1–5, Prague.